HSP70 Multi-Functionality in Cancer
Abstract
1. Introduction
2. The HSP70 Machinery
2.1. HSP70 Structure
2.2. HSP70 Functional Cycle
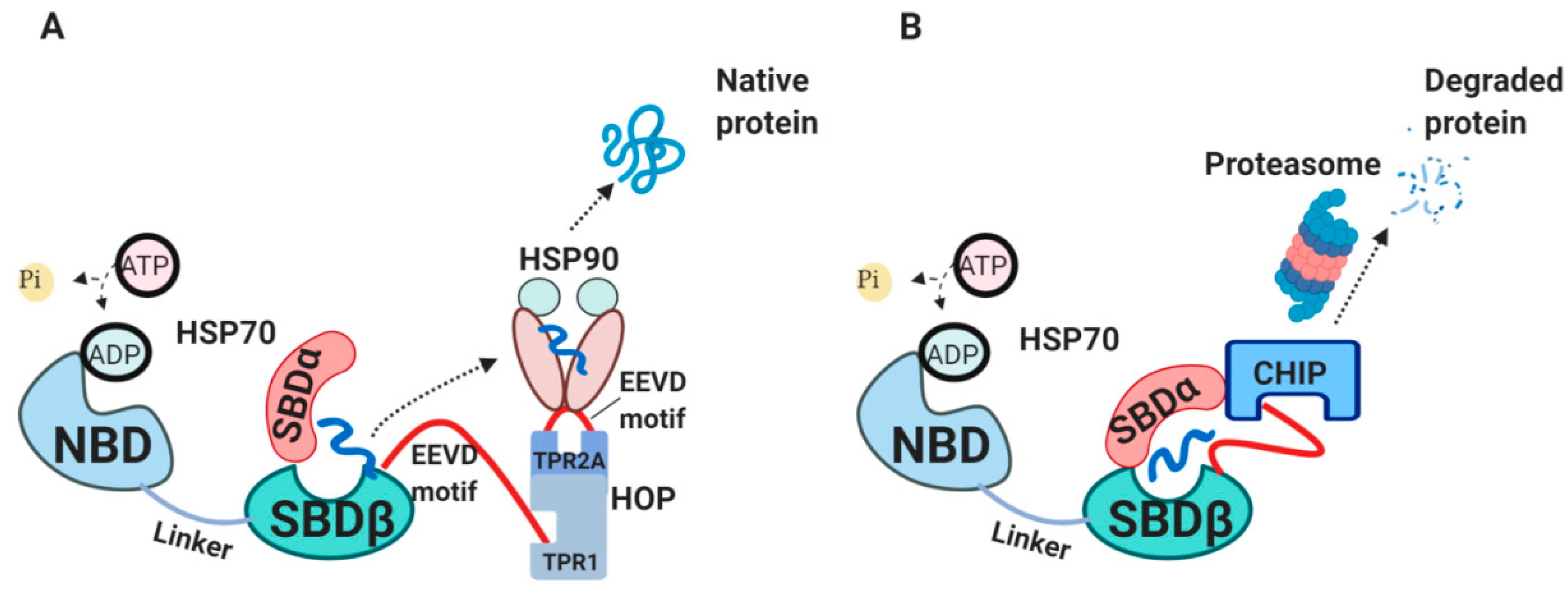
2.3. The Internal HSP70 Network
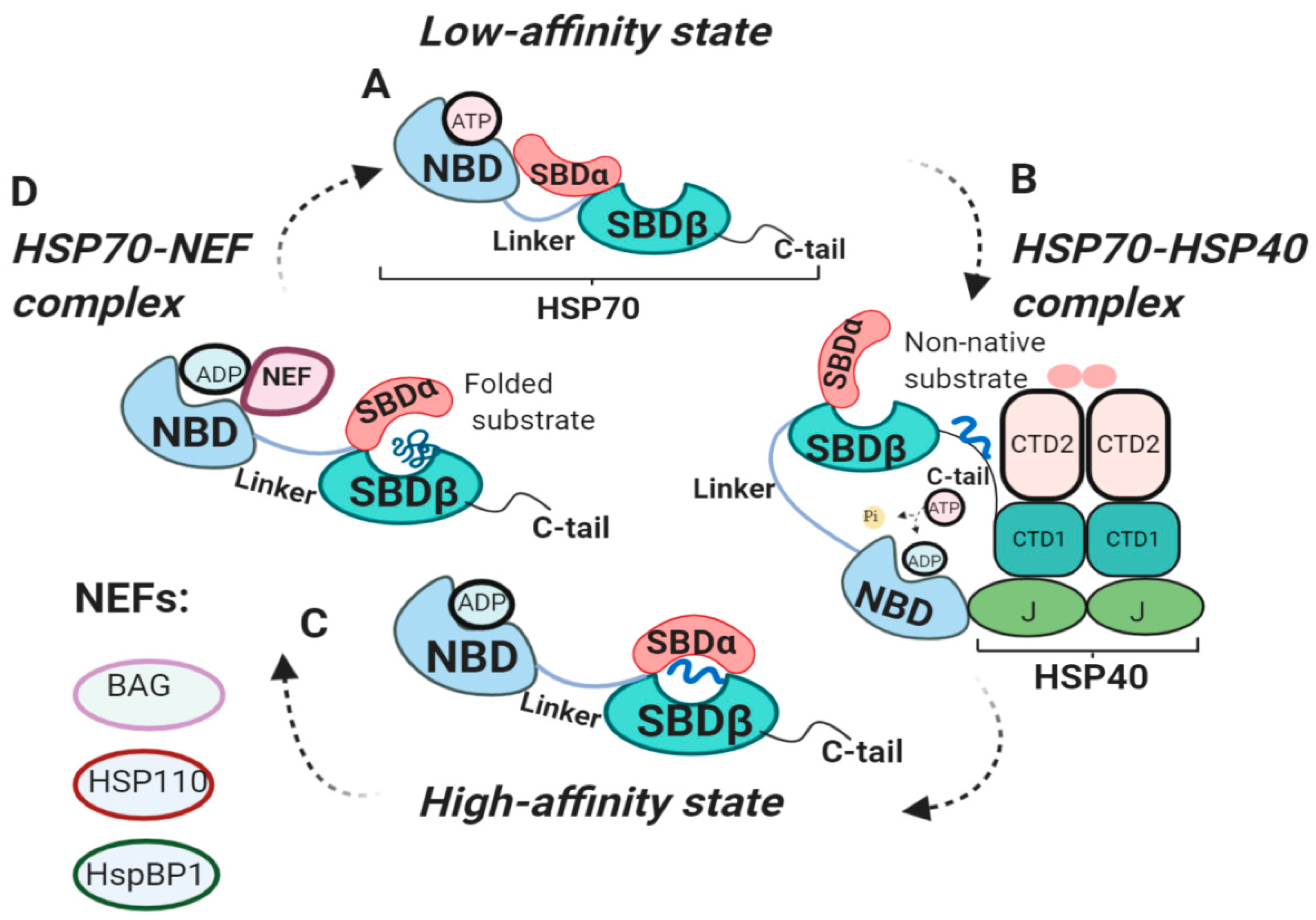
2.4. The External HSP70 Network
3. HSP70 Transport to the Extracellular Milieu
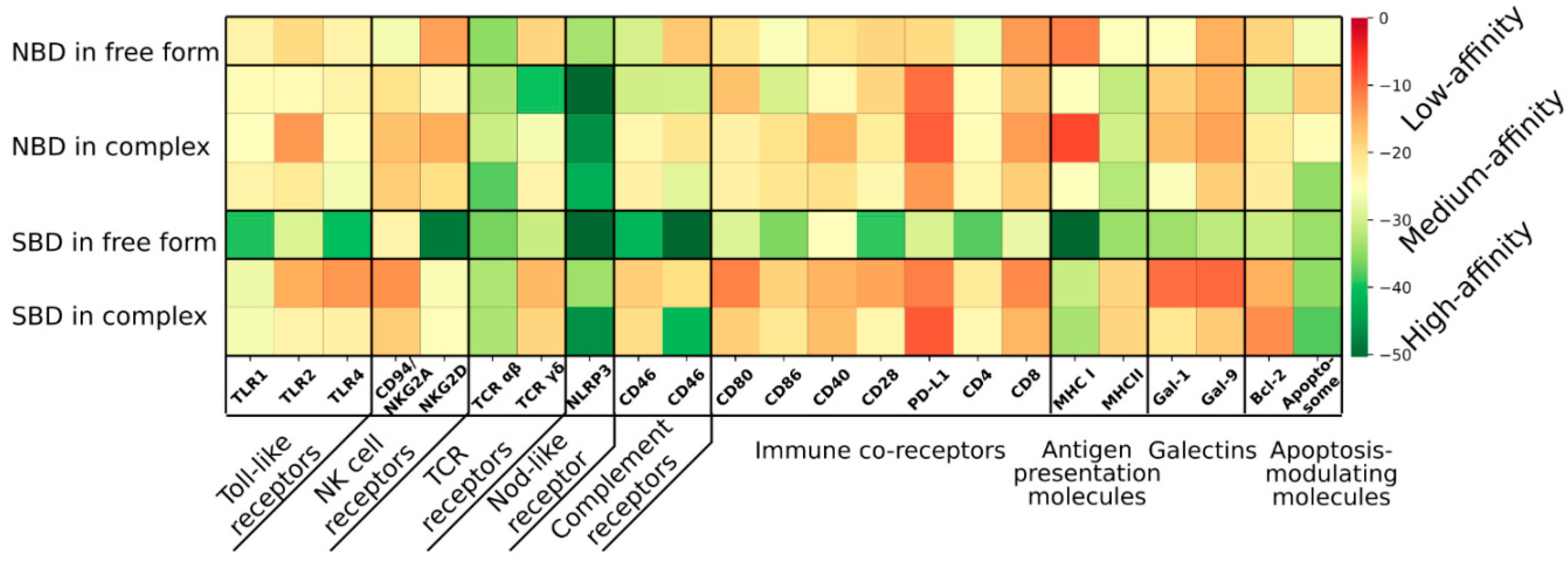
4. HSP70 Receptors
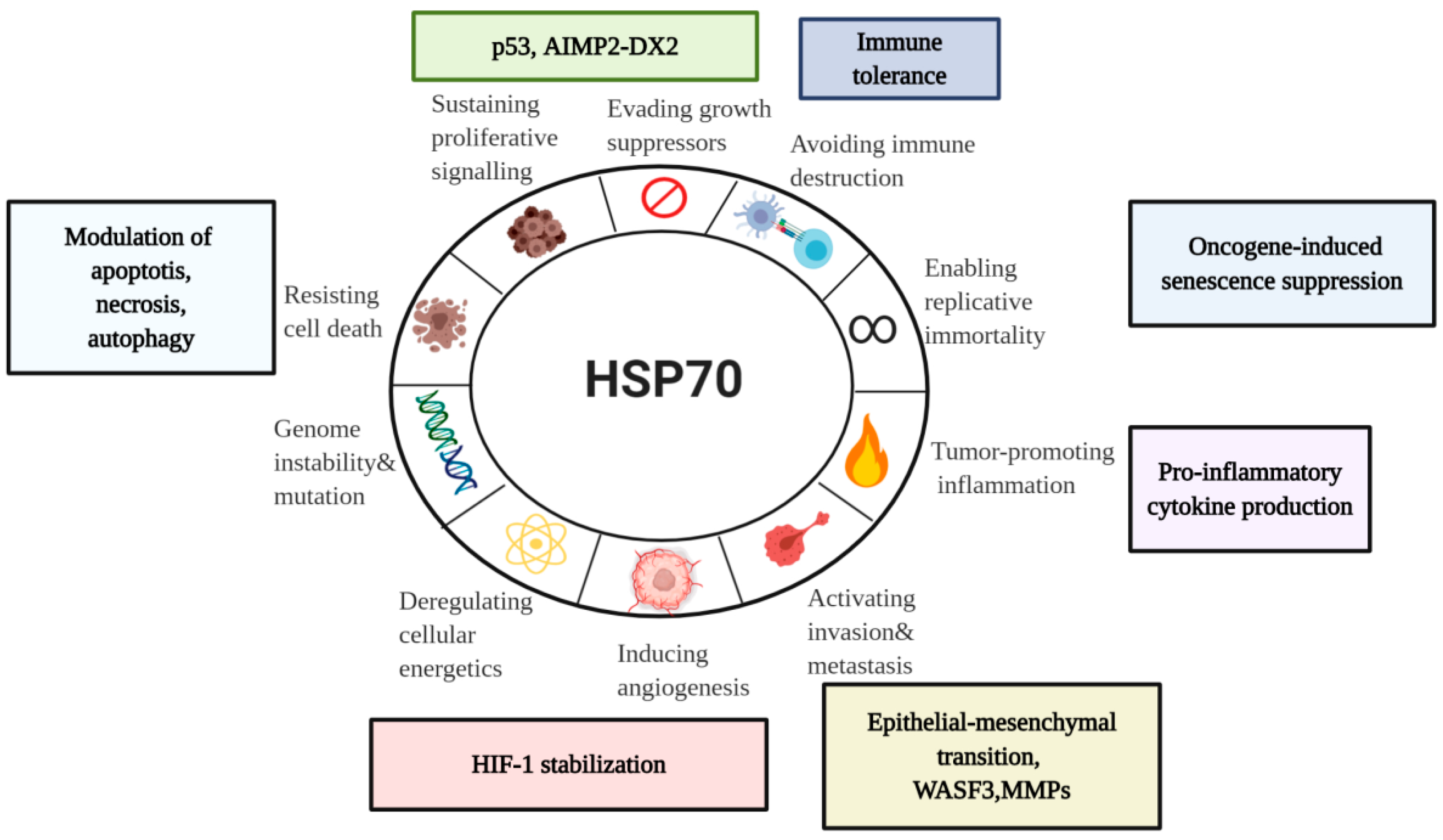
5. Diverse Functions of HSP70 in the Hallmarks of Cancer
5.1. HSP70 and Tumor Immunity
5.2. HSP70 and Tumor Resistance to Cell Death
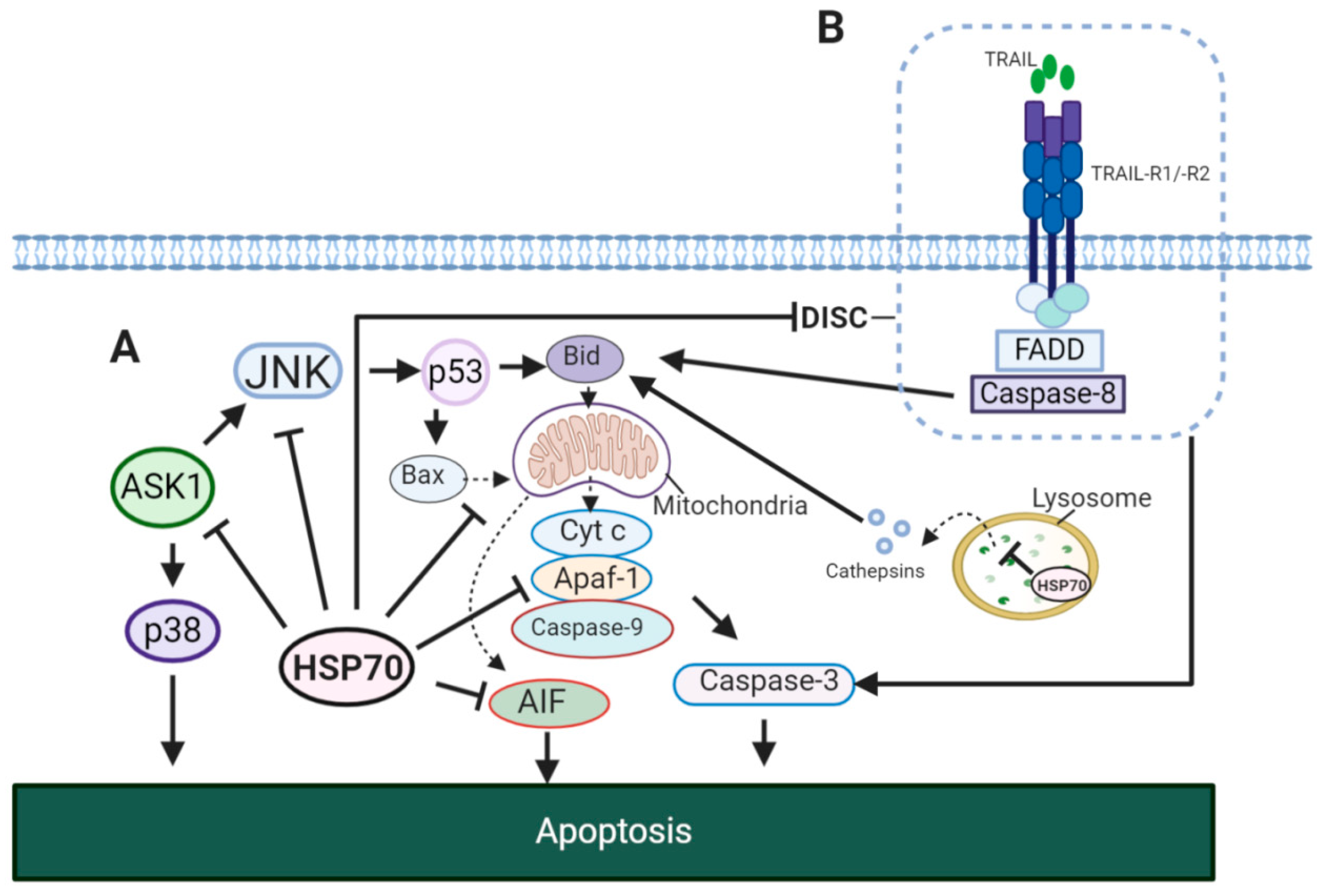
5.3. HSP70 and Senescence Program
5.4. HSP70 in Sustained Proliferation
5.5. HSP70 in Angiogenesis
5.6. HSP70 in Metastasis
6. HSP70 Therapies Targeting Cancer
6.1. Small Molecule Inhibitors of HSP70 Cycle
| HSP70-Targeting Molecules | Effects | Refs. | |
|---|---|---|---|
| SBD-targeting inhibitors | PES (Pifithrin-µ) | Suppressed tumor development in a mouse model of Myc-lymphoma. | [219] |
| ADD70 | Increased sensitivity of colon cancer and melanoma cells to apoptosis; showed antimetastatic effects in vivo. | [220] | |
| Acridizinium derivative 1 | Induced apoptosis in HeLa cells. | [221] | |
| NBD-targeting inhibitors | Synthetic peptide P17 | Decreased melanoma growth in vivo. | [222] |
| JG-98 inhibitor (MKT-077 analog) | Decreased tumor growth in MCF7 xenograft model. | [213] | |
| VER-155008 | Inhibited proliferation of human breast and colon cancer cell lines. | [217] | |
| YK-5 | Induced apoptosis and degradation of HSP70/HSP90 client proteins (HER2,Raf-1, Akt kinases) in breast cancer cells. | [223] | |
| Apoptozole | Induced apoptosis in ovarian, colon and lung cancer cell lines. | [224,225] | |
| HSP70-HSP40 complex inhibitors | MAL3-101 | Inhibited proliferation of multiple myeloma cells derived from patients. | [226] |
| DMT3132 (MAL3-101 analog) | Reduced proliferation in breast cancer cells. | [227,228] | |
| Myricetin | Inhibited tumor growth in pancreatic cancer. | [229] | |
| HSP70-NEF complex inhibitors | YM-1 | Inhibited tumor growth in mammary and melanoma xenograft models. | [168] |
| Allosteric HSP70 inhibitors | HS-72 | Inhibited growth in HER2-positive breast cancel model. | [230] |
6.2. Other HSP70 Inhibitors
6.3. HSP70-Based Therapies in Cancer Clinical Trials
7. Conclusions and Perspectives
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Ritossa, F. A new puffing pattern induced by temperature shock and DNP in drosophila. Cell. Mol. Life Sci. 1962, 18, 571–573. [Google Scholar] [CrossRef]
- Tissières, A.; Mitchell, H.K.; Tracy, U.M. Protein synthesis in salivary glands of Drosophila melanogaster: Relation to chromosome puffs. J. Mol. Biol. 1974, 84, 389–398. [Google Scholar] [CrossRef]
- Moran, L.; Mirault, M.-E.; Arrigo, A.P.; Goldschmidt-Clermont, M.; Tissières, A. Heat shock of Drosophila melanogaster induces the synthesis of new messenger RNAs and proteins. Philos. Trans. R. Soc. B Biol. Sci. 1978, 283, 391–406. [Google Scholar]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [PubMed]
- Beere, H. Stress management—Heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol. 2001, 11, 6–10. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Zhou, Y.; Wang, Y.; Wang, S.; Zhang, W.-Y. Hsp70 promotes chemoresistance by blocking Bax mitochondrial translocation in ovarian cancer cells. Cancer Lett. 2012, 321, 137–143. [Google Scholar] [CrossRef]
- Pelham, H. Hsp70 accelerates the recovery of nucleolar morphology after heat shock. EMBO J. 1984, 3, 3095–3100. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Hunt, C.; Morimoto, R.I. Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of human hsp70. Proc. Natl. Acad. Sci. USA 1985, 82, 6455–6459. [Google Scholar] [CrossRef]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Gupta, R.S.; Singh, B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr. Biol. 1994, 4, 1104–1114. [Google Scholar] [CrossRef]
- Mosser, D.D.; Morimoto, R.I. Molecular chaperones and the stress of oncogenesis. Oncogene 2004, 23, 2907–2918. [Google Scholar] [CrossRef] [PubMed]
- Ciocca, D.R.; Calderwood, S.K. Heat shock proteins in cancer: Diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 2005, 10, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.; Khaleque, A.; Sawyer, U.B.; Ciocca, D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, A.; Handa, N.; Ohsawa, N.; Shida, M.; Kigawa, T.; Hayashi, F.; Shirouzu, M.; Yokoyama, S. The C-Terminal BAG Domain of BAG5 Induces Conformational Changes of the Hsp70 Nucleotide- Binding Domain for ADP-ATP Exchange. Structure 2010, 18, 309–319. [Google Scholar] [CrossRef]
- Arakawa, A.; Handa, N.; Shirouzu, M.; Yokoyama, S. Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci. 2011, 20, 1367–1379. [Google Scholar] [CrossRef]
- Cheeseman, M.; Westwood, I.M.; Barbeau, O.; Rowlands, M.; Dobson, S.; Jones, A.M.; Jeganathan, F.; Burke, R.; Kadi, N.; Workman, P.; et al. Exploiting Protein Conformational Change to Optimize Adenosine-Derived Inhibitors of HSP70. J. Med. Chem. 2016, 59, 4625–4636. [Google Scholar] [CrossRef]
- Gao, X.-C.; Zhou, C.-J.; Zhou, Z.-R.; Wu, M.; Cao, C.-Y.; Hu, H.-Y. The C-terminal Helices of Heat Shock Protein 70 Are Essential for J-domain Binding and ATPase Activation*. J. Biol. Chem. 2012, 287, 6044–6052. [Google Scholar] [CrossRef]
- Hassan, A.Q.; Kirby, C.A.; Zhou, W.; Schuhmann, T.; Kityk, R.; Kipp, D.R.; Baird, J.; Chen, J.; Chen, Y.; Chung, F.; et al. The Novolactone Natural Product Disrupts the Allosteric Regulation of Hsp70. Chem. Biol. 2015, 22, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Westwood, I.M.; Osborne, J.D.; Matthews, T.P.; Cheeseman, M.; Rowlands, M.G.; Jeganathan, F.; Burke, R.; Lee, D.; Kadi, N.; et al. A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms. Sci. Rep. 2016, 6, 34701. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Cho, H.Y.; Kim, D.G.; Roh, Y.; Son, S.-Y.; Mushtaq, A.U.; Kim, M.; Bhattarai, D.; Sivaraman, A.; Lee, Y.; et al. Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development. Nat. Methods 2019, 16, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Osipiuk, J.; Walsh, M.A.; Freeman, B.C.; Morimoto, R.I.; Joachimiak, A. Structure of a new crystal form of human Hsp70 ATPase domain. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 1105–1107. [Google Scholar] [CrossRef]
- Ravalin, M.; Theofilas, P.; Basu, K.; Opoku-Nsiah, K.A.; Assimon, V.A.; Medina-Cleghorn, D.; Chen, Y.-F.; Bohn, M.F.; Arkin, M.; Grinberg, L.T.; et al. Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases. Nat. Methods 2019, 15, 786–794. [Google Scholar] [CrossRef]
