Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Isolation and Culture
2.2. RNA Isolation
2.3. RNA Sequencing and Hybridization
2.4. RT-qPCR for Validation of Gene Expression from Total RNA and Linear RNA-Depleted Samples
2.5. Validation of Dexamethasone-Dependent Targets
2.6. Validation of miRNA and piRNA Expression
2.7. Statistical Analysis
3. Results
3.1. MSC Immunophenotype and Osteo/Chondrogenic Differentiation Potential
3.2. Differential Expression of circRNA in Early Osteo- and Chondrogenesis
3.3. Validation of Gene Expression from Total RNA and Linear RNA-Depleted Samples
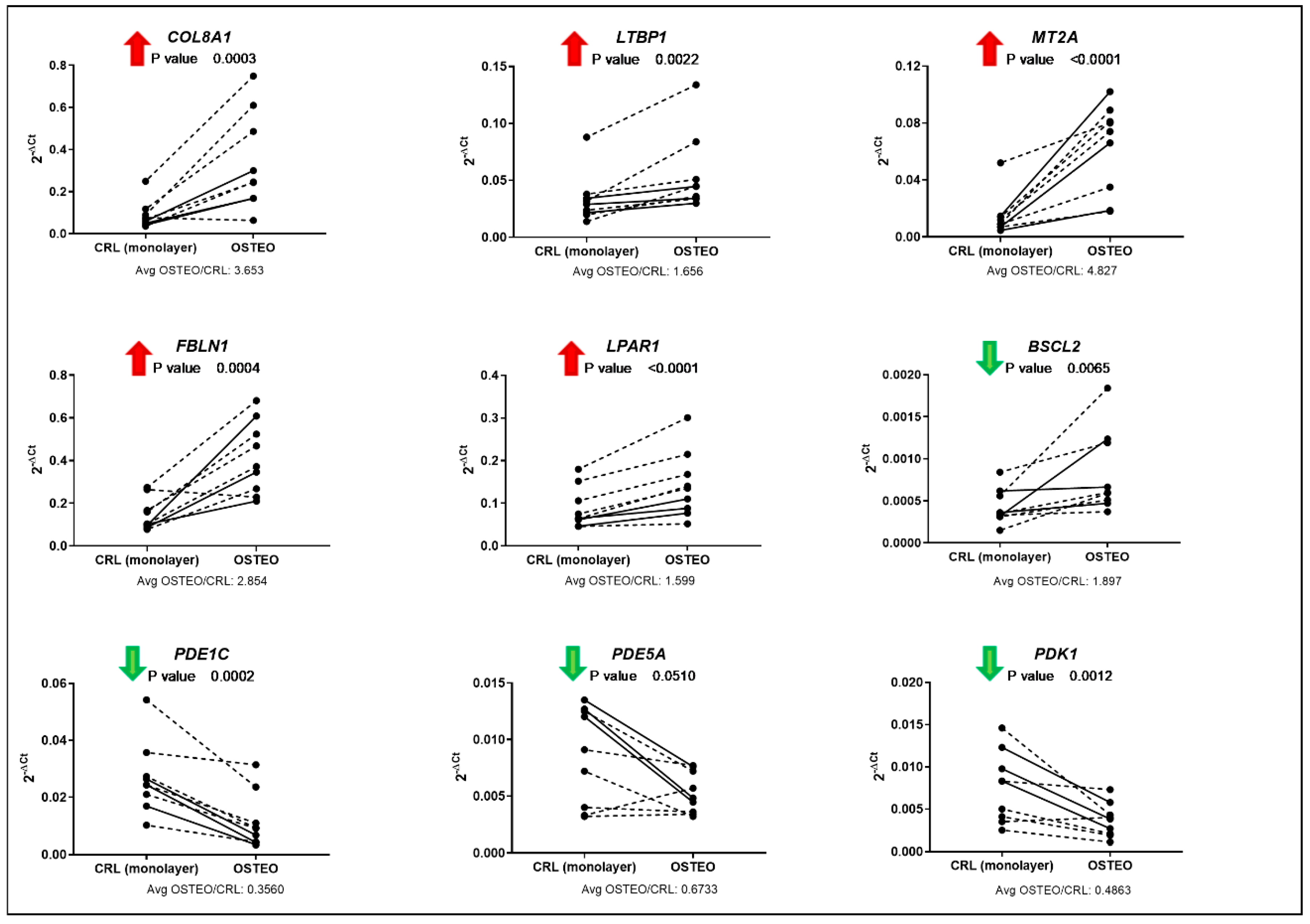
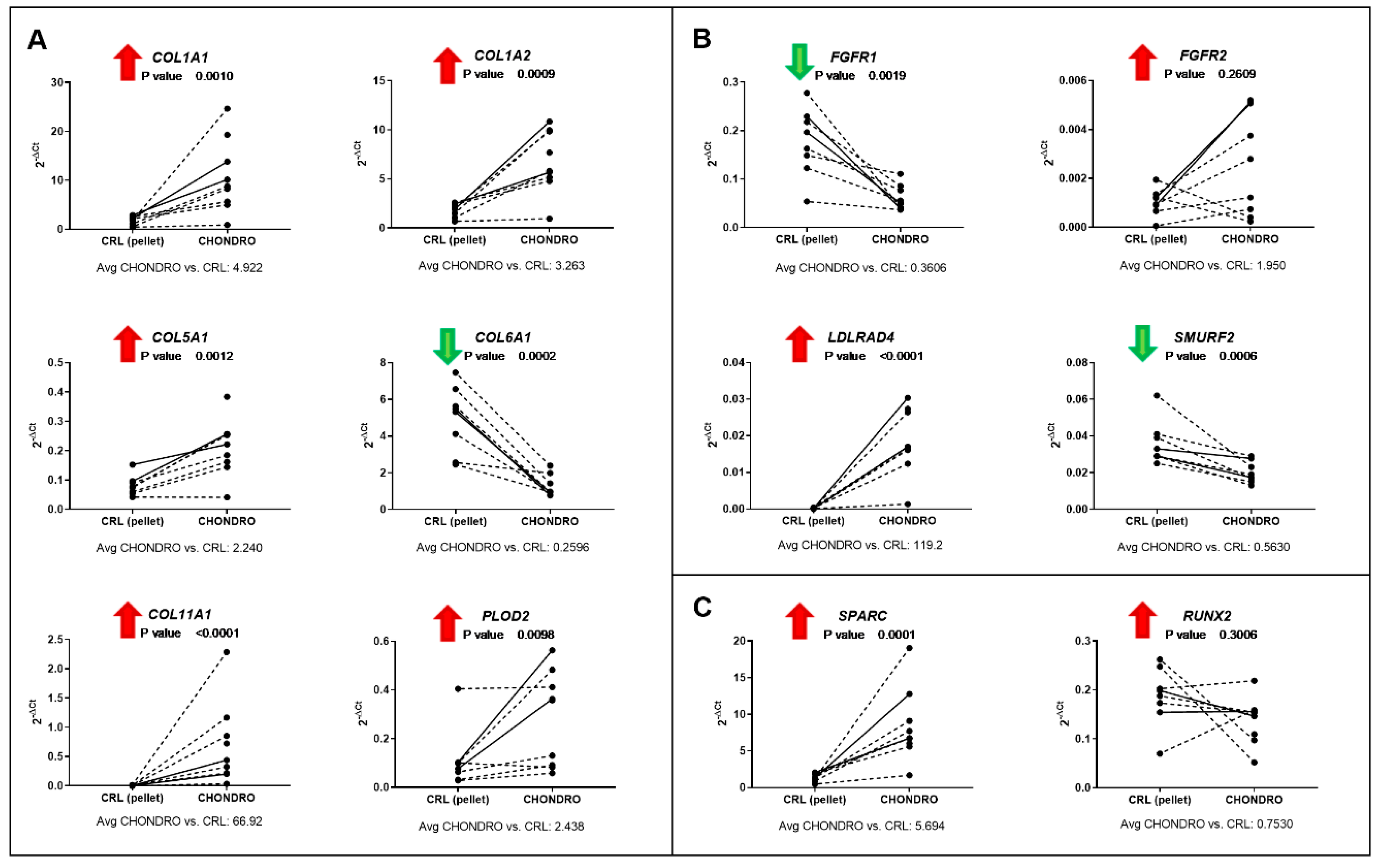
3.4. Validation of Total Gene Expression for Specific Osteo- or Chondrogenesis Targets
3.5. Differential Expression of microRNA in Early Osteo- and Chondrogenesis
3.6. Differential Expression of Piwi-Interacting RNA in Early Osteo- and Chondrogenesis
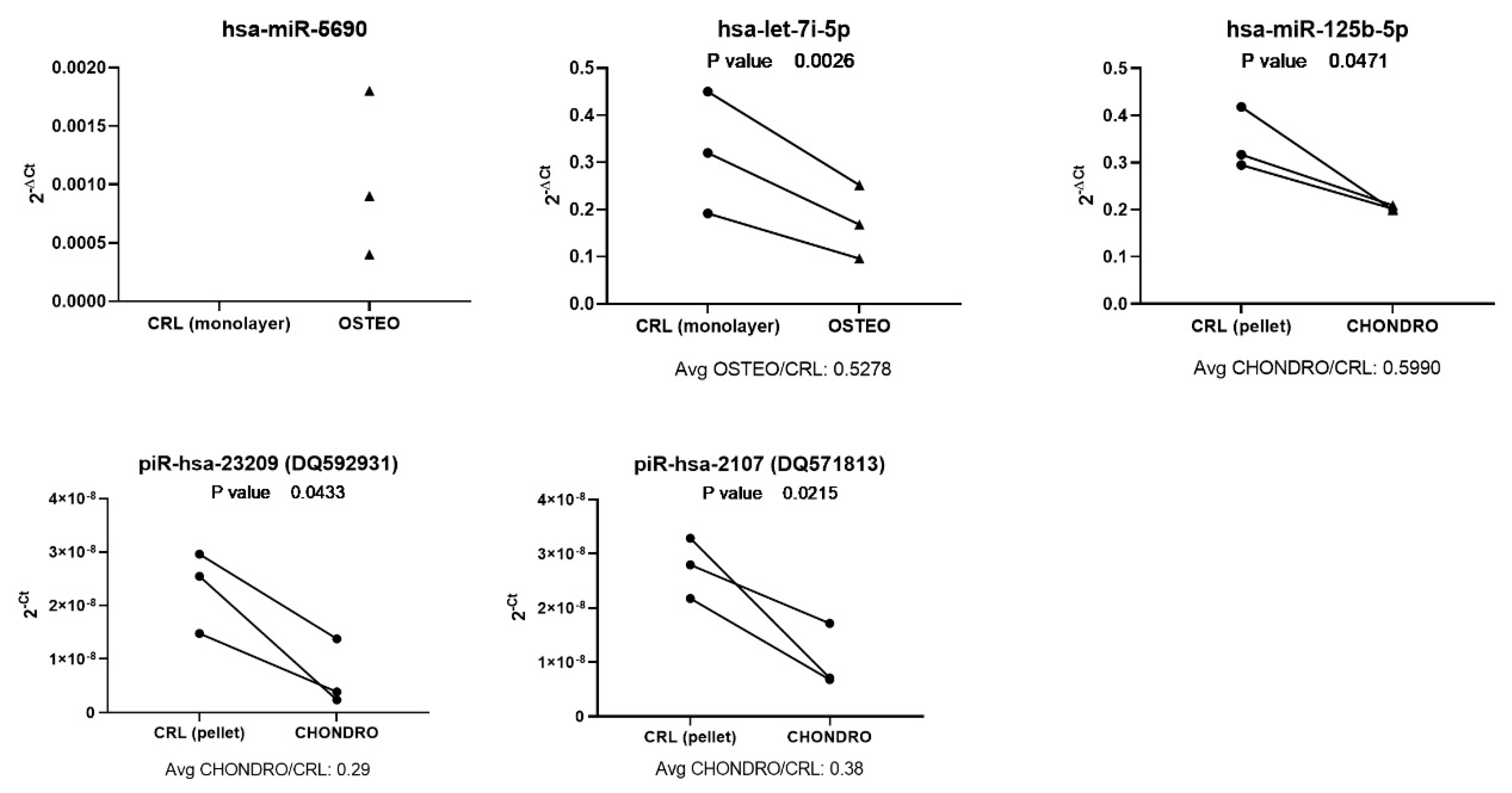
3.7. Validation of miRNA and piRNA Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Armiento, A.R.; Alini, M.; Stoddart, M.J. Articular fibrocartilage—Why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2018, 146, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Bara, J.J.; Richards, R.G.; Alini, M.; Stoddart, M.J. Concise review: Bone marrow-derived mesenchymal stem cells change phenotype following in vitro culture: Implications for basic research and the clinic. Stem Cells 2014, 32, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.A.; Kalomoiris, S.; Nolta, J.A.; Fierro, F.A. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells 2014, 32, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Gou, S.; Wang, L.; Huang, K.; Liu, L.; Zhao, W.; Zheng, L.; Xiao, J. MicroRNAs and Peroxisome Proliferator-Activated Receptors Governing the Differentiation of Mesenchymal Stem Cells. Curr. Stem Cell Res. Ther. 2016, 11, 197–207. [Google Scholar] [CrossRef]
- Li, N.; Long, B.; Han, W.; Yuan, S.; Wang, K. microRNAs: Important regulators of stem cells. Stem Cell. Res. Ther. 2017, 8, 110. [Google Scholar] [CrossRef]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef]
- Han, Y.N.; Li, Y.; Xia, S.Q.; Zhang, Y.Y.; Zheng, J.H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem. 2017, 44, 1–20. [Google Scholar] [CrossRef]
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199. [Google Scholar] [CrossRef]
