Molecular Diagnostics in Human Papillomavirus-Related Head and Neck Squamous Cell Carcinoma
Abstract
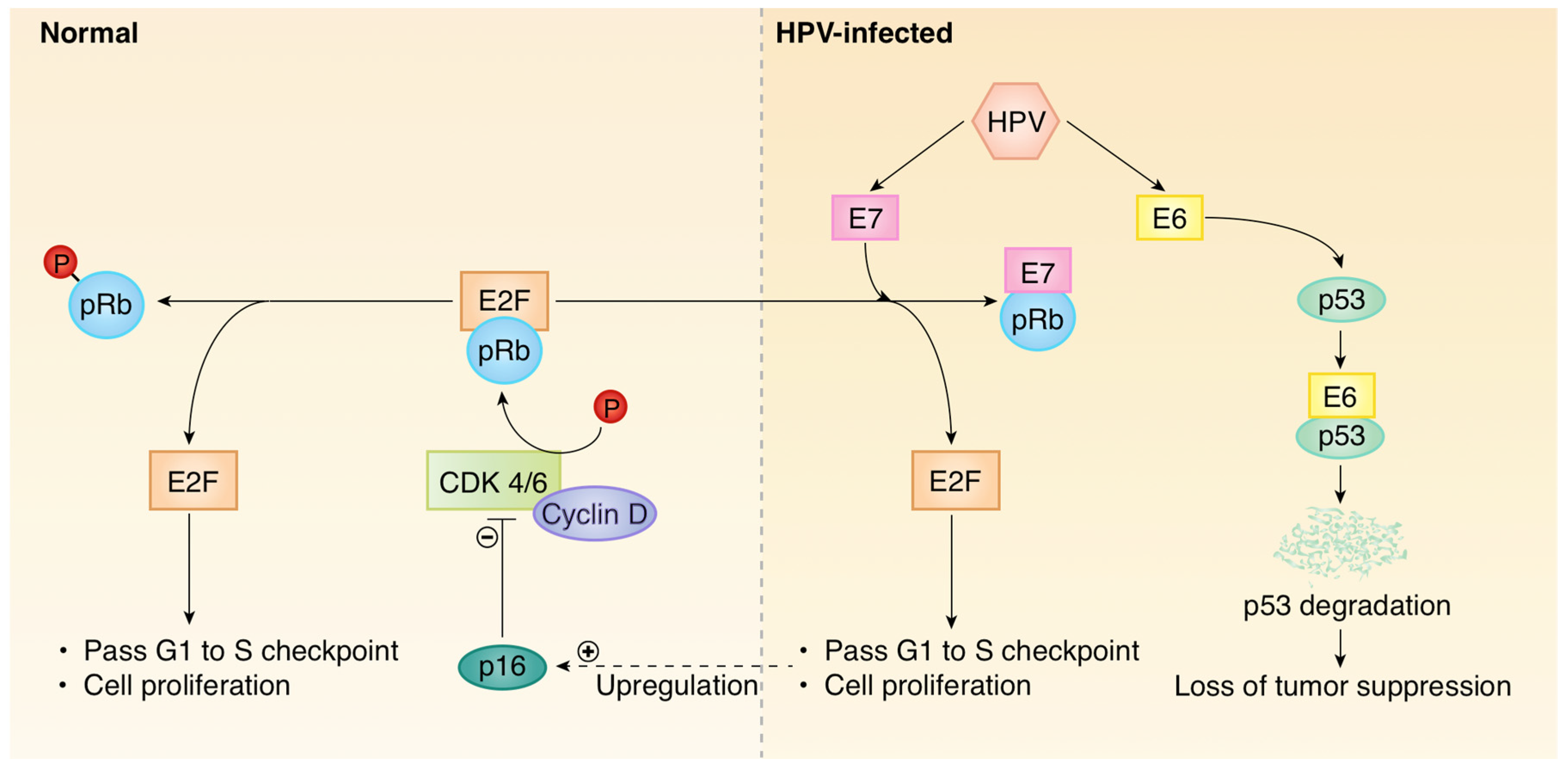
1. Introduction
2. Detection of HPV in Tissue Biopsies
2.1. p16 Staining of Tissue Specimens
2.2. HPV ISH in Tissue Specimens
2.3. HPV PCR in Tissue Specimens
3. Detection of HPV in Fine-Needle Aspiration Biopsy Specimens
3.1. p16 Staining of FNA Biopsies
3.2. HPV ISH on FNA Biopsies
3.3. HPV PCR on FNA Biopsies
4. Detection of HPV in the Blood
4.1. Detection of HPV DNA in Blood Samples
4.2. Circulating HPV DNA as a Biomarker
4.3. Circulating Antibodies against HPV
5. Detection of HPV in Saliva
5.1. Salivary HPV DNA
5.2. Salivary microRNA
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chaturvedi, A.K.; Engels, E.A.; Pfeiffer, R.M.; Hernandez, B.Y.; Xiao, W.; Kim, E.; Jiang, B.; Goodman, M.T.; Sibug-Saber, M.; Cozen, W.; et al. Human papillomavirus and rising oropharyngeal cancer incidence in the united states. J. Clin. Oncol. 2011, 29, 4294–4301. [Google Scholar] [CrossRef]
- Windon, M.J.; D’Souza, G.; Rettig, E.M.; Westra, W.H.; van Zante, A.; Wang, S.J.; Ryan, W.R.; Mydlarz, W.K.; Ha, P.K.; Miles, B.A.; et al. Increasing prevalence of human papillomavirus-positive oropharyngeal cancers among older adults. Cancer 2018, 124, 2993–2999. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, G.; Kreimer, A.R.; Viscidi, R.; Pawlita, M.; Fakhry, C.; Koch, W.M.; Westra, W.H.; Gillison, M.L. Case-control study of human papillomavirus and oropharyngeal cancer. N. Engl. J. Med. 2007, 356, 1944–1956. [Google Scholar] [CrossRef] [PubMed]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Nulton, T.J.; Olex, A.L.; Dozmorov, M.; Morgan, I.M.; Windle, B. Analysis of the cancer genome atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget 2017, 8, 17684–17699. [Google Scholar] [CrossRef]
- Werness, B.A.; Levine, A.J.; Howley, P.M. Association of human papillomavirus types 16 and 18 e6 proteins with p53. Science 1990, 248, 76–79. [Google Scholar] [CrossRef]
- Psyrri, A.; DeFilippis, R.A.; Edwards, A.P.; Yates, K.E.; Manuelidis, L.; DiMaio, D. Role of the retinoblastoma pathway in senescence triggered by repression of the human papillomavirus e7 protein in cervical carcinoma cells. Cancer Res. 2004, 64, 3079–3086. [Google Scholar] [CrossRef]
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin d/cdk4. Nature 1993, 366, 704–707. [Google Scholar] [CrossRef]
- Li, Y.; Nichols, M.A.; Shay, J.W.; Xiong, Y. Transcriptional repression of the d-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product prb. Cancer Res. 1994, 54, 6078–6082. [Google Scholar]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in notch1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef]
- Poeta, M.L.; Manola, J.; Goldwasser, M.A.; Forastiere, A.; Benoit, N.; Califano, J.A.; Ridge, J.A.; Goodwin, J.; Kenady, D.; Saunders, J.; et al. Tp53 mutations and survival in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2007, 357, 2552–2561. [Google Scholar] [CrossRef] [PubMed]