- Shida, M.; Arakawa, A.; Ishii, R.; Kishishita, S.; Takagi, T.; Kukimoto-Niino, M.; Sugano, S.; Tanaka, A.; Shirouzu, M.; Yokoyama, S. Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 223–232. [Google Scholar] [CrossRef]
- Sriram, M.; Osipiuk, J.; Freeman, B.; Morimoto, R.; Joachimiak, A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 1997, 5, 403–414. [Google Scholar] [CrossRef]
- Umehara, K.; Hoshikawa, M.; Tochio, N.; Tate, S.-I. Substrate Binding Switches the Conformation at the Lynchpin Site in the Substrate-Binding Domain of Human Hsp70 to Enable Allosteric Interdomain Communication. Molecules 2018, 23, 528. [Google Scholar] [CrossRef]
- Zhang, P.; Leu, J.I.-J.; Murphy, M.E.; George, D.L.; Marmorstein, R. Crystal Structure of the Stress-Inducible Human Heat Shock Protein 70 Substrate-Binding Domain in Complex with Peptide Substrate. PLoS ONE 2014, 9, e103518. [Google Scholar] [CrossRef]
- Murphy, M.E. The HSP70 family and cancer. Carcinogen 2013, 34, 1181–1188. [Google Scholar] [CrossRef]
- Ha, J.-H.; McKay, D.B. Kinetics of Nucleotide-Induced Changes in the Tryptophan Fluorescence of the Molecular Chaperone Hsc70 and Its Subfragments Suggest the ATP-Induced Conformational Change Follows Initial ATP Binding. Biochemistry 1995, 34, 11635–11644. [Google Scholar] [CrossRef] [PubMed]
- Palleros, D.R.; Raid, K.L.; Shi, L.; Welch, W.J.; Fink, A.L. ATP-induced protein Hsp70 complex dissociation requires K+ but not ATP hydrolysis. Nature 1993, 365, 664–666. [Google Scholar] [CrossRef] [PubMed]
- Schmid, D.; Baici, A.; Gehring, H.; Christen, P. Kinetics of molecular chaperone action. Science 1994, 263, 971–973. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K.; Heuser, J.; Eisenberg, E.; Greene, L. Complex formation between clathrin and uncoating ATPase. J. Biol. Chem. 1994, 269, 6931–6939. [Google Scholar]
- Hartl, F.U. Molecular chaperones in cellular protein folding. Nature 1996, 381, 571–580. [Google Scholar] [CrossRef]
- Laufen, T.; Mayer, M.P.; Beisel, C.; Klostermeier, D.; Mogk, A.; Reinstein, J.; Bukau, B. Mechanism of regulation of Hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. USA 1999, 96, 5452–5457. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef]
- Suzuki, H.; Noguchi, S.; Arakawa, H.; Tokida, T.; Hashimoto, M.; Satow, Y. Peptide-Binding Sites As Revealed by the Crystal Structures of the Human Hsp40 Hdj1 C-Terminal Domain in Complex with the Octapeptide from Human Hsp70. Biochemistry 2010, 49, 8577–8584. [Google Scholar] [CrossRef]
- Jiang, Y.; Rossi, P.; Kalodimos, C.G. Structural basis for client recognition and activity of Hsp40 chaperones. Science 2019, 365, 1313–1319. [Google Scholar] [CrossRef]
- Craig, E.; Huang, P.; Aron, R.; Andrew, A. The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Ergeb. der Physiol. 2006, 156. [Google Scholar] [CrossRef]
- Hageman, J.; Kampinga, H.H. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 2008, 14, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Diamant, S.; Goloubinoff, P. Temperature-Controlled Activity of DnaK−DnaJ−GrpE Chaperones: Protein-Folding Arrest and Recovery during and after Heat Shock Depends on the Substrate Protein and the GrpE Concentration†. Biochemistry 1998, 37, 9688–9694. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Shevde, L.A.; Samant, R.S. Multi-faceted role of HSP40 in cancer. Clin. Exp. Metastasis 2009, 26, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Bracher, A.; Verghese, J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front. Mol. Biosci. 2015, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, J.L.; Bracher, A. Nucleotide Exchange Factors for HSP70 Molecular Chaperones. In Networking of Chaperones by Co-Chaperones; Molecular Biology Intelligence Unit; Springer: New York, NY, USA, 2007; pp. 1–12. [Google Scholar]
- Li, Z.; Hartl, F.U.; Bracher, A. Structure and function of Hip, an attenuator of the Hsp70 chaperone cycle. Nat. Struct. Mol. Biol. 2013, 20, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.M.; Miyata, Y.; Klinedinst, S.; Peng, H.-M.; Chua, J.P.; Komiyama, T.; Li, X.; Morishima, Y.; Merry, D.E.; Pratt, W.B.; et al. Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Methods 2012, 9, 112–118. [Google Scholar] [CrossRef]
- Rousaki, A.; Miyata, Y.; Jinwal, U.K.; Dickey, C.A.; Gestwicki, J.E.; Zuiderweg, E.R.P. Allosteric Drugs: The Interaction of Antitumor Compound MKT-077 with Human Hsp70 Chaperones. J. Mol. Biol. 2011, 411, 614–632. [Google Scholar] [CrossRef]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.; Moarefi, I. Structure of TPR Domain–Peptide Complexes. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef]
- Smith, D.F.; Sullivan, W.P.; Marion, T.N.; Zaitsu, K.; Madden, B.; McCormick, D.J.; O Toft, D. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol. Cell. Biol. 1993, 13, 869–876. [Google Scholar] [CrossRef]
- Honoré, B.; Leffers, H.; Madsen, P.; Rasmussen, H.H.; Vandekerckhove, J.; E Celis, J. Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J. Biol. Chem. 1992, 267, 8485–8491. [Google Scholar]
- Stankiewicz, M.; Nikolay, R.; Rybin, V.; Mayer, M.P. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J. 2010, 277, 3353–3367. [Google Scholar] [CrossRef] [PubMed]
- Höhfeld, J.; Cyr, D.M.; Patterson, C. From the cradle to the grave: Molecular chaperones that may choose between folding and degradation. EMBO Rep. 2001, 2, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Connell, P.; Ballinger, C.A.; Jiang, J.; Wu, Y.; Thompson, L.J.; Höhfeld, J.; Patterson, C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nature 2000, 3, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Meacham, G.C.; Patterson, C.; Zhang, W.; Younger, J.M.; Cyr, D.M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nature 2000, 3, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, C.A.; Connell, P.; Wu, Y.; Hu, Z.; Thompson, L.J.; Yin, L.-Y.; Patterson, C. Identification of CHIP, a Novel Tetratricopeptide Repeat-Containing Protein That Interacts with Heat Shock Proteins and Negatively Regulates Chaperone Functions. Mol. Cell. Biol. 1999, 19, 4535–4545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Amick, J.; Chakravarti, R.; Santarriaga, S.; Schlanger, S.; McGlone, C.; Dare, M.; Nix, J.C.; Scaglione, K.M.; Stuehr, D.J.; et al. A Bipartite Interaction between Hsp70 and CHIP Regulates Ubiquitination of Chaperoned Client Proteins. Structure 2015, 23, 472–482. [Google Scholar] [CrossRef]
- Neckers, L.; Workman, P. Hsp90 molecular chaperone inhibitors: Are we there yet? Clin. Cancer Res. 2012, 18, 64–76. [Google Scholar] [CrossRef]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell. Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef]