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; Chuva de Sousa Lopes, S.M.; van der Westerlaken, L.A.J.; Zischler, H.; Butter, F.; Roelen, B.A.J.; et al. Piwi Proteins and piRNAs in Mammalian Oocytes and Early Embryos. Cell Rep. 2015, 10, 2069–2082. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.-Y.; Wan, F.-C.; Liu, F.-J.; Liu, J.; Zhang, N.; Jin, S.-H.; Li, J.-Y. Deep sequencing analysis of small non-coding RNAs reveals the diversity of microRNAs and piRNAs in the human epididymis. Gene 2012, 497, 330–335. [Google Scholar] [CrossRef]
- Huang, G.; Hu, H.; Xue, X.; Shen, S.; Gao, E.; Guo, G.; Shen, X.; Zhang, X. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin. Transl. Oncol. 2013, 15, 563–568. [Google Scholar] [CrossRef]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr. Biol. 2009, 19, 2066–2076. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Hui, G.; Yuan, L.; Shi, D.; Wang, Y.; Du, M.; Zhong, D.; Ma, L.; Tong, N.; Qin, C.; et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015, 356, 561–567. [Google Scholar] [CrossRef] [PubMed]
- van Wolfswinkel, J.C. Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integr. Comp. Biol. 2014, 54, 700–713. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, X.; Gao, H.; Jin, J.M.; Li, A.X.; Kim, Y.S.; Pal, S.K.; Nelson, R.A.; Lau, C.M.; Guo, C.; et al. Piwi-Interacting RNAs (piRNAs) Are Dysregulated in Renal Cell Carcinoma and Associated with Tumor Metastasis and Cancer-Specific Survival. Mol. Med. 2015, 21, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Busch, J.; Ralla, B.; Jung, M.; Wotschofsky, Z.; Trujillo-Arribas, E.; Schwabe, P.; Kilic, E.; Fendler, A.; Jung, K. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J. Exp. Clin. Cancer Res. 2015, 34, 61. [Google Scholar] [CrossRef]
- Juliano, C.; Wang, J.; Lin, H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef]
- Hashim, A.; Rizzo, F.; Marchese, G.; Ravo, M.; Tarallo, R.; Nassa, G.; Giurato, G.; Santamaria, G.; Cordella, A.; Cantarella, C.; et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget 2014, 5, 9901–9910. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef]
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef]
