Human Papilloma Virus-Associated Cervical Cancer and Health Disparities
Abstract
1. Introduction
1.1. Cervical Cancer Epidemiology and Risk Factors
1.2. Cervical Cancer Health Disparities
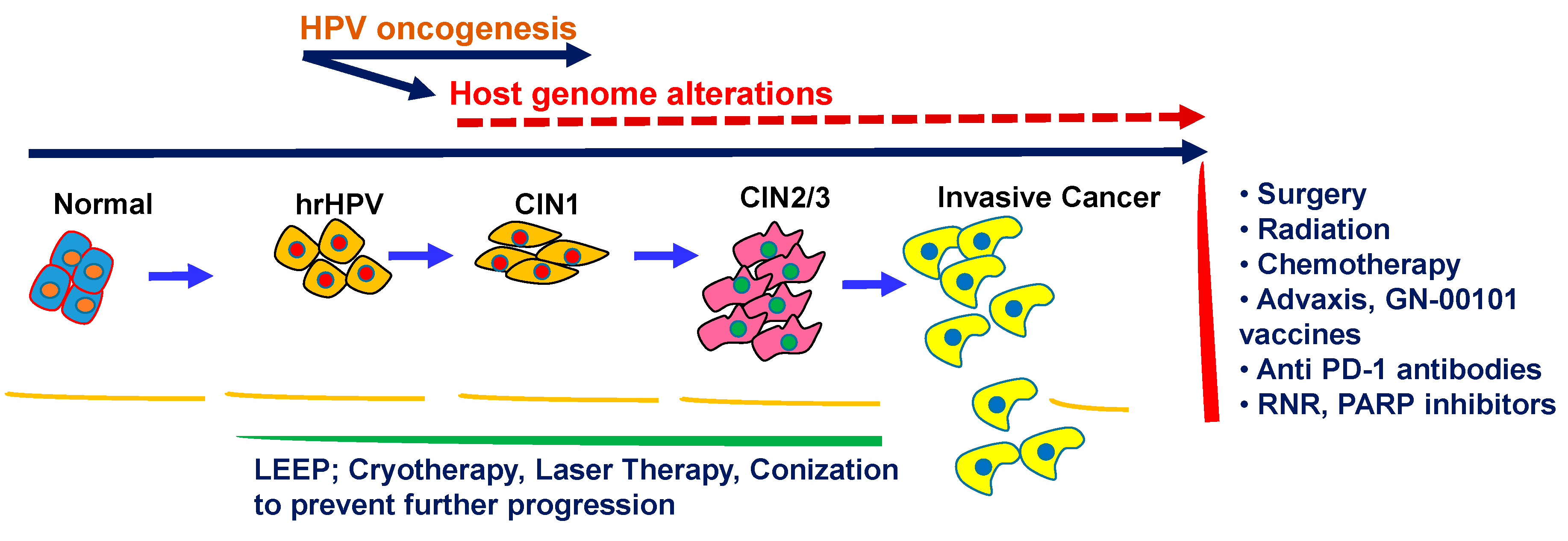
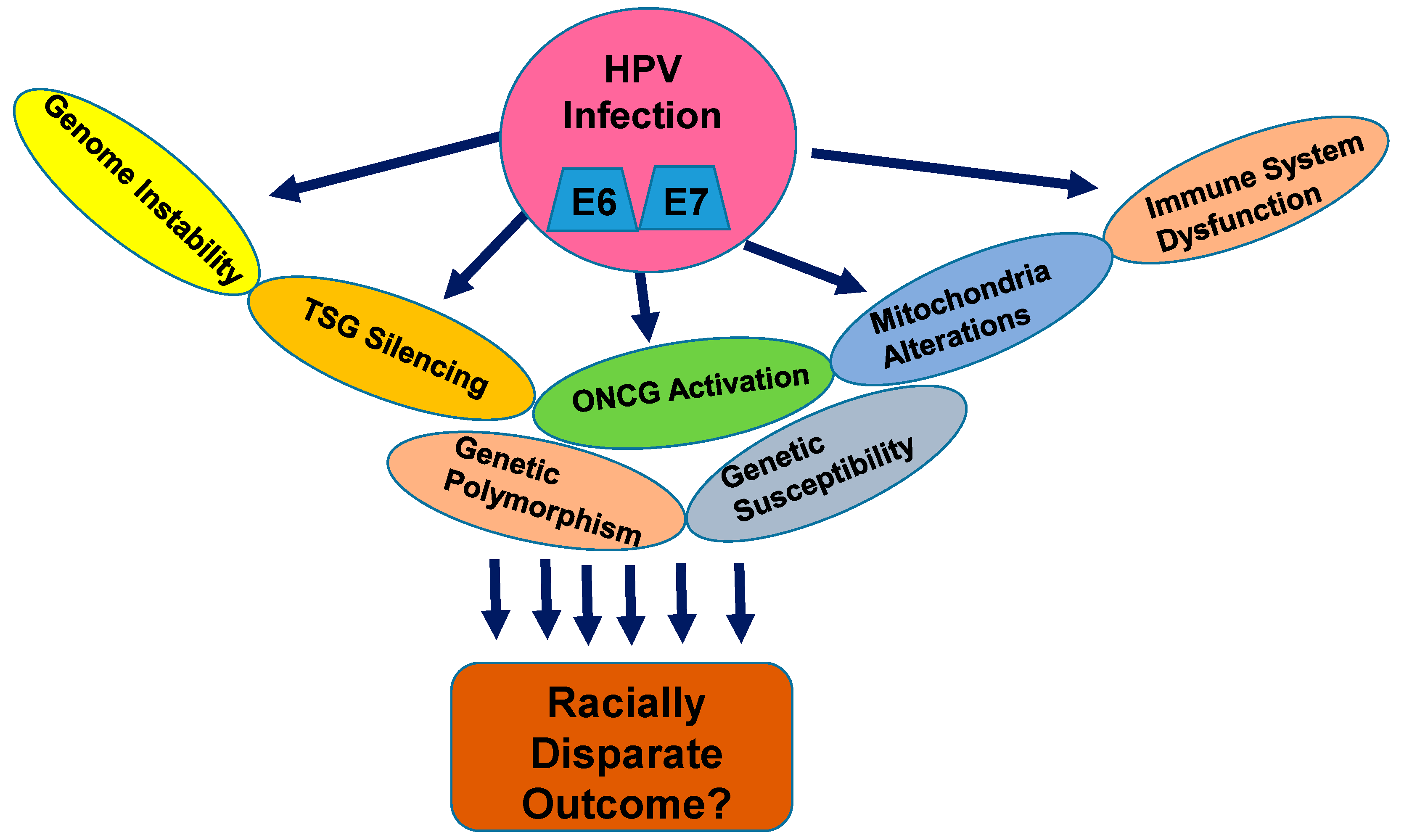
1.3. Cervical Cancer Biology and Progression with HPV
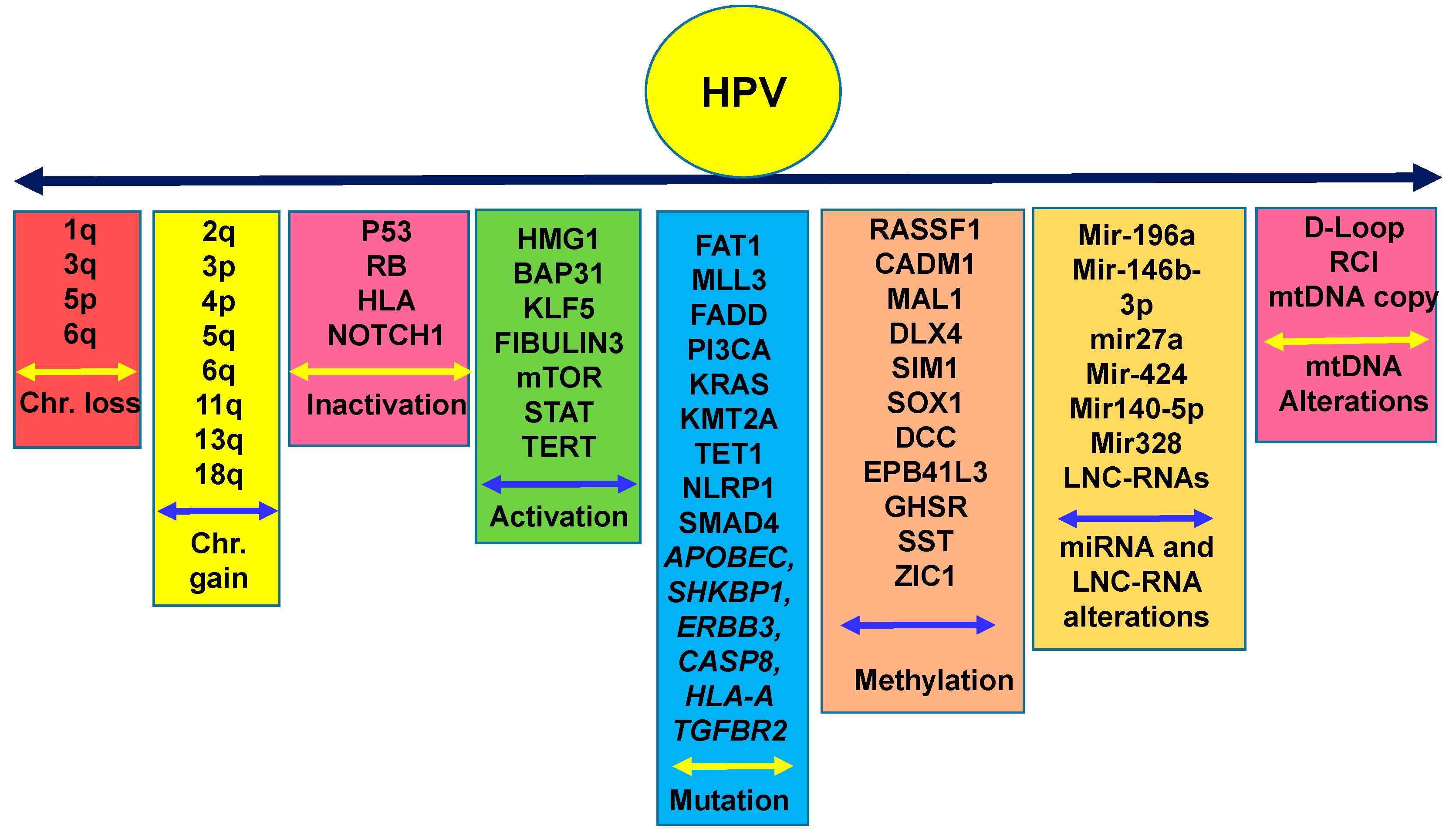
1.4. Nuclear Genetic Alterations in Cervical Cancer and Racial Disparities: How Far are We?
1.5. Epigenetic Alterations in CC in Various Disparate Populations
1.6. Mitochondrial Genomic Alterations in CC
1.7. Genetic Polymorphism and CC Risk
1.8. Early Cervical Cancer Detection, Prevention, and Treatment