- Miyata, Y.; Nakamoto, H.; Neckers, L. The therapeutic target Hsp90 and cancer hallmarks. Curr. Pharm. Des. 2013, 19, 347–365. [Google Scholar] [CrossRef]
- Rodina, A.; Wang, T.; Yan, P.; Gomes, E.D.; Dunphy, M.P.S.; Pillarsetty, N.; Koren, J.; Gerecitano, J.F.; Taldone, T.; Zong, H.; et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature 2016, 538, 397–401. [Google Scholar] [CrossRef]
- Bromberg, Z.; Weiss, Y.G. The Role of the Membrane-Initiated Heat Shock Response in Cancer. Front. Mol. Biosci. 2016, 3, 545. [Google Scholar] [CrossRef] [PubMed]
- Shevtsov, M.; Huile, G.; Multhoff, G. Membrane heat shock protein 70: A theranostic target for cancer therapy. Philos. Trans. R. Soc. B Biol. Sci. 2017, 373, 20160526. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.K.; Wang, H.; Yim, A.M.; Le Naour, F.; Brichory, F.; Jang, J.H.; Zhao, R.; Puravs, E.; Tra, J.; Michael, C.W.; et al. Global Profiling of the Cell Surface Proteome of Cancer Cells Uncovers an Abundance of Proteins with Chaperone Function. J. Biol. Chem. 2002, 278, 7607–7616. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Botzler, C.; Wiesnet, M.; Müller, E.; Meier, T.; Wilmanns, W.; Issels, R.D. A stress-inducible 72-kDa heat-shock protein (HSP72) is expressed on the surface of human tumor cells, but not on normal cells. Int. J. Cancer 1995, 61, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Lauwers, E.; Wang, Y.-C.; Gallardo, R.; Van Der Kant, R.; Michiels, E.; Swerts, J.; Baatsen, P.; Zaiter, S.S.; McAlpine, S.R.; Gounko, N.; et al. Hsp90 Mediates Membrane Deformation and Exosome Release. Mol. Cell 2018, 71, 689–702.e9. [Google Scholar] [CrossRef] [PubMed]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005, 65, 5238–5247. [Google Scholar] [CrossRef] [PubMed]
- Guzhova, I.V.; Shevtsov, M.; Abkin, S.; Pankratova, K.M.; Margulis, B. Intracellular and extracellular Hsp70 chaperone as a target for cancer therapy. Int. J. Hyperth. 2013, 29, 399–408. [Google Scholar] [CrossRef]
- Clayton, A.; Laurino, L.; Wang, X.X.; De La Houssaye, B.A.; Sosa, L.; Dupraz, S.; Cáceres, A.; Pfenninger, K.H.; Quiroga, S. Induction of heat shock proteins in B-cell exosomes. J. Cell Sci. 2005, 118, 3631–3638. [Google Scholar] [CrossRef]
- Lancaster, G.I.; Febbraio, M.A. Exosome-dependent trafficking of HSP70: A novel secretory pathway for cellular stress proteins. J. Biol. Chem. 2005, 280, 23349–23355. [Google Scholar] [CrossRef]
- Mambula, S.S.; Stevenson, M.A.; Ogawa, K.; Calderwood, S. Mechanisms for Hsp70 secretion: Crossing membranes without a leader. Methods 2007, 43, 168–175. [Google Scholar] [CrossRef]
- Juhasz, K.; Thuenauer, R.; Spachinger, A.; Duda, E.; Horvàth, I.; Vigh, L.; Sonnleitner, A.; Balogi, Z. Lysosomal Rerouting of Hsp70 Trafficking as a Potential Immune Activating Tool for Targeting Melanoma. Curr. Pharm. Des. 2013, 19, 430–440. [Google Scholar] [CrossRef]
- Balogi, Z.; Multhoff, G.; Jensen, T.K.; Lloyd-Evans, E.; Yamashima, T.; Jäättelä, M.; Harwood, J.L.; Vígh, L.; Multhoff, G. Hsp70 interactions with membrane lipids regulate cellular functions in health and disease. Prog. Lipid Res. 2019, 74, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Murakami, N.; Kühnel, A.; Schmid, T.E.; Ilicic, K.; Stangl, S.; Braun, I.S.; Gehrmann, M.; Molls, M.; Itami, J.; Multhoff, G. Role of membrane Hsp70 in radiation sensitivity of tumor cells. Radiat. Oncol. 2015, 10, 149. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, M.; Liebisch, G.; Schmitz, G.; Anderson, R.; Steinem, C.; De Maio, A.; Pockley, A.G.; Multhoff, G. Tumor-Specific Hsp70 Plasma Membrane Localization Is Enabled by the Glycosphingolipid Gb3. PLoS ONE 2008, 3, e1925. [Google Scholar] [CrossRef] [PubMed]
- Vega, V.L.; Rodríguez-Silva, M.; Frey, T.; Gehrmann, M.; Diaz, J.C.; Steinem, C.; Multhoff, G.; Arispe, N.; De Maio, A. Hsp70 Translocates into the Plasma Membrane after Stress and Is Released into the Extracellular Environment in a Membrane-Associated Form that Activates Macrophages. J. Immunol. 2008, 180, 4299–4307. [Google Scholar] [CrossRef] [PubMed]
- Arispe, N.; Doh, M.; Simakova, O.; Kurganov, B.; De Maio, A. Hsc70 and Hsp70 interact with phosphatidylserine on the surface of PC12 cells resulting in a decrease of viability. FASEB J. 2004, 18, 1636–1645. [Google Scholar] [CrossRef]
- Mamelak, D.; Lingwood, C. The ATPase Domain of hsp70 Possesses a Unique Binding Specificity for 3′-Sulfogalactolipids. J. Biol.Chem. 2001, 276, 449–456. [Google Scholar] [CrossRef]
- Asea, A.; Kraeft, S.-K.; Kurt-Jones, E.A.; Stevenson, M.A.; Chen, L.B.; Finberg, R.W.; Koo, G.C.; Calderwood, S. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat. Med. 2000, 6, 435–442. [Google Scholar] [CrossRef]
- Asea, A. Novel Signal Transduction Pathway Utilized by Extracellular HSP70. Role of Toll-Like Receptor (TLR) 2 and TLR4. J. Biol. Chem. 2002, 277, 15028–15034. [Google Scholar] [CrossRef]
- Vabulas, R.M. HSP70 as Endogenous Stimulus of the Toll/Interleukin-1 Receptor Signal Pathway. J. Biol. Chem. 2002, 277, 15107–15112. [Google Scholar] [CrossRef]
- Delneste, Y.; Magistrelli, G.; Gauchat, J.-F.; Haeuw, J.-F.; Aubry, J.-P.; Nakamura, K.; Kawakami-Honda, N.; Goetsch, L.; Sawamura, T.; Bonnefoy, J.-Y.; et al. Involvement of LOX-1 in Dendritic Cell-Mediated Antigen Cross-Presentation. Immunity 2002, 17, 353–362. [Google Scholar] [CrossRef]
- Thériault, J.R.; Mambula, S.S.; Sawamura, T.; Stevenson, M.A.; Calderwood, S.K. Extracellular HSP70 binding to surface receptors present on antigen presenting cells and endothelial/epithelial cells. FEBS Lett. 2005, 579, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Binder, R.J.; Ramalingam, T.; Srivastava, P.K. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 2001, 14, 303–313. [Google Scholar] [CrossRef]
- Becker, T.; Hartl, F.-U.; Wieland, F.T. CD40, an extracellular receptor for binding and uptake of Hsp70–peptide complexes. J. Cell Biol. 2002, 158, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Somensi, N.; Brum, P.O.; Ramos, V.D.M.; Gasparotto, J.; Zanotto-Filho, A.; Rostirolla, D.C.; Morrone, M.D.S.; Moreira, J.C.F.; Gelain, D.P. Extracellular HSP70 Activates ERK1/2, NF-kB and Pro-Inflammatory Gene Transcription Through Binding with RAGE in A549 Human Lung Cancer Cells. Cell. Physiol. Biochem. 2017, 42, 2507–2522. [Google Scholar] [CrossRef] [PubMed]
- Hromadníková, I.; Li, S.; Kotlabova, K.; Dickinson, A.M. Influence of In Vitro IL-2 or IL-15 Alone or in Combination with Hsp 70 Derived 14-Mer Peptide (TKD) on the Expression of NK Cell Activatory and Inhibitory Receptors on Peripheral Blood T Cells, B Cells and NKT Cells. PLoS ONE 2016, 11, e0151535. [Google Scholar] [CrossRef]