- Zheng, F.; Wang, M.; Li, Y.; Huang, C.; Tao, D.; Xie, F.; Zhang, H.; Sun, J.; Zhang, C.; Gu, C.; et al. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019, 460, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Liu, Z.; Ni, W.; Yao, R.; Xu, Y.; Quan, R.; Zhang, M.; Li, H.; Liu, L.; et al. Circular RNA circ-FoxO3 Inhibits Myoblast Cells Differentiation. Cells 2019, 8, 616. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.; Luo, Q.; Song, Y.; Yang, F.; Wang, Y.; Jin, M.; Zhang, D. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1alpha regulation. J. Cell. Biochem. 2019, 120, 19019–19030. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Liu, Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem. Biophys. Res. Commun. 2019, 516, 546–550. [Google Scholar] [CrossRef]
- Chen, H.; Mao, M.; Jiang, J.; Zhu, D.; Li, P. Circular RNA CDR1as acts as a sponge of miR-135b-5p to suppress ovarian cancer progression. Onco Targets Ther. 2019, 12, 3869–3879. [Google Scholar] [CrossRef]
- Lai, Z.; Yang, Y.; Wang, C.; Yang, W.; Yan, Y.; Wang, Z.; Xu, J.; Jiang, K. Circular RNA 0047905 acts as a sponge for microRNA4516 and microRNA1227-5p, initiating gastric cancer progression. Cell Cycle 2019, 18. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, J.; Huang, R.; Wang, Y.; Nie, X.; Gui, R. Circular RNA expression profiles are significantly altered in mice bone marrow stromal cells after total body irradiation. Leuk. Res. 2018, 70, 67–73. [Google Scholar] [CrossRef]
- Ren, W.; Yang, L.; Deng, T.; Wu, C.; Li, Y.; Wu, J.; Huang, Z.; Du, F.; Guo, L. Calcitonin gene-related peptide regulates FOSL2 expression and cell proliferation of BMSCs via mmu_circRNA_003795. Mol. Med. Report. 2019, 19, 3732–3742. [Google Scholar] [CrossRef]
- Sun, B.; Shi, L.; Shi, Q.; Jiang, Y.; Su, Z.; Yang, X.; Zhang, Y. Circular RNAs are abundantly expressed and upregulated during repair of the damaged endometrium by Wharton’s jelly-derived mesenchymal stem cells. Stem Cell. Res. Ther. 2018, 9, 314. [Google Scholar] [CrossRef]
- Ruan, Z.-B.; Chen, G.-C.; Zhang, R.; Zhu, L. Circular RNA expression profiles during the differentiation of human umbilical cord–derived mesenchymal stem cells into cardiomyocyte-like cells. J. Cell. Physiol. 2019, 234, 16412–16423. [Google Scholar] [CrossRef]
- Sun, X.; Jin, Y.; Liang, Q.; Tang, J.; Chen, J.; Yu, Q.; Li, F.; Li, Y.; Wu, J.; Wu, S. Altered expression of circular RNAs in human placental chorionic plate-derived mesenchymal stem cells pretreated with hypoxia. J. Clin. Lab. Anal. 2019, 33, e22825. [Google Scholar] [CrossRef]