1.8.1. Human Papilloma Virus Detection and Vaccination
1.8.2. Pap Testing
1.8.3. Colposcopic Testing
1.8.4. Treatment for Cervical Cancer and Therapeutic Vaccines
1.9. The Road Ahead
1.9.1. Screening and Preventive Strategies to Reduce Disparate Outcome
1.9.2. The Molecular Biological Basis of CC Racial Health Disparities
2. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schiffman, M.; Solomon, D. Clinical practice. Cervical-cancer screening with human papillomavirus and cytologic cotesting. N. Engl. J. Med. 2013, 369, 2324–2331. [Google Scholar] [CrossRef] [PubMed]
- Tewari, K.S.; Sill, M.W.; Long, H.J., 3rd; Penson, R.T.; Huang, H.; Ramondetta, L.M.; Landrum, L.M.; Oaknin, A.; Reid, T.J.; Leitao, M.M.; et al. Improved survival with bevacizumab in advanced cervical cancer. N. Engl. J. Med. 2014, 370, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, D.K.; Hashibe, M.; Kepka, D.; Maurer, K.A.; Werner, T.L. Too many women are dying from cervix cancer: Problems and solutions. Gynecol. Oncol. 2018, 151, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Cervical Cancer-cancer Stat facts- Surveillance, Epidemiology, and End Resulrs (SEER) Program- National Cancer Institute. Available online: https://seer.cancer.gov/statfacts/html/cervix.html (accessed on 7 May 2019).