- Stangl, S.; Gross, C.; Pockley, A.G.; Asea, A.; Multhoff, G. Influence of Hsp70 and HLA-E on the killing of leukemic blasts by cytokine/Hsp70 peptide-activated human natural killer (NK) cells. Cell Stress Chaperones 2008, 13, 221–230. [Google Scholar] [CrossRef]
- Gross, C.; Hansch, D.; Gastpar, R.; Multhoff, G. Interaction of Heat Shock Protein 70 Peptide with NK Cells Involves the NK Receptor CD94. Biol. Chem. 2003, 384. [Google Scholar] [CrossRef]
- An, H.-J.; Kim, Y.J.; Song, N.H.; Park, B.S.; Kim, H.M.; Lee, J.D.; Paik, S.-G.; Lee, J.-O.; Lee, H. Crystallographic and Mutational Analysis of the CD40-CD154 Complex and Its Implications for Receptor Activation*. J. Biol. Chem. 2011, 286, 11226–11235. [Google Scholar] [CrossRef]
- Argiriadi, M.A.; Benatuil, L.; Dubrovska, I.; Egan, D.A.; Gao, L.; Greischar, A.; Hardman, J.; Harlan, J.; Iyer, R.B.; Judge, R.A.; et al. CD40/anti-CD40 antibody complexes which illustrate agonist and antagonist structural switches. BMC Mol. Cell Biol. 2019, 20, 13–29. [Google Scholar] [CrossRef]
- Basu, S.; Yang, J.; Xu, B.; Magiera-Mularz, K.; Skalniak, L.; Musielak, B.; Kholodovych, V.; Holak, T.A.; Hu, L. Design, Synthesis, Evaluation, and Structural Studies of C2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction. J. Med. Chem. 2019, 62, 7250–7263. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.C.; Hong, C.; Akey, I.V.; Yuan, S.; Akey, C. A near atomic structure of the active human apoptosome. eLife 2016, 5, 423. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Langley, R.J.; Brown, P.H.; Xu, G.; Teng, L.; Wang, Q.; I Gonzales, M.; Callender, G.G.; I Nishimura, M.; Topalian, S.L.; et al. Structural basis for the recognition of mutant self by a tumor-specific, MHC class II–restricted T cell receptor. Nat. Immunol. 2007, 8, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Zheng, L.; Lin, J.; Zhang, B.; Zhu, Y.; Li, N.; Xie, S.; Wang, Y.; Gao, N.; Huang, Z. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 2019, 573, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.J.; Esnouf, R.M.; Manso-Sancho, R.; Gilbert, R.J.C.; James, J.R.; Yu, C.; A Fennelly, J.; Vowles, C.; Hanke, T.; Walse, B.; et al. Crystal structure of a soluble CD28-Fab complex. Nat. Immunol. 2005, 6, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Forneris, F.; Wu, J.; Xue, X.; Ricklin, D.; Lin, Z.; Sfyroera, G.; Tzekou, A.; Volokhina, E.; Granneman, J.C.; Hauhart, R.; et al. Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 2016, 35, 1133–1149. [Google Scholar] [CrossRef]
- Ikemizu, S.; Gilbert, R.J.C.; Fennelly, J.A.; Collins, A.V.; Harlos, K.; Jones, Y.; Stuart, D.I.; Davis, S.J. Structure and dimerization of a soluble form of B7-1. Immunity 2000, 12, 51–60. [Google Scholar] [CrossRef]
- Leahy, D.J.; Axel, R.; Hendrickson, W.A. Crystal structure of a soluble form of the human T cell coreceptor CD8 at 2.6 Å resolution. Cell 1992, 68, 1145–1162. [Google Scholar] [CrossRef]
- López-Lucendo, M.F.; Solís, D.; André, S.; Hirabayashi, J.; Kasai, K.-I.; Kaltner, H.; Gabius, H.-J.; Romero, A. Growth-regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-site Mutations and their Impact on the Thermodynamics of Ligand Binding. J. Mol. Biol. 2004, 343, 957–970. [Google Scholar] [CrossRef]
- Madden, D. The antigenic identity of peptide-MHC complexes: A comparison of the conformations of five viral peptides presented by HLA-A2. Cell 1993, 75, 693–708. [Google Scholar] [CrossRef]
- McFarland, B.J.; Kortemme, T.; Yu, S.F.; Baker, D.; Strong, R. Symmetry recognizing asymmetry: Analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure 2003, 11. [Google Scholar]
- Park, B.S.; Song, N.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.; Payne, A.; De Candole, B.; Ford, D.; Hutchinson, B.; Trevitt, G.; Turner, J.; Edwards, C.; Watkins, C.; Whitcombe, I.; et al. Tetrahydroisoquinoline amide substituted phenyl pyrazoles as selective Bcl-2 inhibitors. Bioorganic Med. Chem. Lett. 2009, 19, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Santiago, C.; Celma, M.L.; Stehle, T.; Casasnovas, J.M. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 2009, 17, 124–129. [Google Scholar] [CrossRef]
- Schulze, M.-S.E.D.; Anders, A.-K.; Sethi, D.K.; Call, M. Disruption of Hydrogen Bonds between Major Histocompatibility Complex Class II and the Peptide N-Terminus Is Not Sufficient to Form a Human Leukocyte Antigen-DM Receptive State of Major Histocompatibility Complex Class II. PLoS ONE 2013, 8, e69228. [Google Scholar] [CrossRef]
- Sharif, H.; Wang, L.; Wang, W.L.; Magupalli, V.G.; Andreeva, L.; Qiao, Q.; Hauenstein, A.V.; Wu, Z.; Núñez, G.; Mao, Y.J.; et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 2019, 570, 338–343. [Google Scholar] [CrossRef]
- Solís, D.; Maté, M.J.; Lohr, M.; Ribeiro, J.P.; López-Merino, L.; André, S.; Buzamet, E.; Cañada, F.J.; Kaltner, H.; Lensch, M.; et al. N-domain of human adhesion/growth-regulatory galectin-9: Preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution. Int. J. Biochem. Cell Biol. 2010, 42, 1019–1029. [Google Scholar] [CrossRef]
- Su, L.; Wang, Y.; Wang, J.; Mifune, Y.; Morin, M.D.; Jones, B.T.; Moresco, E.M.Y.; Boger, D.L.; Beutler, B.; Zhang, H. Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim. J. Med. Chem. 2019, 62, 2938–2949. [Google Scholar] [CrossRef]
- Sullivan, L.C.; Clements, C.S.; Beddoe, T.; Johnson, D.; Hoare, H.L.; Lin, J.; Huyton, T.; Hopkins, E.J.; Reid, H.H.; Wilce, M.C.; et al. The Heterodimeric Assembly of the CD94-NKG2 Receptor Family and Implications for Human Leukocyte Antigen-E Recognition. Immunity 2007, 27, 900–911. [Google Scholar] [CrossRef]
- Wu, H.; Myszka, D.G.; Tendian, S.W.; Brouillette, C.G.; Sweet, R.W.; Chaiken, I.M.; Hendrickson, W.A. Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding. Proc. Natl. Acad. Sci. USA 1996, 93, 15030–15035. [Google Scholar] [CrossRef]
- Yoshida, H.; Teraoka, M.; Nishi, N.; Nakakita, S.-I.; Nakamura, T.; Hirashima, M.; Kamitori, S. X-ray Structures of Human Galectin-9 C-terminal Domain in Complexes with a Biantennary Oligosaccharide and Sialyllactose*. J. Biol. Chem. 2010, 285, 36969–36976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Schwartz, J.-C.D.; Almo, S.C.; Nathenson, S.G. Crystal structure of the receptor-binding domain of human B7-2: Insights into organization and signaling. Proc. Natl. Acad. Sci. USA 2003, 100, 2586–2591. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Fedorov, E.; Samanta, D.; Hillerich, B.; Seidel, R.; Almo, S. Crystal Structure of T Cell Costimulatory Ligand B7-1 (CD80). 2014. Available online: http://www.rcsb.org/structure/4RWH (accessed on 1 January 2020).