- Jordan, R.C.; Lingen, M.W.; Perez-Ordonez, B.; He, X.; Pickard, R.; Koluder, M.; Jiang, B.; Wakely, P.; Xiao, W.; Gillison, M.L. Validation of methods for oropharyngeal cancer hpv status determination in US cooperative group trials. Am. J. Surg. Pathol. 2012, 36, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S., Jr.; Beadle, B.; Bishop, J.A.; Chernock, R.D.; Colasacco, C.; Lacchetti, C.; Moncur, J.T.; Rocco, J.W.; Schwartz, M.R.; Seethala, R.R.; et al. Human papillomavirus testing in head and neck carcinomas: Guideline from the college of american pathologists. Arch. Pathol. Lab. Med. 2018, 142, 559–597. [Google Scholar] [CrossRef] [PubMed]
- Fakhry, C.; Lacchetti, C.; Rooper, L.M.; Jordan, R.C.; Rischin, D.; Sturgis, E.M.; Bell, D.; Lingen, M.W.; Harichand-Herdt, S.; Thibo, J.; et al. Human papillomavirus testing in head and neck carcinomas: Asco clinical practice guideline endorsement of the college of american pathologists guideline. J. Clin. Oncol. 2018, 36, 3152–3161. [Google Scholar] [CrossRef]
- Prigge, E.S.; Arbyn, M.; von Knebel Doeberitz, M.; Reuschenbach, M. Diagnostic accuracy of p16(ink4a) immunohistochemistry in oropharyngeal squamous cell carcinomas: A systematic review and meta-analysis. Int. J. Cancer 2017, 140, 1186–1198. [Google Scholar] [CrossRef]
- Rietbergen, M.M.; Snijders, P.J.; Beekzada, D.; Braakhuis, B.J.; Brink, A.; Heideman, D.A.; Hesselink, A.T.; Witte, B.I.; Bloemena, E.; Baatenburg-De Jong, R.J.; et al. Molecular characterization of p16-immunopositive but hpv DNA-negative oropharyngeal carcinomas. Int. J. Cancer 2014, 134, 2366–2372. [Google Scholar] [CrossRef]
- Coordes, A.; Lenz, K.; Qian, X.; Lenarz, M.; Kaufmann, A.M.; Albers, A.E. Meta-analysis of survival in patients with hnscc discriminates risk depending on combined hpv and p16 status. Eur. Arch. Otorhinolaryngol. 2016, 273, 2157–2169. [Google Scholar] [CrossRef]
- Albers, A.E.; Qian, X.; Kaufmann, A.M.; Coordes, A. Meta analysis: Hpv and p16 pattern determines survival in patients with hnscc and identifies potential new biologic subtype. Sci. Rep. 2017, 7, 16715. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Zhang, Q.; Kong, C.S.; Harris, J.; Fertig, E.J.; Harari, P.M.; Wang, D.; Redmond, K.P.; Shenouda, G.; Trotti, A.; et al. P16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J. Clin. Oncol. 2014, 32, 3930–3938. [Google Scholar] [CrossRef]
- Fakhry, C.; Westra, W.H.; Wang, S.J.; van Zante, A.; Zhang, Y.; Rettig, E.; Yin, L.X.; Ryan, W.R.; Ha, P.K.; Wentz, A.; et al. The prognostic role of sex, race, and human papillomavirus in oropharyngeal and nonoropharyngeal head and neck squamous cell cancer. Cancer 2017, 123, 1566–1575. [Google Scholar] [CrossRef]
- Lewis, J.S., Jr.; Thorstad, W.L.; Chernock, R.D.; Haughey, B.H.; Yip, J.H.; Zhang, Q.; El-Mofty, S.K. P16 positive oropharyngeal squamous cell carcinoma: An entity with a favorable prognosis regardless of tumor hpv status. Am. J. Surg. Pathol. 2010, 34, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Rischin, D.; Young, R.J.; Fisher, R.; Fox, S.B.; Le, Q.T.; Peters, L.J.; Solomon, B.; Choi, J.; O’Sullivan, B.; Kenny, L.M.; et al. Prognostic significance of p16ink4a and human papillomavirus in patients with oropharyngeal cancer treated on trog 02.02 phase iii trial. J. Clin. Oncol. 2010, 28, 4142–4148. [Google Scholar] [CrossRef] [PubMed]