- Crosbie, E.J.; Einstein, M.H.; Franceschi, S.; Kitchener, H.C. Human papillomavirus and cervical cancer. Lancet 2013, 382, 889–899. [Google Scholar] [CrossRef]
- Panda, C.K. Differential deletions of chromosome 3p are associated with the development of uterine cervical carcinoma in Indian patients. Cancer Genet. Cytogenet. 2003, 56, 263–269. [Google Scholar]
- Tsu, V.; Jeronimo, J. Saving the World’s Women from Cervical Cancer. N. Engl. J. Med. 2016, 374, 2509–2511. [Google Scholar] [CrossRef] [PubMed]
- Kahn, J.A. HPV vaccination for the prevention of cervical intraepithelial neoplasia. N. Engl. J. Med. 2009, 361, 271–278. [Google Scholar] [CrossRef]
- Akinlotan, M.; Bolin, J.N.; Helduser, J.; Ojinnaka, C.; Lichorad, A.; McClellan, D. Cervical Cancer Screening Barriers and Risk Factor Knowledge Among Uninsured Women. J. Community Health 2017, 42, 770–778. [Google Scholar] [CrossRef]
- Owusu, G.A.; Eve, S.B.; Cready, C.M.; Koelln, K.; Trevino, F.; Urrutia-Rojas, X.; Baumer, J. Race and ethnic disparities in cervical cancer screening in a safety-net system. Matern. Child Health J. 2005, 9, 285–295. [Google Scholar] [CrossRef]
- Pierce Campbell, C.M.; Menezes, L.J.; Paskett, E.D.; Giuliano, A.R. Prevention of invasive cervical cancer in the United States: Past, present, and future. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1402–1408. [Google Scholar] [CrossRef]
- Khan, H.M.; Gabbidon, K.; Abdool-Ghany, F.; Saxena, A.; Gomez, E.; Stewart, T.S. Health disparities between Black Hispanic and Black non-Hispanic cervical cancer cases in the USA. Asian Pac. J. Cancer Prev. 2014, 15, 9719–9723. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Gupta, D.; Caputo, T.A.; Holcomb, K. Disparities in Gynecological Malignancies. Front. Oncol. 2016, 6, 36. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, J.; Hatch, B.; Coronado, G.; Ezekiel, D.; Cowburn, S.; Escamilla-Sanchez, O.; Marino, M. Role of Race/Ethnicity, Language, and Insurance in Use of Cervical Cancer Prevention Services Among Low-Income Hispanic Women, 2009–2013. Prev. Chronic Dis. 2018, 15, E25. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, S.D.; Vanderford, N.L.; Huang, B.; Vanderpool, R.C. A Social-Ecological Review of Cancer Disparities in Kentucky. South. Med. J. 2018, 111, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Maguire, R.L.; Vidal, A.C.; Murphy, S.K.; Hoyo, C. Disparities in Cervical Cancer Incidence and Mortality: Can Epigenetics Contribute to Eliminating Disparities? Adv. Cancer Res. 2017, 133, 129–156. [Google Scholar] [PubMed]
- de Peralta, M.A.; Holaday, B.; Hadoto, I.M. Cues to Cervical Cancer Screening Among U.S. Hispanic Women. Hisp. Health Care Int. 2017, 15, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Mann, L.; Foley, K.L.; Tanner, A.E.; Sun, C.J.; Rhodes, S.D. Increasing Cervical Cancer Screening Among US Hispanics/Latinas: A Qualitative Systematic Review. J. Cancer Educ. 2015, 30, 374–387. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health. Human papilloma virus subtypes. In Human papilloma Virus Database; NIH: Bethesda, MA, USA, 2019. [Google Scholar]
- Kuguyo, O.; Tsikai, N.; Thomford, N.E.; Magwali, T.; Madziyire, M.G.; Nhachi, C.F.B.; Matimba, A.; Dandara, C. Genetic Susceptibility for Cervical Cancer in African Populations: What Are the Host Genetic Drivers? Omics 2018, 22, 468–483. [Google Scholar] [CrossRef]
- Tewari, K.S.; Sill, M.W.; Long, H.J. Improved Survival with Bevacizumab in Advanced Cervical Cancer. N. Engl. J. Med. 2017, 377, 702. [Google Scholar]
- Fu, F.; Wang, T.; Wu, Z.; Feng, Y.; Wang, W.; Zhou, S.; Ma, X.; Wang, S. HMGA1 exacerbates tumor growth through regulating the cell cycle and accelerates migration/invasion via targeting miR-221/222 in cervical cancer. Cell Death Dis. 2018, 9, 594. [Google Scholar] [CrossRef]