- Le Nours, J.; Rossjohn, J. Recognition of MHC-Like Molecule 2019. Available online: https://www.rcsb.org/structure/6MWR (accessed on 1 January 2020).
- Pierce, B.; Hourai, Y.; Weng, Z. Accelerating Protein Docking in ZDOCK Using an Advanced 3D Convolution Library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.; Weng, Z. ZRANK: Reranking protein docking predictions with an optimized energy function. Proteins Struct. Funct. Bioinform. 2007, 67, 1078–1086. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Srivastava, P.K. Interaction of Heat Shock Proteins with Peptides and Antigen Presenting Cells: Chaperoning of the Innate and Adaptive Immune Responses. Annu. Rev. Immunol. 2002, 20, 395–425. [Google Scholar] [CrossRef]
- Blachere, N.E.; Li, Z.; Chandawarkar, R.Y.; Suto, R.; Jaikaria, N.S.; Basu, S.; Udono, H.; Srivastava, P.K. Heat Shock Protein–Peptide Complexes, Reconstituted In Vitro, Elicit Peptide-specific Cytotoxic T Lymphocyte Response and Tumor Immunity. J. Exp. Med. 1997, 186, 1315–1322. [Google Scholar] [CrossRef]
- Multhoff, G.; Pfister, K.; Gehrmann, M.; Hantschel, M.; Gross, C.; Hafner, M.; Hiddemann, W. A 14-mer Hsp70 peptide stimulates natural killer (NK) cell activity. Cell Stress Chaperons 2001, 6, 337–344. [Google Scholar] [CrossRef]
- Multhoff, G.; Botzler, C.; Jennen, L.; Schmidt, J.; Ellwart, J.; Issels, R. Heat shock protein 72 on tumor cells: A recognition structure for natural killer cells. J. Immunol. 1997, 158, 4341–4350. [Google Scholar]
- Multhoff, G. Heat shock protein 70 (Hsp70) stimulates proliferation and cytolytic activity of natural killer cells. Exp. Hematol. 1999, 27, 1627–1636. [Google Scholar] [CrossRef]
- Specht, H.M.; Ahrens, N.; Blankenstein, C.; Duell, T.; Fietkau, R.; Gaipl, U.S.; Günther, C.; Gunther, S.; Habl, G.; Hautmann, H.; et al. Heat Shock Protein 70 (Hsp70) Peptide Activated Natural Killer (NK) Cells for the Treatment of Patients with Non-Small Cell Lung Cancer (NSCLC) after Radiochemotherapy (RCTx) – From Preclinical Studies to a Clinical Phase II Trial. Front. Immunol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Rückert, M.; Deloch, L.; Fietkau, R.; Frey, B.; Hecht, M.; Gaipl, U.S. Immune modulatory effects of radiotherapy as basis for well-reasoned radioimmunotherapies. Strahlenther. und Onkol. 2018, 194, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Günther, S.; Ostheimer, C.; Stangl, S.; Specht, H.M.; Mozes, P.; Jesinghaus, M.; Vordermark, D.; Combs, S.E.; Peltz, F.; Jung, M.P.; et al. Correlation of Hsp70 Serum Levels with Gross Tumor Volume and Composition of Lymphocyte Subpopulations in Patients with Squamous Cell and Adeno Non-Small Cell Lung Cancer. Front. Immunol. 2015, 6, 128. [Google Scholar] [CrossRef] [PubMed]
- Ostheimer, C.; Gunther, S.; Bache, M.; Vordermark, D.; Multhoff, G. Dynamics of Heat Shock Protein 70 Serum Levels As a Predictor of Clinical Response in Non-Small-Cell Lung Cancer and Correlation with the Hypoxia-Related Marker Osteopontin. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- A Daniels, G.; Sanchez-Perez, L.; Diaz, R.M.; Kottke, T.; Thompson, J.; Lai, M.; Gough, M.; Karim, M.; Bushell, A.; Chong, H.; et al. A simple method to cure established tumors by inflammatory killing of normal cells. Nat. Biotechnol. 2004, 22, 1125–1132. [Google Scholar] [CrossRef]
- Breloer, M.; Fleischer, B.; Von Bonin, A. In vivo and in vitro activation of T cells after administration of Ag-negative heat shock proteins. J. Immunol. 1999, 162. [Google Scholar]
- Rothammer, A.; Sage, E.K.; Werner, C.; Combs, S.E.; Multhoff, G. Increased heat shock protein 70 (Hsp70) serum levels and low NK cell counts after radiotherapy - potential markers for predicting breast cancer recurrence? Radiat. Oncol. 2019, 14, 78. [Google Scholar] [CrossRef]
- Pfister, K.; Radons, J.; Busch, R.; Tidball, J.G.; Pfeifer, M.; Freitag, L.; Feldmann, H.-J.; Milani, V.; Issels, R.; Multhoff, G. Patient survival by Hsp70 membrane phenotype. Cancer 2007, 110, 926–935. [Google Scholar] [CrossRef]
- Zininga, T.; Ramatsui, L.; Shonhai, A. Heat Shock Proteins as Immunomodulants. Molecules 2018, 23, 2846. [Google Scholar] [CrossRef]
- Wachstein, J.; Tischer, S.; Figueiredo, C.; Limbourg, A.; Falk, C.; Immenschuh, S.; Blasczyk, R.; Eiz-Vesper, B. HSP70 Enhances Immunosuppressive Function of CD4+CD25+FoxP3+ T Regulatory Cells and Cytotoxicity in CD4+CD25− T Cells. PLoS ONE 2012, 7, e51747. [Google Scholar] [CrossRef]
- Asea, A. Initiation of the Immune Response by Extracellular Hsp72: Chaperokine Activity of Hsp72. Curr. Immunol. Rev. 2006, 2, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Asea, A. Chaperokine-induced signal transduction pathways. Exerc. Immunol. Rev. 2003, 9, 25–33. [Google Scholar] [PubMed]
- Martine, P.; Chevriaux, A.; Derangère, V.; Apetoh, L.; Garrido, C.; Ghiringhelli, F.; Rebe, C. HSP70 is a negative regulator of NLRP3 inflammasome activation. Cell Death Dis. 2019, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, M.; Parsamanesh, N.; Bahrami, A.; Atkin, S.; Sahebkar, A. Role of the NLRP3 inflammasome in cancer. Mol. Cancer 2018, 17, 158. [Google Scholar] [CrossRef] [PubMed]
- Yashin, D.V.; Romanova, E.A.; Ivanova, O.K.; Sashchenko, L.P. The Tag7–Hsp70 cytotoxic complex induces tumor cell necroptosis via permeabilisation of lysosomes and mitochondria. Biochimie 2016, 123, 32–36. [Google Scholar] [CrossRef]
- Sashchenko, L.P.; Dukhanina, E.A.; Yashin, D.V.; Shatalov, Y.V.; Romanova, E.A.; Korobko, E.V.; Demin, A.V.; Lukyanova, T.I.; Kabanova, O.D.; Khaidukov, S.V.; et al. Peptidoglycan Recognition Protein Tag7 Forms a Cytotoxic Complex with Heat Shock Protein 70 in Solution and in Lymphocytes. J. Biol. Chem. 2003, 279, 2117–2124. [Google Scholar] [CrossRef]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.-P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef]
- Bausero, M.A.; Gastpar, R.; Multhoff, G.; Asea, A. Alternative Mechanism by which IFN-γ Enhances Tumor Recognition: Active Release of Heat Shock Protein 721. J. Immunol. 2005, 175, 2900–2912. [Google Scholar] [CrossRef]
- De Vita, F.; Orditura, M.; Galizia, G.; Romano, C.; Infusino, S.; Auriemma, A.; Lieto, E.; Catalano, G. Serum interleukin-10 levels in patients with advanced gastrointestinal malignancies. Cancer 1999, 86, 1936–1943. [Google Scholar] [CrossRef]
- Lebrecht, A.; Hefler, L.; Tempfer, C.B.; Koelbl, H. Serum Cytokine Concentrations in Patients with Cervical Cancer: Interleukin-4, Interferon-γ, and Monocyte Chemoattractant Protein-1. Gynecol. Oncol. 2001, 83, 170–171. [Google Scholar] [CrossRef]
- Udono, H.; Srivastava, P.K. Comparison of tumor-specific immunogenicities of stress-induced proteins gp96, hsp90, and hsp70. J. Immunol. 1994, 152, 5398–5403. [Google Scholar]
- Weng, D.; Calderwood, S.K.; Gong, J. Preparation of a heat-shock protein 70-based vaccine from DC-tumor fusion cells. Breast Cancer 2011, 787, 255–265. [Google Scholar]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Murshid, A. Molecular Chaperone Accumulation in Cancer and Decrease in Alzheimer’s Disease: The Potential Roles of HSF1. Front. Neurosci. 2017, 11, 192. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.D.; Caron, A.W.; Bourget, L.; Denis-Larose, C.; Massie, B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol. Cell. Biol. 1997, 17, 5317–5327. [Google Scholar] [CrossRef]
- Nylandsted, J.; Rohde, M.; Brand, K.; Bastholm, L.; Elling, F.; Jäättelä, M. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. USA 2000, 97, 7871–7876. [Google Scholar] [CrossRef]
- Gao, Y.; Han, C.; Huang, H.; Xin, Y.; Xu, Y.; Luo, L.; Yin, Z. Heat shock protein 70 together with its co-chaperone CHIP inhibits TNF-α induced apoptosis by promoting proteasomal degradation of apoptosis signal-regulating kinase1. Apoptosis 2010, 15, 822–833. [Google Scholar] [CrossRef]
- Tournier, C. Requirement of JNK for Stress- Induced Activation of the Cytochrome c-Mediated Death Pathway. Science 2000, 288, 870–874. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Reddy, E.P. JNK signaling in apoptosis. Oncogene 2008, 27, 6245–6251. [Google Scholar] [CrossRef]
- Sax, J.K.; Fei, P.; Murphy, M.E.; Bernhard, E.; Korsmeyer, S.J.; El-Deiry, W.S. BID regulation by p53 contributes to chemosensitivity. Nature 2002, 4, 842–849. [Google Scholar] [CrossRef]
- Ravagnan, L.; Gurbuxani, S.; Susin, S.A.; Maisse, C.; Daugas, E.; Zamzami, N.; Mak, T.; Jäättelä, M.; Penninger, J.M.; Garrido, C.; et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nature 2001, 3, 839–843. [Google Scholar] [CrossRef]