- Jung, A.C.; Briolat, J.; Millon, R.; de Reynies, A.; Rickman, D.; Thomas, E.; Abecassis, J.; Clavel, C.; Wasylyk, B. Biological and clinical relevance of transcriptionally active human papillomavirus (hpv) infection in oropharynx squamous cell carcinoma. Int. J. Cancer 2010, 126, 1882–1894. [Google Scholar] [CrossRef] [PubMed]
- Ukpo, O.C.; Flanagan, J.J.; Ma, X.J.; Luo, Y.; Thorstad, W.L.; Lewis, J.S., Jr. High-risk human papillomavirus e6/e7 mrna detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am. J. Surg. Pathol. 2011, 35, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.A.; Ma, X.J.; Wang, H.; Luo, Y.; Illei, P.B.; Begum, S.; Taube, J.M.; Koch, W.M.; Westra, W.H. Detection of transcriptionally active high-risk hpv in patients with head and neck squamous cell carcinoma as visualized by a novel e6/e7 mrna in situ hybridization method. Am. J. Surg. Pathol. 2012, 36, 1874–1882. [Google Scholar] [CrossRef]
- Kerr, D.A.; Arora, K.S.; Mahadevan, K.K.; Hornick, J.L.; Krane, J.F.; Rivera, M.N.; Ting, D.T.; Deshpande, V.; Faquin, W.C. Performance of a branch chain rna in situ hybridization assay for the detection of high-risk human papillomavirus in head and neck squamous cell carcinoma. Am. J. Surg. Pathol. 2015, 39, 1643–1652. [Google Scholar] [CrossRef]
- Schache, A.G.; Liloglou, T.; Risk, J.M.; Jones, T.M.; Ma, X.J.; Wang, H.; Bui, S.; Luo, Y.; Sloan, P.; Shaw, R.J.; et al. Validation of a novel diagnostic standard in hpv-positive oropharyngeal squamous cell carcinoma. Br. J. Cancer 2013, 108, 1332–1339. [Google Scholar] [CrossRef]
- de Roda Husman, A.M.; Walboomers, J.M.; van den Brule, A.J.; Meijer, C.J.; Snijders, P.J. The use of general primers gp5 and gp6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by pcr. J. Gen. Virol. 1995, 76 Pt 4, 1057–1062. [Google Scholar] [CrossRef]
- Kleter, B.; van Doorn, L.J.; ter Schegget, J.; Schrauwen, L.; van Krimpen, K.; Burger, M.; ter Harmsel, B.; Quint, W. Novel short-fragment pcr assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 1998, 153, 1731–1739. [Google Scholar] [CrossRef]
- Ritchie, J.M.; Smith, E.M.; Summersgill, K.F.; Hoffman, H.T.; Wang, D.; Klussmann, J.P.; Turek, L.P.; Haugen, T.H. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int. J. Cancer 2003, 104, 336–344. [Google Scholar] [CrossRef]
- Li, W.; Thompson, C.H.; O’Brien, C.J.; McNeil, E.B.; Scolyer, R.A.; Cossart, Y.E.; Veness, M.J.; Walker, D.M.; Morgan, G.J.; Rose, B.R. Human papillomavirus positivity predicts favourable outcome for squamous carcinoma of the tonsil. Int. J. Cancer 2003, 106, 553–558. [Google Scholar] [CrossRef]
- Fukushima, K.; Ogura, H.; Watanabe, S.; Yabe, Y.; Masuda, Y. Human papillomavirus type 16 DNA detected by the polymerase chain reaction in non-cancer tissues of the head and neck. Eur. Arch. Otorhinolaryngol. 1994, 251, 109–112. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Sdek, P.; Cao, J.; Chen, W.T. Human papillomavirus type 16 and 18 DNA in oral squamous cell carcinoma and normal mucosa. Int. J. Oral Maxillofac. Surg. 2004, 33, 71–74. [Google Scholar] [CrossRef]