- Liu, R.; Chang, W.; Li, J.; Cheng, Y.; Dang, E.; Yang, X.; Wang, Q.; Wang, G.; Li, X.; Zhang, K. Mesenchymal stem cells in psoriatic lesions affect the skin microenvironment through circular RNA. Exp. Dermatol. 2019, 28, 292–299. [Google Scholar] [CrossRef]
- Li, X.; Peng, B.; Zhu, X.; Wang, P.; Xiong, Y.; Liu, H.; Sun, K.; Wang, H.; Ou, L.; Wu, Z.; et al. Changes in related circular RNAs following ERβ knockdown and the relationship to rBMSC osteogenesis. Biochem. Biophys. Res. Commun. 2017, 493, 100–107. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, X.; Huang, Y.; Jia, L.; Li, W. The Circular RNA Landscape of Periodontal Ligament Stem Cells During Osteogenesis. J. Periodontol. 2017, 88, 906–914. [Google Scholar] [CrossRef]
- Gu, X.; Li, M.; Jin, Y.; Liu, D.; Wei, F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, Y.; Zheng, Y.; Huang, Y.; Zhang, Y.; Jia, L.; Li, W. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell. Res. Ther. 2018, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Zhu, S.; Chen, J.; Wang, J.; Rong, Q.; Chen, S. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed. Pharmacother. 2019, 109, 1709–1717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jia, L.; Zheng, Y. circRNA Expression Profiles in Human Bone Marrow Stem Cells Undergoing Osteoblast Differentiation. Stem Cell Rev. 2019, 15, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Maass, P.G.; Glažar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Gardner, O.F.W.; Alini, M.; Stoddart, M.J. Mesenchymal Stem Cells Derived from Human Bone Marrow. In Cartilage Tissue Engineering: Methods and Protocols; Doran, P.M., Ed.; Springer: New York, NY, USA, 2015; pp. 41–52. [Google Scholar] [CrossRef]
- Herrmann, M.; Hildebrand, M.; Menzel, U.; Fahy, N.; Alini, M.; Lang, S.; Benneker, L.; Verrier, S.; Stoddart, J.M.; Bara, J.J. Phenotypic Characterization of Bone Marrow Mononuclear Cells and Derived Stromal Cell Populations from Human Iliac Crest, Vertebral Body and Femoral Head. Int. J. Mol. Sci. 2019, 20, 3454. [Google Scholar] [CrossRef]
- Della Bella, E.; Stoddart, M.J. Cell detachment rapidly induces changes in non-coding RNA expression in human mesenchymal stromal cells. Biotechniques. in press. [CrossRef]