- Guo, F.; Sigua, C.; Bali, P.; George, P.; Fiskus, W.; Scuto, A.; Annavarapu, S.; Mouttaki, A.; Sondarva, G.; Wei, S.; et al. Mechanistic role of heat shock protein 70 in Bcr-Abl–mediated resistance to apoptosis in human acute leukemia cells. Blood 2005, 105, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Gabai, V.; Mabuchi, K.; Mosser, D.D.; Sherman, M. Hsp72 and Stress Kinase c-jun N-Terminal Kinase Regulate the Bid-Dependent Pathway in Tumor Necrosis Factor-Induced Apoptosis. Mol. Cell. Biol. 2002, 22, 3415–3424. [Google Scholar] [CrossRef] [PubMed]
- Nylandsted, J.; Gyrd-Hansen, M.; Danielewicz, A.; Fehrenbacher, N.; Lademann, U.; Høyer-Hansen, M.; Weber, E.; Multhoff, G.; Rohde, M.; Jäättelä, M. Heat Shock Protein 70 Promotes Cell Survival by Inhibiting Lysosomal Membrane Permeabilization. J. Exp. Med. 2004, 200, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Bivik, C.; Rosdahl, I.; Öllinger, K. Hsp70 protects against UVB induced apoptosis by preventing release of cathepsins and cytochrome c in human melanocytes. Carcinogen 2006, 28, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, M.; Kirkegaard-Sorensen, T.; Ostenfeld, M.S.; Aaboe, M.; Hoyer-Hansen, M.; Orntoft, T.F.; Rohde, M.; Jäättelä, M. Lens Epithelium-Derived Growth Factor Is an Hsp70-2 Regulated Guardian of Lysosomal Stability in Human Cancer. Cancer Res. 2007, 67, 2559–2567. [Google Scholar] [CrossRef]
- Park, S.-H.; Baek, K.-H.; Shin, I.; Shin, I. Subcellular Hsp70 Inhibitors Promote Cancer Cell Death via Different Mechanisms. Cell Chem. Biol. 2018, 25, 1242–1254.e8. [Google Scholar] [CrossRef]
- Yaglom, J.A.; Ekhterae, D.; Gabai, V.; Sherman, M. Regulation of Necrosis of H9c2 Myogenic Cells upon Transient Energy Deprivation. J. Biol. Chem. 2003, 278, 50483–50496. [Google Scholar] [CrossRef]
- Karsch-Bluman, A.; Feiglin, A.; Arbib, E.; Stern, T.; Shoval, H.; Schwob, O.; Berger, M.; Benny, O. Tissue necrosis and its role in cancer progression. Oncogene 2018, 38, 1920–1935. [Google Scholar] [CrossRef]
- Stankiewicz, A.R.; Lachapelle, G.; Foo, C.P.Z.; Radicioni, S.M.; Mosser, D.D. Hsp70 Inhibits Heat-induced Apoptosis Upstream of Mitochondria by Preventing Bax Translocation. J. Biol. Chem. 2005, 280, 38729–38739. [Google Scholar] [CrossRef]
- Beere, H.M.; Wolf, B.B.; Cain, K.; Mosser, D.D.; Mahboubi, A.; Kuwana, T.; Tailor, P.; Morimoto, R.I.; Cohen, G.M.; Green, D.R. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nature 2000, 2, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Yaglom, J.A.; Gabai, V.; Sherman, M. High Levels of Heat Shock Protein Hsp72 in Cancer Cells Suppress Default Senescence Pathways. Cancer Res. 2007, 67, 2373–2381. [Google Scholar] [CrossRef]
- Gabai, V.; Yaglom, J.A.; Waldman, T.; Sherman, M.Y. Heat Shock Protein Hsp72 Controls Oncogene-Induced Senescence Pathways in Cancer Cells. Mol. Cell. Biol. 2008, 29, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Hunt, C.; Yaglom, J.A.; Gabai, V.; Sherman, M.Y. Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene 2011, 30, 2836–2845. [Google Scholar] [CrossRef] [PubMed]
- Colvin, T.A.; Gabai, V.; Gong, J.; Calderwood, S.K.; Li, H.; Gummuluru, S.; Matchuk, O.; Smirnova, S.G.; Orlova, N.V.; Zamulaeva, I.A.; et al. Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014, 74, 4731–4740. [Google Scholar] [CrossRef]
- Meng, L.; Gabai, V.; Sherman, M.Y. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 2010, 29, 5204–5213. [Google Scholar] [CrossRef]
- Oda, T.; Sekimoto, T.; Kurashima, K.; Fujimoto, M.; Nakai, A.; Yamashita, T. Acute HSF1 depletion induces cellular senescence through the MDM2-p53-p21 pathway in human diploid fibroblasts. J. Cell Sci. 2018, 131, jcs210724. [Google Scholar] [CrossRef]
- Boysen, M.; Kityk, R.; Mayer, M.P. Hsp70- and Hsp90-Mediated Regulation of the Conformation of p53 DNA Binding Domain and p53 Cancer Variants. Mol. Cell 2019, 74, 831–843.e4. [Google Scholar] [CrossRef]
- Dahiya, V.; Agam, G.; Lawatscheck, J.; Rutz, D.A.; Lamb, D.C.; Buchner, J. Coordinated Conformational Processing of the Tumor Suppressor Protein p53 by the Hsp70 and Hsp90 Chaperone Machineries. Mol. Cell 2019, 74, 816–830.e7. [Google Scholar] [CrossRef]
- Han, J.M.; Park, B.-J.; Park, S.G.; Oh, Y.S.; Choi, S.J.; Lee, S.W.; Hwang, S.-K.; Chang, S.-H.; Cho, M.-H.; Kim, S. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. USA 2008, 105, 11206–11211. [Google Scholar] [CrossRef]
- Huang, L.E.; Bunn, H.F. Hypoxia-inducible Factor and Its Biomedical Relevance. J. Biol. Chem. 2003, 278, 19575–19578. [Google Scholar] [CrossRef] [PubMed]
- Ryan, H.E.; Lo, J.; Johnson, R.S. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998, 17, 3005–3015. [Google Scholar] [CrossRef] [PubMed]
- Gordan, J.D.; Simon, M.C. Hypoxia-inducible factors: Central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 2007, 17, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Schmid, T.; Frank, R.; Brüne, B. PI3K/Akt Is Required for Heat Shock Proteins to Protect Hypoxia-inducible Factor 1 from pVHL-independent Degradation. J. Biol. Chem. 2004, 279, 13506–13513. [Google Scholar] [CrossRef]
- Gabai, V.; Meng, L.; Kim, G.; Mills, T.A.; Benjamin, I.J.; Sherman, M.Y. Heat Shock Transcription Factor Hsf1 Is Involved in Tumor Progression via Regulation of Hypoxia-Inducible Factor 1 and RNA-Binding Protein HuR. Mol. Cell. Biol. 2012, 32, 929–940. [Google Scholar] [CrossRef]
- De Silanes, I.L.; Fan, J.; Yang, X.; Zonderman, A.B.; Potapova, O.; Pizer, E.S.; Gorospe, M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003, 22, 7146–7154. [Google Scholar] [CrossRef]
- Kim, T.-K.; Na, H.-J.; Lee, W.R.; Jeoung, M.H.; Lee, S. Heat shock protein 70-1A is a novel angiogenic regulator. Biochem. Biophys. Res. Commun. 2016, 469, 222–228. [Google Scholar] [CrossRef]
- Park, S.L.; Chung, T.-W.; Kim, S.; Hwang, B.; Kim, J.M.; Lee, H.M.; Cha, H.-J.; Seo, Y.; Choe, S.Y.; Ha, K.-T.; et al. HSP70-1 is required for interleukin-5-induced angiogenic responses through eNOS pathway. Sci. Rep. 2017, 7, 44687. [Google Scholar] [CrossRef]
- Kluger, H.M.; Goel, H.L.; Breen, M.; Zhang, J.; Das, I.; Aznavoorian-Cheshire, S.; Greenberg, N.M.; Elgavish, A.; Languino, L. Using a Xenograft Model of Human Breast Cancer Metastasis to Find Genes Associated with Clinically Aggressive Disease. Cancer Res. 2005, 65, 5578–5587. [Google Scholar] [CrossRef]
- Teng, Y.; Ngoka, L.; Mei, Y.; Lesoon, L.; Cowell, J. HSP90 and HSP70 Proteins Are Essential for Stabilization and Activation of WASF3 Metastasis-promoting Protein*. J. Biol. Chem. 2012, 287, 10051–10059. [Google Scholar] [CrossRef]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Kasioumi, P.; Vrazeli, P.; Vezyraki, P.; Zerikiotis, S.; Katsouras, C.; Damalas, A.; Angelidis, C. Hsp70 (HSP70A1A) downregulation enhances the metastatic ability of cancer cells. Int. J. Oncol. 2018, 54, 821–832. [Google Scholar] [CrossRef]
- Mao, H.; Li, F.; Ruchalski, K.; Mosser, D.D.; Schwartz, J.; Wang, Y.; Borkan, S.C.; Stortchevoi, A.; Varshney, U.; Rajbhandary, U.L. hsp72 Inhibits Focal Adhesion Kinase Degradation in ATP-depleted Renal Epithelial Cells. J. Biol. Chem. 2003, 278, 18214–18220. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.W.; Lee, J.E.; Kim, S.Y.; Kang, M.-W.; Na, M.H.; Lee, C.S.; Song, K.S.; Lim, S.P. The C-terminus of Hsp70-Interacting Protein Promotes Met Receptor Degradation. J. Thorac. Oncol. 2011, 6, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Iii, J.K.; Jinwal, U.K.; Jin, Y.; O’Leary, J.; Jones, J.; Johnson, A.G.; Blair, L.J.; Abisambra, J.F.; Chang, L.; Miyata, Y.; et al. Facilitating Akt Clearance via Manipulation of Hsp70 Activity and Levels*. J. Biol. Chem. 2009, 285, 2498–2505. [Google Scholar]
- Li, H.; Li, Y.; Liu, D.; Sun, H.; Su, N.; Yang, F.; Liu, J. Extracellular HSP70/HSP70-PCs Promote Epithelial-Mesenchymal Transition of Hepatocarcinoma Cells. PLoS ONE 2013, 8, e84759. [Google Scholar] [CrossRef]
- Komarova, E.; Marchenko, L.; Zhakhov, A.; Nikotina, A.; Aksenov, N.; Suezov, R.; Ischenko, A.; Margulis, B.; Guzhova, I.V. Extracellular Hsp70 Reduces the Pro-Tumor Capacity of Monocytes/Macrophages Co-Cultivated with Cancer Cells. Int. J. Mol. Sci. 2019, 21, 59. [Google Scholar] [CrossRef]
- Mao, H.; Li, Z.; Zhou, Y.; Li, Z.; Zhuang, S.; An, X.; Zhang, B.; Chen, W.; Nie, J.; Wang, Z.; et al. HSP72 attenuates renal tubular cell apoptosis and interstitial fibrosis in obstructive nephropathy. Am. J. Physiol. Physiol. 2008, 295, F202–F214. [Google Scholar] [CrossRef]
- Yun, C.-H.; Yoon, S.-Y.; Nguyen, T.T.; Cho, H.-Y.; Kim, T.-H.; Kim, S.-T.; Kim, B.-C.; Hong, Y.-S.; Kim, S.-J.; Lee, H.J. Geldanamycin inhibits TGF-β signaling through induction of Hsp70. Arch. Biochem. Biophys. 2010, 495, 8–13. [Google Scholar] [CrossRef]