- van Houten, V.M.; Snijders, P.J.; van den Brekel, M.W.; Kummer, J.A.; Meijer, C.J.; van Leeuwen, B.; Denkers, F.; Smeele, L.E.; Snow, G.B.; Brakenhoff, R.H. Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int. J. Cancer 2001, 93, 232–235. [Google Scholar] [CrossRef]
- Smeets, S.J.; Hesselink, A.T.; Speel, E.J.; Haesevoets, A.; Snijders, P.J.; Pawlita, M.; Meijer, C.J.; Braakhuis, B.J.; Leemans, C.R.; Brakenhoff, R.H. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int. J. Cancer 2007, 121, 2465–2472. [Google Scholar] [CrossRef]
- Yang, Z.; Gomez-Fernandez, C.; Lora Gonzalez, M.; Esebua, M.; Kerr, D.A. Hpv testing through p16 immunocytochemistry in neck-mass fna and its correlation with tissue samples. Cancer Cytopathol. 2019, 127, 458–464. [Google Scholar] [CrossRef]
- Jalaly, J.B.; Lewis, J.S., Jr.; Collins, B.T.; Wu, X.; Ma, X.J.; Luo, Y.; Bernadt, C.T. Correlation of p16 immunohistochemistry in fna biopsies with corresponding tissue specimens in hpv-related squamous cell carcinomas of the oropharynx. Cancer Cytopathol. 2015, 123, 723–731. [Google Scholar] [CrossRef]
- Xu, B.; Ghossein, R.; Lane, J.; Lin, O.; Katabi, N. The utility of p16 immunostaining in fine needle aspiration in p16-positive head and neck squamous cell carcinoma. Hum. Pathol. 2016, 54, 193–200. [Google Scholar] [CrossRef]
- Buonocore, D.J.; Fowle, E.; Lin, O.; Xu, B.; Katabi, N.; Cohen, J.M. Cytologic evaluation of p16 staining in head and neck squamous cell carcinoma in cytolyt versus formalin-fixed material. Cancer Cytopathol. 2019. [Google Scholar] [CrossRef]
- Holmes, B.J.; Maleki, Z.; Westra, W.H. The fidelity of p16 staining as a surrogate marker of human papillomavirus status in fine-needle aspirates and core biopsies of neck node metastases: Implications for hpv testing protocols. Acta Cytol. 2015, 59, 97–103. [Google Scholar] [CrossRef]
- Begum, S.; Gillison, M.L.; Nicol, T.L.; Westra, W.H. Detection of human papillomavirus-16 in fine-needle aspirates to determine tumor origin in patients with metastatic squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2007, 13, 1186–1191. [Google Scholar] [CrossRef]
- Wong, K.S.; Krane, J.F.; Jo, V.Y. Heterogeneity of p16 immunohistochemistry and increased sensitivity of rna in situ hybridization in cytology specimens of hpv-related head and neck squamous cell carcinoma. Cancer Cytopathol. 2019, 127, 632–642. [Google Scholar] [CrossRef]
- Kerr, D.A.; Pitman, M.B.; Sweeney, B.; Arpin, R.N., 3rd; Wilbur, D.C.; Faquin, W.C. Performance of the roche cobas 4800 high-risk human papillomavirus test in cytologic preparations of squamous cell carcinoma of the head and neck. Cancer Cytopathol. 2014, 122, 167–174. [Google Scholar] [CrossRef]
- Guo, M.; Khanna, A.; Dhillon, J.; Patel, S.J.; Feng, J.; Williams, M.D.; Bell, D.M.; Gong, Y.; Katz, R.L.; Sturgis, E.M.; et al. Cervista hpv assays for fine-needle aspiration specimens are a valid option for human papillomavirus testing in patients with oropharyngeal carcinoma. Cancer Cytopathol. 2014, 122, 96–103. [Google Scholar] [CrossRef]