- Wang, J.; Zhang, P.; Chen, R.; He, S.; Li, Y.; Zheng, Y.; Lu, Y.; Kan, Y. piRBase: A comprehensive database of piRNA sequences. Nucleic Acids Res. 2018, 47, D175–D180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Si, X.; Skogerbø, G.; Wang, J.; Cui, D.; Li, Y.; Sun, X.; Liu, L.; Sun, B.; Chen, R.; et al. piRBase: A web resource assisting piRNA functional study. Database 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Loebel, C.; Czekanska, E.M.; Bruderer, M.; Salzmann, G.; Alini, M.; Stoddart, M.J. In vitro osteogenic potential of human mesenchymal stem cells is predicted by Runx2/Sox9 ratio. Tissue Eng. Part A 2015, 21, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Zhao, R.; Li, Y.; Lin, Z.; Wan, J.; Xu, C.; Zeng, Y.; Zhu, Y. miR-199b-5p modulates BMSC osteogenesis via suppressing GSK-3beta/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2016, 477, 749–754. [Google Scholar] [CrossRef]
- Gennero, I.; Laurencin-Dalicieux, S.; Conte-Auriol, F.; Briand-Mesange, F.; Laurencin, D.; Rue, J.; Beton, N.; Malet, N.; Mus, M.; Tokumura, A.; et al. Absence of the lysophosphatidic acid receptor LPA1 results in abnormal bone development and decreased bone mass. Bone 2011, 49, 395–403. [Google Scholar] [CrossRef]
- Cherubini, A.; Barilani, M.; Rossi, R.L.; Jalal, M.M.K.; Rusconi, F.; Buono, G.; Ragni, E.; Cantarella, G.; Simpson, H.; Peault, B.; et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019, 47, 5325–5340. [Google Scholar] [CrossRef]
- Schäcke, H.; Schottelius, A.; Döcke, W.-D.; Strehlke, P.; Jaroch, S.; Schmees, N.; Rehwinkel, H.; Hennekes, H.; Asadullah, K. Dissociation of transactivation from transrepression by a selective glucocorticoid receptor agonist leads to separation of therapeutic effects from side effects. Proc. Natl. Acad. Sci. USA 2004, 101, 227–232. [Google Scholar] [CrossRef]
- Jiang, W.; Cazacu, S.; Xiang, C.; Zenklusen, J.C.; Fine, H.A.; Berens, M.; Armstrong, B.; Brodie, C.; Mikkelsen, T. FK506 binding protein mediates glioma cell growth and sensitivity to rapamycin treatment by regulating NF-kappaB signaling pathway. Neoplasia 2008, 10, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Egeland, M.; Zunszain, P.A.; Pariante, C.M. Molecular mechanisms in the regulation of adult neurogenesis during stress. Nat. Rev. Neurosci. 2015, 16, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Binder, E.B. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 2009, 34, S186–S195. [Google Scholar] [CrossRef] [PubMed]