- Li, Y.; Kang, X.; Wang, Q. HSP70 decreases receptor-dependent phosphorylation of Smad2 and blocks TGF-β-induced epithelial-mesenchymal transition. J. Genet. Genom. 2011, 38, 111–116. [Google Scholar] [CrossRef]
- Liu, J.; Bao, J.; Hao, J.; Peng, Y.; Hong, F. HSP70 inhibits high glucose-induced Smad3 activation and attenuates epithelial-to-mesenchymal transition of peritoneal mesothelial cells. Mol. Med. Rep. 2014, 10, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.-M.; Lee, J. MET: Roles in epithelial-mesenchymal transition and cancer stemness. Ann. Transl. Med. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Lai, C.-H.; Tsai, C.-L.; Hsueh, S.; Hsueh, C.; Lin, C.-Y.; Chou, H.-H.; Lin, Y.-J.; Chen, H.-W.; Chang, T.-C.; et al. Tumor Stress-Induced Phosphoprotein1 (STIP1) as a Prognostic Biomarker in Ovarian Cancer. PLoS ONE 2013, 8, e57084. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, A.C.C.; Wang, H.; Fan, H.; Chen, X.; Zhang, I.; Zhang, L.; Lima, F.R.S.; Badie, B. Increased expression of stress inducible protein 1 in glioma-associated microglia/macrophages. J. Neuroimmunol. 2014, 274, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Willmer, T.; Contu, L.; Blatch, G.; Edkins, A.L. Knockdown of Hop downregulates RhoC expression, and decreases pseudopodia formation and migration in cancer cell lines. Cancer Lett. 2013, 328, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Walsh, N.; Larkin, A.; Swan, N.; Conlon, K.; Dowling, P.; McDermott, R.; Clynes, M. RNAi knockdown of Hop (Hsp70/Hsp90 organising protein) decreases invasion via MMP-2 down regulation. Cancer Lett. 2011, 306, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Iwasaki, M.; Sugio, A.; Hishiya, A.; Tanaka, R.; Endo, T.; Takayama, S.; Saito, T. BAG3 (BCL2-associated athanogene 3) interacts with MMP-2 to positively regulate invasion by ovarian carcinoma cells. Cancer Lett. 2011, 303, 65–71. [Google Scholar] [CrossRef]
- Lee, K.-J.; Kim, Y.M.; Kim, D.Y.; Jeoung, O.; Han, K.; Lee, S.-T.; Lee, Y.-S.; Park, K.H.; Park, J.H.; Kim, D.J.; et al. Release of heat shock protein 70 (Hsp70) and the effects of extracellular Hsp70 on matric metalloproteinase-9 expression in human monocytic U937 cells. Exp. Mol. Med. 2006, 38, 364–374. [Google Scholar] [CrossRef]
- Chetty, C.; Vanamala, S.K.; Gondi, C.; Dinh, D.H.; Gujrati, M.; Rao, J.S. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell. Signal. 2011, 24, 549–559. [Google Scholar] [CrossRef]
- Sossey-Alaoui, K.; Li, X.; Ranalli, T.A.; Cowell, J. WAVE3-mediated Cell Migration and Lamellipodia Formation Are Regulated Downstream of Phosphatidylinositol 3-Kinase*. J. Biol. Chem. 2005, 280, 21748–21755. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, M.Q.; Cheney, R.; Sharma, S.; Cowell, J. Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br. J. Cancer 2010, 103, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Sossey-Alaoui, K.; Safina, A.; Li, X.; Vaughan, M.M.; Hicks, D.G.; Bakin, A.; Cowell, J. Down-Regulation of WAVE3, a Metastasis Promoter Gene, Inhibits Invasion and Metastasis of Breast Cancer Cells. Am. J. Pathol. 2007, 170, 2112–2121. [Google Scholar] [CrossRef] [PubMed]
- Breuninger, S.; Stangl, S.; Werner, C.; Sievert, W.; Lobinger, D.; Foulds, G.; Wagner, S.; Pickhard, A.; Piontek, G.; Kokowski, K.; et al. Membrane Hsp70—A Novel Target for the Isolation of Circulating Tumor Cells After Epithelial-to-Mesenchymal Transition. Front. Oncol. 2018, 8, 497. [Google Scholar] [CrossRef] [PubMed]
- Farkas, B.; Hantschel, M.; Magyarlaki, M.; Becker, B.; Scherer, K.; Landthaler, M.; Pfister, K.; Gehrmann, M.; Gross, C.; Mackensen, A.; et al. Heat shock protein 70 membrane expression and melanoma-associated marker phenotype in primary and metastatic melanoma. Melanoma Res. 2003, 13, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Botzler, C.; Schmidt, J.; Luz, A.; Jennen, L.; Issels, R.; Multhoff, G. Differential Hsp70 plasma-membrane expression on primary human tumors and metastases in mice with severe combined immunodeficiency. Int. J. Cancer 1998, 77, 942–948. [Google Scholar] [CrossRef]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Ben-Jacob, E.; Onuchic, J.N.; Levine, H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef]
- Stangl, S.; Gehrmann, M.; Riegger, J.; Kuhs, K.; Riederer, I.; Sievert, W.; Hube, K.; Mocikat, R.; Dressel, R.; Kremmer, E.; et al. Targeting membrane heat-shock protein 70 (Hsp70) on tumors by cmHsp70.1 antibody. Proc. Natl. Acad. Sci. USA USA 2010, 108, 733–738. [Google Scholar] [CrossRef]
- Williamson, D.; Borgognoni, J.; Clay, A.; Daniels, Z.; Dokurno, P.; Drysdale, M.J.; Foloppe, N.; Francis, G.L.; Graham, C.J.; Howes, R.; et al. Novel Adenosine-Derived Inhibitors of 70 kDa Heat Shock Protein, Discovered Through Structure-Based Design†. J. Med. Chem. 2009, 52, 1510–1513. [Google Scholar] [CrossRef]
- Macias, A.; Williamson, D.; Allen, N.; Borgognoni, J.; Clay, A.; Daniels, Z.; Dokurno, P.; Drysdale, M.J.; Francis, G.L.; Graham, C.J.; et al. Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity. J. Med. Chem. 2011, 54, 4034–4041. [Google Scholar] [CrossRef]
- Li, X.; Colvin, T.; Rauch, J.N.; Acosta-Alvear, D.; Kampmann, M.; Dunyak, B.; Hann, B.; Aftab, B.T.; Murnane, M.; Cho, M.; et al. Validation of the Hsp70-Bag3 protein-protein interaction as a potential therapeutic target in cancer. Mol. Cancer Ther. 2015, 14, 642–648. [Google Scholar] [CrossRef]
- Yaglom, J.A.; Wang, Y.; Li, A.; Li, Z.; Monti, S.; Alexandrov, I.; Lu, X.; Sherman, M.Y. Cancer cell responses to Hsp70 inhibitor JG-98: Comparison with Hsp90 inhibitors and finding synergistic drug combinations. Sci. Rep. 2018, 8, 3010. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.; Gabai, V. Hsp70 in cancer: Back to the future. Oncogene 2014, 34, 4153–4161. [Google Scholar] [CrossRef] [PubMed]
- Koren, J.; Miyata, Y.; Kiray, J.; O’Leary, J.C.; Nguyen, L.; Guo, J.; Blair, L.J.; Li, X.; Jinwal, U.K.; Cheng, J.Q.; et al. Correction: Rhodacyanine Derivative Selectively Targets Cancer Cells and Overcomes Tamoxifen Resistance. PLoS ONE 2012, 7. [Google Scholar]
- Massey, A.J.; Williamson, D.; Browne, H.; Murray, J.B.; Dokurno, P.; Shaw, T.; Macias, A.; Daniels, Z.; Geoffroy, S.; Dopson, M.; et al. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 2009, 66, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Deocaris, C.C.; Widodo, N.; Shrestha, B.G.; Kaur, K.; Ohtaka, M.; Yamasaki, K.; Kaul, S.C.; Wadhwa, R. Mortalin sensitizes human cancer cells to MKT-077-induced senescence. Cancer Lett. 2007, 252, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.I.-J.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A Small Molecule Inhibitor of Inducible Heat Shock Protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef]
- Schmitt, E.; Maingret, L.; Puig, P.-E.; Rérole, A.-L.; Hammann, A.; Ghiringhelli, F.; Solary, E.; Kroemer, G.; Garrido, C. Heat Shock Protein 70 Neutralization Exerts Potent Antitumor Effects in Animal Models of Colon Cancer and Melanoma. Cancer Res. 2006, 66, 4191–4197. [Google Scholar] [CrossRef]
- Ernst, K.; Liebscher, M.; Mathea, S.; Granzhan, A.; Schmid, J.; Popoff, M.R.; Ihmels, H.; Barth, H.; Schiene-Fischer, C. A novel Hsp70 inhibitor prevents cell intoxication with the actin ADP-ribosylating Clostridium perfringens iota toxin. Sci. Rep. 2016, 6, 20301. [Google Scholar] [CrossRef]
- Rérole, A.-L.; Gobbo, J.; De Thonel, A.; Schmitt, E.; De Barros, J.P.P.; Hammann, A.; Lanneau, D.; Fourmaux, E.; Demidov, O.; Micheau, O.; et al. Peptides and Aptamers Targeting HSP70: A Novel Approach for Anticancer Chemotherapy. Cancer Res. 2011, 71, 484–495. [Google Scholar] [CrossRef]
- Rodina, A.; Patel, P.D.; Kang, Y.; Patel, Y.; Baaklini, I.; Wong, M.J.; Taldone, T.; Yan, P.; Yang, C.; Maharaj, R.; et al. Identification of an Allosteric Pocket on Human Hsp70 Reveals a Mode of Inhibition of This Therapeutically Important Protein. Chem. Biol. 2013, 20, 1469–1480. [Google Scholar] [CrossRef]
- Williams, D.R.; Park, S.; Shin, I.; Ko, S.-K.; Lee, M.-R. An Apoptosis-Inducing Small Molecule That Binds to Heat Shock Protein 70. Angew. Chem. Int. Ed. 2008, 47, 7466–7469. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.-K.; Kim, J.; Na, D.C.; Park, S.; Park, S.-H.; Hyun, J.Y.; Baek, K.-H.; Kim, N.D.; Kim, N.-K.; Park, Y.N.; et al. A Small Molecule Inhibitor of ATPase Activity of HSP70 Induces Apoptosis and Has Antitumor Activities. Chem. Biol. 2015, 22, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, M.J.; Scott, S.S.; Scott, C.M.; Behrman, S.; Walter, P.; Wipf, P.; Coplan, J.D.; Chrico, W.; Joseph, D.; Brodsky, J.L.; et al. Antimyeloma Effects of the Heat Shock Protein 70 Molecular Chaperone Inhibitor MAL3-101. J. Oncol. 2011, 2011, 1–11. [Google Scholar] [CrossRef]