- Han, M.; Bernadt, C.T.; Murray, B.; Johnson, S.M.; Jalaly, J.B.; Garcia, T.; Adhikari, L.J. Aptima hr-hpv testing from diff-quick-stained fine-needle aspiration smears of oropharyngeal squamous cell carcinoma. J. Am. Soc. Cytopathol. 2016, 5, 221–226. [Google Scholar] [CrossRef]
- Rollo, F.; Dona, M.G.; Pellini, R.; Pichi, B.; Marandino, F.; Covello, R.; Benevolo, M. Cytology and direct human papillomavirus testing on fine needle aspirates from cervical lymph node metastases of patients with oropharyngeal squamous cell carcinoma or occult primary. Cytopathology 2018, 29, 449–454. [Google Scholar] [CrossRef]
- Channir, H.I.; Gronhoj Larsen, C.; Ahlborn, L.B.; van Overeem Hansen, T.; Gerds, T.A.; Charabi, B.W.; Vainer, B.; von Buchwald, C.; Lajer, C.B.; Kiss, K. Validation study of hpv DNA detection from stained fna smears by polymerase chain reaction: Improving the diagnostic workup of patients with a tumor on the neck. Cancer Cytopathol. 2016, 124, 820–827. [Google Scholar] [CrossRef]
- Dockter, J.; Schroder, A.; Hill, C.; Guzenski, L.; Monsonego, J.; Giachetti, C. Clinical performance of the aptima hpv assay for the detection of high-risk hpv and high-grade cervical lesions. J. Clin. Virol. 2009, 45 (Suppl. 1), S55–S61. [Google Scholar] [CrossRef]
- Dahlstrom, K.R.; Li, G.; Hussey, C.S.; Vo, J.T.; Wei, Q.; Zhao, C.; Sturgis, E.M. Circulating human papillomavirus DNA as a marker for disease extent and recurrence among patients with oropharyngeal cancer. Cancer 2015, 121, 3455–3464. [Google Scholar] [CrossRef]
- Mazurek, A.M.; Rutkowski, T.; Fiszer-Kierzkowska, A.; Malusecka, E.; Skladowski, K. Assessment of the total cfdna and hpv16/18 detection in plasma samples of head and neck squamous cell carcinoma patients. Oral Oncol. 2016, 54, 36–41. [Google Scholar] [CrossRef]
- Jensen, K.K.; Gronhoj, C.; Jensen, D.H.; von Buchwald, C. Circulating human papillomavirus DNA as a surveillance tool in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Clin. Otolaryngol. 2018, 43, 1242–1249. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital pcr. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital pcr versus analog real-time pcr. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Damerla, R.R.; Lee, N.Y.; You, D.; Soni, R.; Shah, R.; Reyngold, M.; Katabi, N.; Wu, V.; McBride, S.M.; Tsai, C.J.; et al. Detection of early human papillomavirus-associated cancers by liquid biopsy. JCO Precis. Oncol. 2019, 3, 1–17. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Beaty, B.T.; Marron, D.; Jefferys, S.; Green, R.; Goldman, E.C.; Amdur, R.; Sheets, N.; Dagan, R.; et al. Rapid clearance profile of plasma circulating tumor hpv type 16 DNA during chemoradiotherapy correlates with disease control in hpv-associated oropharyngeal cancer. Clin. Cancer Res. 2019, 25, 4682–4690. [Google Scholar] [CrossRef]
- Economopoulou, P.; Koutsodontis, G.; Avgeris, M.; Strati, A.; Kroupis, C.; Pateras, I.; Kirodimos, E.; Giotakis, E.; Kotsantis, I.; Maragoudakis, P.; et al. Hpv16 e6/e7 expression in circulating tumor cells in oropharyngeal squamous cell cancers: A pilot study. PLoS ONE 2019, 14, e0215984. [Google Scholar] [CrossRef]