- Ma, D.; Chang, L.Y.; Zhao, S.; Zhao, J.J.; Xiong, Y.J.; Cao, F.Y.; Yuan, L.; Zhang, Q.; Wang, X.Y.; Geng, M.L.; et al. KLF5 promotes cervical cancer proliferation, migration and invasion in a manner partly dependent on TNFRSF11a expression. Sci. Rep. 2017, 7, 15683. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Tian, X.; Zhang, J.; Jiang, L. Long Non-coding RNA DLEU1 Promotes Proliferation and Invasion by Interacting With miR-381 and Enhancing HOXA13 Expression in Cervical Cancer. Front. Genet. 2018, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Dang, E.; Yang, S.; Song, C.; Jiang, D.; Li, Z.; Fan, W.; Sun, Y.; Tao, L.; Wang, J.; Liu, T.; et al. BAP31, a newly defined cancer/testis antigen, regulates proliferation, migration, and invasion to promote cervical cancer progression. Cell Death Dis. 2018, 9, 791. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qi, C.; Liu, X.; Li, C.; Chen, J.; Shi, M. Fibulin-3 knockdown inhibits cervical cancer cell growth and metastasis in vitro and in vivo. Sci. Rep. 2018, 8, 10594. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Ou, J.; Zhao, X.; Huang, X.; Huang, Y.; Zhang, Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br. J. Cancer 2014, 110, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Xu, J.; Zhao, K.; Song, P.; Yan, Q.; Fan, W.; Li, W.; Lu, C. Down-regulation of HPGD by miR-146b-3p promotes cervical cancer cell proliferation, migration and anchorage-independent growth through activation of STAT3 and AKT pathways. Cell Death Dis. 2018, 9, 1055. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Huang, B.; Sun, S.; Xiao, M.; Guo, J.; Yi, X.; Cai, J.; Wang, Z. miR-27a inhibits cervical adenocarcinoma progression by downregulating the TGF-βRI signaling pathway. Cell Death Dis. 2018, 9, 395. [Google Scholar] [CrossRef]
- Wang, X.; Xia, Y. microRNA-328 inhibits cervical cancer cell proliferation and tumorigenesis by targeting TCF7L2. Biochem. Biophys. Res. Commun. 2016, 475, 169–175. [Google Scholar] [CrossRef]
- Su, Y.; Xiong, J.; Hu, J.; Wei, X.; Zhang, X.; Rao, L. MicroRNA-140-5p targets insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) to suppress cervical cancer growth and metastasis. Oncotarget 2016, 7, 68397–68411. [Google Scholar] [CrossRef]
- Huang, J.; Qian, Z.; Gong, Y.; Wang, Y.; Guan, Y.; Han, Y.; Yi, X.; Huang, W.; Ji, L.; Xu, J.; et al. Comprehensive genomic variation profiling of cervical intraepithelial neoplasia and cervical cancer identifies potential targets for cervical cancer early warning. J. Med. Genet. 2018, 56, 186–194. [Google Scholar] [CrossRef]
- Nuryadi, E.; Sasaki, Y.; Hagiwara, Y.; Permata, T.B.M.; Sato, H.; Komatsu, S.; Yoshimoto, Y.; Murata, K.; Ando, K.; Kubo, N.; et al. Mutational analysis of uterine cervical cancer that survived multiple rounds of radiotherapy. Oncotarget 2018, 9, 32642–32652. [Google Scholar] [CrossRef] [PubMed]
- Lyng, H.; Beigi, M.; Svendsrud, D.H.; Brustugun, O.T.; Stokke, T.; Kristensen, G.B.; Sundfor, K.; Skjonsberg, A.; De Angelis, P.M. Intratumor chromosomal heterogeneity in advanced carcinomas of the uterine cervix. Int. J. Cancer 2004, 111, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Narayan, G.; Pulido, H.A.; Koul, S.; Lu, X.Y.; Harris, C.P.; Yeh, Y.A.; Vargas, H.; Posso, H.; Terry, M.B.; Gissmann, L.; et al. Genetic analysis identifies putative tumor suppressor sites at 2q35-q36.1 and 2q36.3-q37.1 involved in cervical cancer progression. Oncogene 2003, 22, 3489–3499. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.H.; Arias-Pulido, H.; Lu, X.Y.; Harris, C.P.; Vargas, H.; Zhang, F.F.; Narayan, G.; Schneider, A.; Terry, M.B.; Murty, V.V. Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer 2004, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services; Barretos Cancer Hospital; Baylor College of Medicine; Beckman Research Institute of City of Hope; Buck Institute for Research on Aging; Canada’s Michael Smith Genome Sciences Centre; Harvard Medical School; Helen F. Graham Cancer Center & Research Institute at Christiana Care Health Services; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Saavedra, K.P.; Brebi, P.M.; Roa, J.C. Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin. Epigenet. 2012, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Dong, J.; Chang, W.; Cui, M.; Xu, T. The Progress of Methylation Regulation in Gene Expression of Cervical Cancer. Int. J. Genom. 2018, 2018, 8260652. [Google Scholar] [CrossRef]