- Fewell, S.W.; Smith, C.M.; Lyon, M.A.; Dumitrescu, T.P.; Wipf, P.; Day, B.W.; Brodsky, J.L. Small Molecule Modulators of Endogenous and Co-chaperone-stimulated Hsp70 ATPase Activity. J. Biol. Chem. 2004, 279, 51131–51140. [Google Scholar] [CrossRef]
- Huryn, D.; Brodsky, J.L.; Brummond, K.M.; Chambers, P.G.; Eyer, B.; Ireland, A.W.; Kawasumi, M.; Laporte, M.G.; Lloyd, K.; Manteau, B.; et al. Chemical methodology as a source of small-molecule checkpoint inhibitors and heat shock protein 70 (Hsp70) modulators. Proc. Natl. Acad. Sci. USA USA 2011, 108, 6757–6762. [Google Scholar] [CrossRef]
- Phillips, P.; Sangwan, V.; Borja-Cacho, D.; Dudeja, V.; Vickers, S.; Saluja, A.K. Myricetin induces pancreatic cancer cell death via the induction of apoptosis and inhibition of the phosphatidylinositol 3-kinase (PI3K) signaling pathway. Cancer Lett. 2011, 308, 181–188. [Google Scholar] [CrossRef]
- Howe, M.K.; Bodoor, K.; Carlson, D.A.; Hughes, P.F.; Alwarawrah, Y.; Loiselle, D.R.; Jaeger, A.M.; Darr, D.B.; Jordan, J.L.; Hunter, L.M.; et al. Identification of an allosteric small-molecule inhibitor selective for the inducible form of heat shock protein 70. Chem. Biol. 2014, 21, 1648–1659. [Google Scholar] [CrossRef]
- Phillips, P.A.; Dudeja, V.; McCarroll, J.A.; Borja-Cacho, D.; Dawra, R.K.; Grizzle, W.E.; Vickers, S.M.; Saluja, A.K. Triptolide Induces Pancreatic Cancer Cell Death via Inhibition of Heat Shock Protein 70. Cancer Res. 2007, 67, 9407–9416. [Google Scholar] [CrossRef]
- Banerjee, S.; Thayanithy, V.; Sangwan, V.; MacKenzie, T.N.; Saluja, A.K.; Subramanian, S. Minnelide reduces tumor burden in preclinical models of osteosarcoma. Cancer Lett. 2013, 335, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Antonoff, M.B.; Chugh, R.; Skube, S.J.; Dudeja, V.; Borja-Cacho, D.; Clawson, K.A.; Vickers, S.M.; Saluja, A.K. Role of Hsp-70 in Triptolide-Mediated Cell Death of Neuroblastoma. J. Surg. Res. 2010, 163, 72–78. [Google Scholar] [CrossRef]
- MacKenzie, T.N.; Mujumdar, N.; Banerjee, S.; Sangwan, V.; Sarver, A.; Vickers, S.; Subramanian, S.; Saluja, A.K. Triptolide induces the expression of miR-142-3p: A negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol. Cancer Ther. 2013, 12, 1266–1275. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, B.A.; Chen, E.Z.; Tang, S.; Belgum, H.S.; McCauley, J.A.; Evenson, K.A.; Etchison, R.G.; Jay-Dixon, J.; Patel, M.; Raza, A.; et al. Triptolide and its prodrug minnelide suppress Hsp70 and inhibit in vivo growth in a xenograft model of mesothelioma. Genes Cancer 2015, 6, 144–152. [Google Scholar] [PubMed]
- Rousalova, I.; Banerjee, S.; Sangwan, V.; Evenson, K.; McCauley, J.A.; Kratzke, R.; Vickers, S.M.; Saluja, A.K.; D’Cunha, J. Minnelide: A Novel Therapeutic That Promotes Apoptosis in Non-Small Cell Lung Carcinoma In Vivo. PLoS ONE 2013, 8, e77411. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q. Triptolide and its expanding multiple pharmacological functions. Int. Immunopharmacol. 2011, 11, 377–383. [Google Scholar] [CrossRef]
- Propper, D.; Han, H.; Von Hoff, D.; Borazanci, E.; Reya, T.; Ghergurovich, J.; Pshenichnaya, I.; Antal, C.; Condjella, R.; Sharma, S.; et al. Abstract CT165: Phase II open label trial of minnelide™ in patients with chemotherapy refractory metastatic pancreatic cancer. Clin. Trials 2019. [Google Scholar]
- Hung, C.-M.; Su, Y.-H.; Lin, H.-Y.; Lin, J.-N.; Liu, L.-C.; Ho, C.-T.; Way, T.-D. Demethoxycurcumin Modulates Prostate Cancer Cell Proliferation via AMPK-Induced Down-regulation of HSP70 and EGFR. J. Agric. Food Chem. 2012, 60, 8427–8434. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, J.O.; Kim, J.H.; Lee, S.K.; You, G.Y.; Park, S.H.; Park, J.M.; Kim, E.-K.; Suh, P.-G.; An, J.K.; et al. Quercetin suppresses HeLa cell viability via AMPK-induced HSP70 and EGFR down-regulation. J. Cell. Physiol. 2010, 223, 408–414. [Google Scholar] [CrossRef]
- Choi, J.-A.; Kim, J.-Y.; Lee, J.-Y.; Kang, C.-M.; Kwon, H.-J.; Yoo, Y.D.; Kim, T.-W.; Lee, Y.-S.; Lee, S.-J. Induction of cell cycle arrest and apoptosis in human breast cancer cells by quercetin. Int. J. Oncol. 2001, 19, 837–844. [Google Scholar] [CrossRef]
- Milani, V.; Stangl, S.; Issels, R.; Gehrmann, M.; Wagner, B.; Hube, K.; Mayr, D.; Hiddemann, W.; Molls, M.; Multhoff, G. Anti-tumor activity of patient-derived NK cells after cell-based immunotherapy—A case report. J. Transl. Med. 2009, 7, 50. [Google Scholar] [CrossRef]
- Krause, S.W.; Gastpar, R.; Andreesen, R.; Gross, C.; Ullrich, H.; Thonigs, G.; Pfister, K.; Multhoff, G. Treatment of Colon and Lung Cancer Patients with ex Vivo Heat Shock Protein 70-Peptide-Activated, Autologous Natural Killer Cells: A Clinical Phase I Trial. Clin. Cancer Res. 2004, 10, 3699–3707. [Google Scholar] [CrossRef]
- Trimble, C.L.; Peng, S.; Kos, F.; Gravitt, P.; Viscidi, R.; Sugar, E.; Pardoll, E.; Wu, T. A phase I trial of a human papillomavirus DNA vaccine for HPV16+ cervical intraepithelial neoplasia 2/3. Clin. Cancer Res. 2009, 15, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Yoshimura, K.; Matsui, H.; Shindo, Y.; Tamesa, T.; Tokumitsu, Y.; Hashimoto, N.; Tokuhisa, Y.; Sakamoto, K.; Sakai, K.; et al. Dendritic cells transfected with heat-shock protein 70 messenger RNA for patients with hepatitis C virus-related hepatocellular carcinoma: A phase 1 dose escalation clinical trial. Cancer Immunol. Immunother. 2015, 64, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Kokowski, K.; Stangl, S.; Seier, S.; Hildebrandt, M.; Vaupel, P.; Multhoff, G. Radiochemotherapy combined with NK cell transfer followed by second-line PD-1 inhibition in a patient with NSCLC stage IIIb inducing long-term tumor control: A case study. Strahlenther. und Onkol. 2019, 195, 352–361. [Google Scholar] [CrossRef]
- Li, Z.; Serrano, D.; Baglietto, L.; Johansson, H.; Mariette, F.; Torrisi, R.; Onetto, M.; Paganuzzi, M.; DeCensi, A. Combination of Imatinib Mesylate with Autologous Leukocyte-Derived Heat Shock Protein and Chronic Myelogenous Leukemia. Clin. Cancer Res. 2005, 11, 4460–4468. [Google Scholar] [CrossRef]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/study/NCT00058747?term=hsp70&draw=2&rank=20; (accessed on 18 February 2020).
- Li, J.-L.; Liu, H.-L.; Zhang, X.-R.; Xu, J.-P.; Hu, W.-K.; Liang, M.; Chen, S.-Y.; Hu, F.; Chu, D.-T. A phase I trial of intratumoral administration of recombinant oncolytic adenovirus overexpressing HSP70 in advanced solid tumor patients. Gene Ther. 2008, 16, 376–382. [Google Scholar] [CrossRef]
| HSP70-Based Therapies | Condition | Clinical Trial Phase/Case Study | Refs. |
|---|---|---|---|
| Autologous HSP70-peptide complex in combination with imatinib mesylate | Chronic myeloid leukemia | Phase I | [247] |
| Autologous HSP70-peptide complex (AG-858) in combination with Gleevec | Chronic Myelogenous Leukemia | Phase II | [248] |
| Autologous peripheral blood mononuclear cells (PBMC) pre-activated with TKD peptide/IL-2 | Colon carcinoma | Case study | [242] |
| Autologous NK cells pre-activated with TKD and IL-2 following radiochemotherapy | Advanced colorectal carcinoma and non- small cell lung carcinoma (NSCLC) | Phase I; Phase II | [124,243] |
| DNA vaccine expressing HPV16 E7 mutant form and HSP70 | Cervical intraepithelial neoplasia | Phase I | [244] |
| Dendritic cells transfected with HSP70 mRNA | Hepatocellular carcinoma | Phase I | [245] |
| Autologous NK cells pre-treated with TKD/IL-2 in combination with radiochemotherapy and nivolumab (PD-1 antibody) | NSCLC stage IIIb | Case study | [246] |
| Intratumoral injection of recombinant oncolytic type 2 adenovirus overexpressing HSP70 (H103) | Advanced solid tumors | Phase I | [249] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albakova, Z.; Armeev, G.A.; Kanevskiy, L.M.; Kovalenko, E.I.; Sapozhnikov, A.M. HSP70 Multi-Functionality in Cancer. Cells 2020, 9, 587. https://doi.org/10.3390/cells9030587
Albakova Z, Armeev GA, Kanevskiy LM, Kovalenko EI, Sapozhnikov AM. HSP70 Multi-Functionality in Cancer. Cells. 2020; 9(3):587. https://doi.org/10.3390/cells9030587
Chicago/Turabian StyleAlbakova, Zarema, Grigoriy A. Armeev, Leonid M. Kanevskiy, Elena I. Kovalenko, and Alexander M. Sapozhnikov. 2020. "HSP70 Multi-Functionality in Cancer" Cells 9, no. 3: 587. https://doi.org/10.3390/cells9030587
APA StyleAlbakova, Z., Armeev, G. A., Kanevskiy, L. M., Kovalenko, E. I., & Sapozhnikov, A. M. (2020). HSP70 Multi-Functionality in Cancer. Cells, 9(3), 587. https://doi.org/10.3390/cells9030587