- Hanna, G.J.; Supplee, J.G.; Kuang, Y.; Mahmood, U.; Lau, C.J.; Haddad, R.I.; Janne, P.A.; Paweletz, C.P. Plasma hpv cell-free DNA monitoring in advanced hpv-associated oropharyngeal cancer. Ann. Oncol. 2018, 29, 1980–1986. [Google Scholar] [CrossRef]
- Veyer, D.; Wack, M.; Mandavit, M.; Garrigou, S.; Hans, S.; Bonfils, P.; Tartour, E.; Belec, L.; Wang-Renault, S.F.; Laurent-Puig, P.; et al. Hpv circulating tumoral DNA quantification by droplet-based digital pcr: A promising predictive and prognostic biomarker for hpv-associated oropharyngeal cancers. Int. J. Cancer 2019. [Google Scholar] [CrossRef]
- Lee, J.Y.; Garcia-Murillas, I.; Cutts, R.J.; De Castro, D.G.; Grove, L.; Hurley, T.; Wang, F.; Nutting, C.; Newbold, K.; Harrington, K.; et al. Predicting response to radical (chemo)radiotherapy with circulating hpv DNA in locally advanced head and neck squamous carcinoma. Br. J. Cancer 2017, 117, 876–883. [Google Scholar] [CrossRef]
- Cao, H.; Banh, A.; Kwok, S.; Shi, X.; Wu, S.; Krakow, T.; Khong, B.; Bavan, B.; Bala, R.; Pinsky, B.A.; et al. Quantitation of human papillomavirus DNA in plasma of oropharyngeal carcinoma patients. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, e351–e358. [Google Scholar] [CrossRef]
- Zumbach, K.; Hoffmann, M.; Kahn, T.; Bosch, F.; Gottschlich, S.; Gorogh, T.; Rudert, H.; Pawlita, M. Antibodies against oncoproteins e6 and e7 of human papillomavirus types 16 and 18 in patients with head-and-neck squamous-cell carcinoma. Int. J. Cancer 2000, 85, 815–818. [Google Scholar] [CrossRef]
- Herrero, R.; Castellsague, X.; Pawlita, M.; Lissowska, J.; Kee, F.; Balaram, P.; Rajkumar, T.; Sridhar, H.; Rose, B.; Pintos, J.; et al. Human papillomavirus and oral cancer: The international agency for research on cancer multicenter study. J. Natl. Cancer Inst. 2003, 95, 1772–1783. [Google Scholar] [CrossRef] [PubMed]
- Waterboer, T.; Sehr, P.; Michael, K.M.; Franceschi, S.; Nieland, J.D.; Joos, T.O.; Templin, M.F.; Pawlita, M. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin. Chem. 2005, 51, 1845–1853. [Google Scholar] [CrossRef]
- Kreimer, A.R.; Johansson, M.; Waterboer, T.; Kaaks, R.; Chang-Claude, J.; Drogen, D.; Tjonneland, A.; Overvad, K.; Quiros, J.R.; Gonzalez, C.A.; et al. Evaluation of human papillomavirus antibodies and risk of subsequent head and neck cancer. J. Clin. Oncol. 2013, 31, 2708–2715. [Google Scholar] [CrossRef]
- Holzinger, D.; Wichmann, G.; Baboci, L.; Michel, A.; Hofler, D.; Wiesenfarth, M.; Schroeder, L.; Boscolo-Rizzo, P.; Herold-Mende, C.; Dyckhoff, G.; et al. Sensitivity and specificity of antibodies against hpv16 e6 and other early proteins for the detection of hpv16-driven oropharyngeal squamous cell carcinoma. Int. J. Cancer 2017, 140, 2748–2757. [Google Scholar] [CrossRef]
- Rubenstein, L.M.; Smith, E.M.; Pawlita, M.; Haugen, T.H.; Hamsikova, E.; Turek, L.P. Human papillomavirus serologic follow-up response and relationship to survival in head and neck cancer: A case-comparison study. Infect. Agent. Cancer 2011, 6, 9. [Google Scholar] [CrossRef]