- Haffner, M.C.; Chaux, A.; Meeker, A.K.; Esopi, D.M.; Gerber, J.; Pellakuru, L.G.; Toubaji, A.; Argani, P.; Iacobuzio-Donahue, C.; Nelson, W.G.; et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2011, 2, 627–637. [Google Scholar] [CrossRef]
- Overmeer, R.M.; Louwers, J.A.; Meijer, C.J.; van Kemenade, F.J.; Hesselink, A.T.; Daalmeijer, N.F.; Wilting, S.M.; Heideman, D.A.; Verheijen, R.H.; Zaal, A.; et al. Combined CADM1 and MAL promoter methylation analysis to detect (pre-)malignant cervical lesions in high-risk HPV-positive women. Int. J. Cancer 2011, 129, 2218–2225. [Google Scholar] [CrossRef]
- Bierkens, M.; Hesselink, A.T.; Meijer, C.J.; Heideman, D.A.; Wisman, G.B.; van der Zee, A.G.; Snijders, P.J.; Steenbergen, R.D. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int. J. Cancer 2013, 133, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- De Strooper, L.M.; Hesselink, A.T.; Berkhof, J.; Meijer, C.J.; Snijders, P.J.; Steenbergen, R.D.; Heideman, D.A. Combined CADM1/MAL methylation and cytology testing for colposcopy triage of high-risk HPV-positive women. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1933–1937. [Google Scholar] [CrossRef] [PubMed]
- Sakane, J.; Taniyama, K.; Miyamoto, K.; Saito, A.; Kuraoka, K.; Nishimura, T.; Sentani, K.; Oue, N.; Yasui, W. Aberrant DNA methylation of DLX4 and SIM1 is a predictive marker for disease progression of uterine cervical low-grade squamous intraepithelial lesion. Diagn. Cytopathol. 2015, 43, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Lee, I.H.; Lee, K.H.; Lee, Y.K.; So, K.A.; Hong, S.R.; Hwang, C.S.; Kee, M.K.; Rhee, J.E.; Kang, C.; et al. DNA methylation in human papillomavirus-infected cervical cells is elevated in high-grade squamous intraepithelial lesions and cancer. J. Gynecol. Oncol. 2016, 27, e14. [Google Scholar] [CrossRef]
- Uijterwaal, M.H.; van Zummeren, M.; Kocken, M.; Luttmer, R.; Berkhof, J.; Witte, B.I.; van Baal, W.M.; Graziosi, G.C.M.; Verheijen, R.H.M.; Helmerhorst, T.J.M.; et al. Performance of CADM1/MAL-methylation analysis for monitoring of women treated for high-grade CIN. Gynecol. Oncol. 2016, 143, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.A.; Luhn, P.; Gage, J.C.; Bodelon, C.; Dunn, S.T.; Walker, J.; Zuna, R.; Hewitt, S.; Killian, J.K.; Yan, L.; et al. Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int. J. Cancer 2017, 141, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Verlaat, W.; Snijders, P.J.F.; Novianti, P.W.; Wilting, S.M.; De Strooper, L.M.A.; Trooskens, G.; Vandersmissen, J.; Van Criekinge, W.; Wisman, G.B.A.; Meijer, C.; et al. Genome-wide DNA Methylation Profiling Reveals Methylation Markers Associated with 3q Gain for Detection of Cervical Precancer and Cancer. Clin. Cancer Res. 2017, 23, 3813–3822. [Google Scholar] [CrossRef]
- Hertweck, K.L.; Dasgupta, S. The Landscape of mtDNA Modifications in Cancer: A Tale of Two Cities. Front. Oncol. 2017, 7, 262. [Google Scholar] [CrossRef]
- Dasgupta, S.; Hoque, M.O.; Upadhyay, S.; Sidransky, D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008, 68, 700–706. [Google Scholar] [CrossRef]