- Stojadinovic, O.; Lee, B.; Vouthounis, C.; Vukelic, S.; Pastar, I.; Blumenberg, M.; Brem, H.; Tomic-Canic, M. Novel genomic effects of glucocorticoids in epidermal keratinocytes: inhibition of apoptosis, interferon-gamma pathway, and wound healing along with promotion of terminal differentiation. J. Biol. Chem. 2007, 282, 4021–4034. [Google Scholar] [CrossRef]
- He, C.; Qu, X.; Wan, J.; Rong, R.; Huang, L.; Cai, C.; Zhou, K.; Gu, Y.; Qian, S.Y.; Kang, J.X. Inhibiting Delta-6 Desaturase Activity Suppresses Tumor Growth in Mice. PLoS ONE 2012, 7, e47567. [Google Scholar] [CrossRef]
- Pickering, R.T.; Lee, M.-J.; Karastergiou, K.; Gower, A.; Fried, S.K. Depot Dependent Effects of Dexamethasone on Gene Expression in Human Omental and Abdominal Subcutaneous Adipose Tissues from Obese Women. PLoS ONE 2016, 11, e0167337. [Google Scholar] [CrossRef]
- Saether, T.; Tran, T.N.; Rootwelt, H.; Christophersen, B.O.; Haugen, T.B. Expression and regulation of delta5-desaturase, delta6-desaturase, stearoyl-coenzyme A (CoA) desaturase 1, and stearoyl-CoA desaturase 2 in rat testis. Biol. Reprod. 2003, 69, 117–124. [Google Scholar] [CrossRef]
- Marra, C.A.; de Alaniz, M.J.; Brenner, R.R. Modulation of delta 6 and delta 5 rat liver microsomal desaturase activities by dexamethasone-induced factor. Biochim. Biophys. Acta 1986, 879, 388–393. [Google Scholar] [CrossRef]
- Williams, T.M.; Moolten, D.; Burlein, J.; Romano, J.; Bhaerman, R.; Godillot, A.; Mellon, M.; Rauscher, F.J., 3rd; Kant, J.A. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science 1991, 254, 1791–1794. [Google Scholar] [CrossRef]
- Gubelmann, C.; Schwalie, P.C.; Raghav, S.K.; Röder, E.; Delessa, T.; Kiehlmann, E.; Waszak, S.M.; Corsinotti, A.; Udin, G.; Holcombe, W.; et al. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. eLife 2014, 3, e03346. [Google Scholar] [CrossRef]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Furukawa, Y.; Morita, M.; Iimura, Y.; Silva, F.P.; Li, M.; Yagyu, R.; Nakamura, Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004, 6, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Arikawa, T.; Omura, K.; Morita, I. Regulation of bone morphogenetic protein-2 expression by endogenous prostaglandin E2 in human mesenchymal stem cells. J. Cell. Physiol. 2004, 200, 400–406. [Google Scholar] [CrossRef]