- Fakhry, C.; Qualliotine, J.R.; Zhang, Z.; Agrawal, N.; Gaykalova, D.A.; Bishop, J.A.; Subramaniam, R.M.; Koch, W.M.; Chung, C.H.; Eisele, D.W.; et al. Serum antibodies to hpv16 early proteins warrant investigation as potential biomarkers for risk stratification and recurrence of hpv-associated oropharyngeal cancer. Cancer Prev. Res. (Phila.) 2016, 9, 135–141. [Google Scholar] [CrossRef]
- Schroeder, L.; Wichmann, G.; Willner, M.; Michel, A.; Wiesenfarth, M.; Flechtenmacher, C.; Gradistanac, T.; Pawlita, M.; Dietz, A.; Waterboer, T.; et al. Antibodies against human papillomaviruses as diagnostic and prognostic biomarker in patients with neck squamous cell carcinoma from unknown primary tumor. Int. J. Cancer 2018, 142, 1361–1368. [Google Scholar] [CrossRef]
- Turner, D.O.; Williams-Cocks, S.J.; Bullen, R.; Catmull, J.; Falk, J.; Martin, D.; Mauer, J.; Barber, A.E.; Wang, R.C.; Gerstenberger, S.L.; et al. High-risk human papillomavirus (hpv) screening and detection in healthy patient saliva samples: A pilot study. BMC Oral Health 2011, 11, 28. [Google Scholar] [CrossRef]
- Zhao, M.; Rosenbaum, E.; Carvalho, A.L.; Koch, W.; Jiang, W.; Sidransky, D.; Califano, J. Feasibility of quantitative pcr-based saliva rinse screening of hpv for head and neck cancer. Int. J. Cancer 2005, 117, 605–610. [Google Scholar] [CrossRef]
- Qureishi, A.; Ali, M.; Fraser, L.; Shah, K.A.; Moller, H.; Winter, S. Saliva testing for human papilloma virus in oropharyngeal squamous cell carcinoma: A diagnostic accuracy study. Clin. Otolaryngol. 2018, 43, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, M.; Huang, B.; Katabi, N.; Migliacci, J.; Bryant, R.; Kaplan, S.; Blackwell, T.; Patel, S.; Yang, L.; Pei, Z.; et al. Detection of hpv related oropharyngeal cancer in oral rinse specimens. Oncotarget 2017, 8, 109393–109401. [Google Scholar] [CrossRef]
- SahebJamee, M.; Boorghani, M.; Ghaffari, S.R.; AtarbashiMoghadam, F.; Keyhani, A. Human papillomavirus in saliva of patients with oral squamous cell carcinoma. Med. Oral Patol. Oral Cir. Bucal. 2009, 14, e525–e528. [Google Scholar] [CrossRef]
- Hanna, G.J.; Lau, C.J.; Mahmood, U.; Supplee, J.G.; Mogili, A.R.; Haddad, R.I.; Janne, P.A.; Paweletz, C.P. Salivary hpv DNA informs locoregional disease status in advanced hpv-associated oropharyngeal cancer. Oral Oncol. 2019, 95, 120–126. [Google Scholar] [CrossRef]
- Chuang, A.Y.; Chuang, T.C.; Chang, S.; Zhou, S.; Begum, S.; Westra, W.H.; Ha, P.K.; Koch, W.M.; Califano, J.A. Presence of hpv DNA in convalescent salivary rinses is an adverse prognostic marker in head and neck squamous cell carcinoma. Oral Oncol. 2008, 44, 915–919. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of somatic mutations and hpv in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- Ahn, S.M.; Chan, J.Y.; Zhang, Z.; Wang, H.; Khan, Z.; Bishop, J.A.; Westra, W.; Koch, W.M.; Califano, J.A. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol. Head Neck Surg. 2014, 140, 846–854. [Google Scholar] [CrossRef]