- Dasgupta, S.; Soudry, E.; Mukhopadhyay, N.; Shao, C.; Yee, J.; Lam, S.; Lam, W.; Zhang, W.; Gazdar, A.F.; Fisher, P.B.; et al. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J. Cell. Physiol. 2012, 227, 2451–2460. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Bhat, S.; Mascarenhas, R.; Mallya, S.; Bhat, M.; Pandey, D.; Kushtagi, P.; Thangaraj, K.; Gopinath, P.M.; Satyamoorthy, K. Mitochondrial DNA variation analysis in cervical cancer. Mitochondrion 2014, 16, 73–82. [Google Scholar] [CrossRef]
- Warowicka, A.; Kwasniewska, A.; Gozdzicka-Jozefiak, A. Alterations in mtDNA: A qualitative and quantitative study associated with cervical cancer development. Gynecol. Oncol. 2013, 129, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Goia-Rusanu, C.D.; Iancu, I.V.; Botezatu, A.; Socolov, D.; Huica, I.; Plesa, A.; Anton, G. Mitochondrial DNA mutations in patients with HRHPV-related cervical lesions. Roum. Arch. Microbiol. Immunol. 2011, 70, 5–10. [Google Scholar] [PubMed]
- Paaso, A.; Jaakola, A.; Syrjanen, S.; Louvanto, K. From HPV Infection to Lesion Progression: The Role of HLA Alleles and Host Immunity. Acta Cytol. 2019, 63, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Sun, W.; Han, S.; Cai, Y.; Ren, M.; Shen, Y. Cytochrome P450 1A1 gene polymorphisms and cervical cancer risk: A systematic review and meta-analysis. Medicine 2018, 97, e0210. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Lu, X.; Yang, X.; Xu, N. Association of MBL2 exon1 polymorphisms with high-risk human papillomavirus infection and cervical cancers: A meta-analysis. Arch. Gynecol. Obstet. 2016, 294, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Feldman, S. Making sense of the new cervical-cancer screening guidelines. N. Engl. J. Med. 2011, 365, 2145–2147. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.Z.; Meghani, S.H.; Sommers, M.S.; Hughes, T.L. Health Care-Related Correlates of Cervical Cancer Screening among Sexual Minority Women: An Integrative Review. J. Midwifery Womens Health 2018. [Google Scholar] [CrossRef]
- Rees, I.; Jones, D.; Chen, H.; Macleod, U. Interventions to improve the uptake of cervical cancer screening among lower socioeconomic groups: A systematic review. Prev. Med. 2018, 111, 323–335. [Google Scholar] [CrossRef]
- Wentzensen, N.; Walker, J.; Smith, K.; Gold, M.A.; Zuna, R.; Massad, L.S.; Liu, A.; Silver, M.I.; Dunn, S.T.; Schiffman, M. A prospective study of risk-based colposcopy demonstrates improved detection of cervical precancers. Am. J. Obstet. Gynecol. 2018, 218, e601–e604. [Google Scholar] [CrossRef]
- Ge, Y.; Christensen, P.; Luna, E.; Armylagos, D.; Xu, J.; Schwartz, M.R.; Mody, D.R. Role of HPV genotyping in risk assessment among cytology diagnosis categories: Analysis of 4562 cases with cytology-HPV cotesting and follow-up biopsies. Int. J. Gynecol. Cancer 2019. [Google Scholar] [CrossRef]
- Markowitz, L.E.; Gee, J.; Chesson, H.; Stokley, S. Ten Years of Human Papillomavirus Vaccination in the United States. Acad. Pediatr. 2018, 18, S3–S10. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.M.; DeMars, L.R. HPV vaccines—A review of the first decade. Gynecol. Oncol. 2017, 146, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Draper, E.; Bissett, S.L.; Howell-Jones, R.; Waight, P.; Soldan, K.; Jit, M.; Andrews, N.; Miller, E.; Beddows, S. A randomized, observer-blinded immunogenicity trial of Cervarix((R)) and Gardasil((R)) Human Papillomavirus vaccines in 12-15 year old girls. PLoS ONE 2013, 8, e61825. [Google Scholar] [CrossRef] [PubMed]
- Petrosky, E.; Bocchini, J.A., Jr.; Hariri, S.; Chesson, H.; Curtis, C.R.; Saraiya, M.; Unger, E.R.; Markowitz, L.E. Use of 9-valent human papillomavirus (HPV) vaccine: Updated HPV vaccination recommendations of the advisory committee on immunization practices. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 300–304. [Google Scholar] [PubMed]
- Iavazzo, C.; Boutas, I.; Grigoriadis, C.; Vrachnis, N.; Salakos, N. Management of ASCUS findings in Papanicolaou smears. A retrospective study. Eur. J. Gynaecol. Oncol. 2012, 33, 605–609. [Google Scholar] [PubMed]