- Casado-Diaz, A.; Santiago-Mora, R.; Dorado, G.; Quesada-Gomez, J.M. The omega-6 arachidonic fatty acid, but not the omega-3 fatty acids, inhibits osteoblastogenesis and induces adipogenesis of human mesenchymal stem cells: potential implication in osteoporosis. Osteoporos. Int. 2013, 24, 1647–1661. [Google Scholar] [CrossRef] [PubMed]
- Van Balkom, B.W.; Eisele, A.S.; Pegtel, D.M.; Bervoets, S.; Verhaar, M.C. Quantitative and qualitative analysis of small RNAs in human endothelial cells and exosomes provides insights into localized RNA processing, degradation and sorting. J. Extr. Vesicles 2015, 4, 26760. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, T.; Sakai, T.; Yonezawa, T.; Hiraiwa, H.; Hamada, T.; Nakashima, M.; Ono, Y.; Ishizuka, S.; Nakahara, H.; Lotz, M.K.; et al. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res. Ther. 2013, 15, R28. [Google Scholar] [CrossRef]
- Rasheed, Z.; Rasheed, N.; Abdulmonem, W.A.; Khan, M.I. MicroRNA-125b-5p regulates IL-1β induced inflammatory genes via targeting TRAF6-mediated MAPKs and NF-κB signaling in human osteoarthritic chondrocytes. Sci. Rep. 2019, 9, 6882. [Google Scholar] [CrossRef]
- Jia, J.; Wang, J.; Zhang, J.; Cui, M.; Sun, X.; Li, Q.; Zhao, B. MiR-125b Inhibits LPS-Induced Inflammatory Injury via Targeting MIP-1alpha in Chondrogenic Cell ATDC5. Cell. Physiol. Biochem. 2018, 45, 2305–2316. [Google Scholar] [CrossRef]
- Goettsch, C.; Rauner, M.; Pacyna, N.; Hempel, U.; Bornstein, S.R.; Hofbauer, L.C. miR-125b regulates calcification of vascular smooth muscle cells. Am. J. Pathol. 2011, 179, 1594–1600. [Google Scholar] [CrossRef]
- Eguchi, T.; Watanabe, K.; Hara, E.S.; Ono, M.; Kuboki, T.; Calderwood, S.K. OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS ONE 2013, 8, e58796. [Google Scholar] [CrossRef]
- Chen, D.; Liu, L.; Luo, X.; Mu, A.; Yan, L.; Chen, X.; Wang, L.; Wang, N.; He, H.; Zhou, H.; et al. Effect of SMYD3 on the microRNA expression profile of MCF-7 breast cancer cells. Oncol. Lett. 2017, 14, 1831–1840. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torres, A.G.; Reina, O.; Stephan-Otto Attolini, C.; Ribas de Pouplana, L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 8451–8456. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Sarkar, A.; Parida, S.; Ghosh, Z.; Mallick, B. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s disease and their probable role in pathogenesis. Mol. Biosyst. 