- Liu, C.J.; Kao, S.Y.; Tu, H.F.; Tsai, M.M.; Chang, K.W.; Lin, S.C. Increase of microrna mir-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010, 16, 360–364. [Google Scholar] [CrossRef]
- Duz, M.B.; Karatas, O.F.; Guzel, E.; Turgut, N.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. Identification of mir-139-5p as a saliva biomarker for tongue squamous cell carcinoma: A pilot study. Cell. Oncol. (Dordr.) 2016, 39, 187–193. [Google Scholar] [CrossRef]
- Greither, T.; Vorwerk, F.; Kappler, M.; Bache, M.; Taubert, H.; Kuhnt, T.; Hey, J.; Eckert, A.W. Salivary mir-93 and mir-200a as post-radiotherapy biomarkers in head and neck squamous cell carcinoma. Oncol. Rep. 2017, 38, 1268–1275. [Google Scholar] [CrossRef]
- Wan, Y.; Vagenas, D.; Salazar, C.; Kenny, L.; Perry, C.; Calvopina, D.; Punyadeera, C. Salivary mirna panel to detect hpv-positive and hpv-negative head and neck cancer patients. Oncotarget 2017, 8, 99990–100001. [Google Scholar] [CrossRef] [PubMed]
- Schache, A.G.; Liloglou, T.; Risk, J.M.; Filia, A.; Jones, T.M.; Sheard, J.; Woolgar, J.A.; Helliwell, T.R.; Triantafyllou, A.; Robinson, M.; et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: Sensitivity, specificity, and prognostic discrimination. Clin. Cancer Res. 2011, 17, 6262–6271. [Google Scholar] [CrossRef] [PubMed]
| Specimen Source | Diagnostic Test | Sensitivity | Specificity | Reference |
|---|---|---|---|---|
| Tissue specimen | ||||
| p16 IHC | 94% | 83% | Prigge et al. [15] | |
| DNA ISH | 65% | 94% | Kerr et al. [26] Schache et al. [27] | |
| RNA ISH | 91–97% | 93–94% | Kerr et al. [26] | |
| DNA PCR | 97% | 87% | Schache et al. [82] | |
| Fine-needle aspiration | ||||
| p16 IHC | 39–70% * | 100% | Yang et al. [36] Xu et al. [38] | |
| DNA ISH | - | - | - | |
| RNA ISH | 97% | - | Wong et al. [42] | |
| PCR | 94% | 100% | Channir et al. [47] | |
| Blood/serum | ||||
| Circulating tumor DNA by PCR | 54–92% | 97–100% | Jensen et al. [51] Damerla et al. [54] Chera et al. [55] | |
| E6 antibody (oropharynx) | 96% | 98% | Holzinger et al. [65] | |
| E6 antibody (other subsites) | 50% | 100% | Holzinger et al. [65] | |
| Saliva | ||||
| DNA PCR | 72–79% | 90% | Qureishi et al. [71] Rosenthal et al. [72] | |
| microRNA | 65% | 95% | Wan et al. [81] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wai, K.C.; Strohl, M.P.; van Zante, A.; Ha, P.K. Molecular Diagnostics in Human Papillomavirus-Related Head and Neck Squamous Cell Carcinoma. Cells 2020, 9, 500. https://doi.org/10.3390/cells9020500
Wai KC, Strohl MP, van Zante A, Ha PK. Molecular Diagnostics in Human Papillomavirus-Related Head and Neck Squamous Cell Carcinoma. Cells. 2020; 9(2):500. https://doi.org/10.3390/cells9020500
Chicago/Turabian StyleWai, Katherine C., Madeleine P. Strohl, Annemieke van Zante, and Patrick K. Ha. 2020. "Molecular Diagnostics in Human Papillomavirus-Related Head and Neck Squamous Cell Carcinoma" Cells 9, no. 2: 500. https://doi.org/10.3390/cells9020500
APA StyleWai, K. C., Strohl, M. P., van Zante, A., & Ha, P. K. (2020). Molecular Diagnostics in Human Papillomavirus-Related Head and Neck Squamous Cell Carcinoma. Cells, 9(2), 500. https://doi.org/10.3390/cells9020500