- Oberlin, A.M.; Rahangdale, L.; Chinula, L.; Fuseini, N.M.; Chibwesha, C.J. Making HPV vaccination available to girls everywhere. Int. J. Gynaecol. Obstet. 2018, 143, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Fader, A.N. Surgery in Cervical Cancer. N. Engl. J. Med. 2018, 379, 1955–1957. [Google Scholar] [CrossRef]
- Cory, L.; Chu, C. ADXS-HPV: A therapeutic Listeria vaccination targeting cervical cancers expressing the HPV E7 antigen. Hum. Vaccines Immunother. 2014, 10, 3190–3195. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Reimers, L.L.; Studentsov, Y.Y.; Einstein, M.H.; Burk, R.D. Serological response to an HPV16 E7 based therapeutic vaccine in women with high-grade cervical dysplasia. Gynecol. Oncol. 2010, 116, 208–212. [Google Scholar] [CrossRef]
- Maldonado, L.; Teague, J.E.; Morrow, M.P.; Jotova, I.; Wu, T.C.; Wang, C.; Desmarais, C.; Boyer, J.D.; Tycko, B.; Robins, H.S.; et al. Intramuscular therapeutic vaccination targeting HPV16 induces T cell responses that localize in mucosal lesions. Sci. Transl. Med. 2014, 6, 221ra213. [Google Scholar] [CrossRef]
- Saglam, O.; Conejo-Garcia, J. PD-1/PD-L1 immune checkpoint inhibitors in advanced cervical cancer. Integr. Cancer Sci. Ther. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Dyer, B.A.; Zamarin, D.; Eskandar, R.N.; Mayadev, J.M. Role of Immunotherapy in the Management of Locally Advanced and Recurrent/Metastatic Cervical Cancer. J. Natl. Compr. Cancer Netw. 2019, 17, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.P.; Zhu, Y.L.; Ratner, E.S. Targeting Cyclin-Dependent Kinases for Treatment of Gynecologic Cancers. Front. Oncol. 2018, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Markt, S.C.; Tang, T.; Cronin, A.M.; Katz, I.T.; Howitt, B.E.; Horowitz, N.S.; Lee, L.J.; Wright, A.A. Insurance status and cancer treatment mediate the association between race/ethnicity and cervical cancer survival. PLoS ONE 2018, 13, e0193047. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.Y.; Tseng, M. Ethnic density and cancer: A review of the evidence. Cancer 2018, 124, 1877–1903. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.M.; Gabbidon, K.; Saxena, A.; Abdool-Ghany, F.; Dodge, J.M., 3rd; Lenzmeier, T. Disparities in Cervical Cancer Characteristics and Survival Between White Hispanics and White Non-Hispanic Women. J. Womens Health 2016, 25, 1052–1058. [Google Scholar] [CrossRef]
- Olaku, O.O.; Taylor, E.A. Cancer in the Medically Underserved Population. Prim. Care 2017, 44, 87–97. [Google Scholar] [CrossRef]
- Corey, L.; Valente, A.; Wade, K. Personalized Medicine in Gynecologic Cancer: Fact or Fiction? Obstet. Gynecol. Clin. N. Am. 2019, 46, 155–163. [Google Scholar] [CrossRef]
- Garralda, E.; Dienstmann, R.; Piris, A.; Brana, I.; Rodon, J.; Tabernero, J. New clinical trial designs in the era of Precision Medicine. Mol. Oncol. 2019, 13, 549–557. [Google Scholar] [CrossRef]
- Loomans-Kropp, H.A.; Umar, A. Cancer prevention and screening: The next step in the era of precision medicine. NPJ Precis. Oncol. 2019, 3, 3. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olusola, P.; Banerjee, H.N.; Philley, J.V.; Dasgupta, S. Human Papilloma Virus-Associated Cervical Cancer and Health Disparities. Cells 2019, 8, 622. https://doi.org/10.3390/cells8060622
Olusola P, Banerjee HN, Philley JV, Dasgupta S. Human Papilloma Virus-Associated Cervical Cancer and Health Disparities. Cells. 2019; 8(6):622. https://doi.org/10.3390/cells8060622
Chicago/Turabian StyleOlusola, Patti, Hirendra Nath Banerjee, Julie V. Philley, and Santanu Dasgupta. 2019. "Human Papilloma Virus-Associated Cervical Cancer and Health Disparities" Cells 8, no. 6: 622. https://doi.org/10.3390/cells8060622
APA StyleOlusola, P., Banerjee, H. N., Philley, J. V., & Dasgupta, S. (2019). Human Papilloma Virus-Associated Cervical Cancer and Health Disparities. Cells, 8(6), 622. https://doi.org/10.3390/cells8060622