2017, 13, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Krawetz, S.A.; Kruger, A.; Lalancette, C.; Tagett, R.; Anton, E.; Draghici, S.; Diamond, M.P. A survey of small RNAs in human sperm. Hum. Reprod. 2011, 26, 3401–3412. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, Q.; Shen, J.; Yang, B.B.; Ding, X. Circbank: A comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019, 16, 899–905. [Google Scholar] [CrossRef]
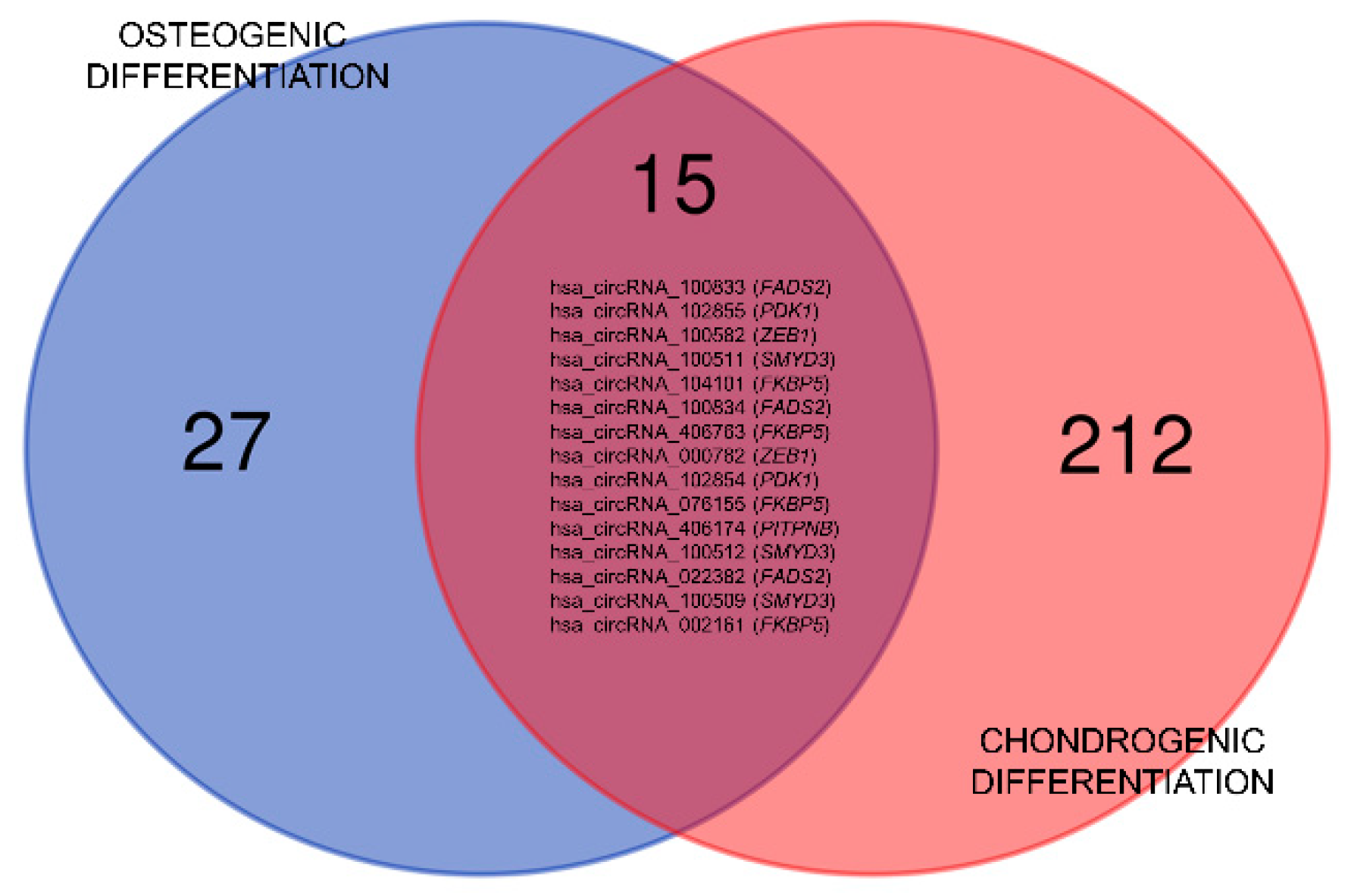

| Rank | probeID | P-value | FDR | FC (abs) | Reg | circRNA | Gene Symbol | miRNA Response Elements |
|---|---|---|---|---|---|---|---|---|
| #1 | ASCRP3005562 | 6.047E-08 | 0.000720802 | 5.4280811 | up | hsa_circRNA_104101 | FKBP5 | hsa-miR-1468-5p hsa-miR-642a-5p hsa-miR-561-5p hsa-miR-29b-1-5p hsa-miR-708-5p |
| #2 | ASCRP3000836 | 1.1812E-06 | 0.007039952 | 3.9089404 | up | hsa_circRNA_002161 | FKBP5 | hsa-miR-619-5p hsa-miR-20b-3p hsa-miR-367-3p hsa-miR-153-5p hsa-miR-665 |
| #3 | ASCRP3000835 | 8.88526E-06 | 0.02118246 | 3.3616827 | up | hsa_circRNA_076155 | FKBP5 | hsa-miR-6845-3p hsa-miR-1468-5p hsa-miR-642a-5p hsa-miR-6834-3p hsa-miR-561-5p |
| #4 | ASCRP3000434 | 0.004935888 | 0.960907885 | 2.8301448 | up | hsa_circRNA_022382 | FADS2 | hsa-miR-5586-5p hsa-miR-4726-5p hsa-miR-4640-5p hsa-miR-3138 hsa-miR-8080 |
| #5 | ASCRP3003129 | 0.019088529 | 0.960907885 | 2.8255694 | up | hsa_circRNA_100833 | FADS2 | hsa-miR-765 hsa-miR-495-3p hsa-miR-665 hsa-miR-193b-5p hsa-miR-124-5p |
| #6 | ASCRP3001707 | 1.86216E-06 | 0.007398982 | 2.6965752 | up | hsa_circRNA_406763 | FKBP5 | hsa-miR-1273g-3p hsa-miR-1285-3p hsa-miR-619-5p hsa-miR-1183 hsa-miR-512-5p |
| #7 | ASCRP3013118 | 0.003143209 | 0.960907885 | 2.304461 | up | hsa_circRNA_100834 | FADS2 | hsa-miR-873-5p hsa-miR-23b-5p hsa-miR-181a-2-3p hsa-miR-93-3p hsa-miR-299-3p |
| #8 | ASCRP3005871 | 0.046552395 | 0.960907885 | 2.2820422 | up | hsa_circRNA_103249 | FBLN1 | hsa-miR-671-5p hsa-let-7b-5p hsa-let-7e-5p hsa-let-7i-5p hsa-miR-185-3p |
| #9 | ASCRP3001839 | 0.000150493 | 0.172024477 | 2.0442825 | up | hsa_circRNA_406308 | COL8A1 | hsa-miR-153-5p hsa-miR-126-5p hsa-miR-1277-5p hsa-miR-548az-5p hsa-miR-3607-3p |
| #10 | ASCRP3013548 | 0.004719482 | 0.960907885 | 2.0061379 | up | hsa_circRNA_405468 | MT2A | hsa-miR-6791-5p hsa-miR-6859-5p hsa-miR-450a-1-3p hsa-miR-4257 hsa-miR-1236-5p |
| #12 | ASCRP3009465 | 0.007384491 | 0.960907885 | 1.9128242 | up | hsa_circRNA_057186 | AGPS | hsa-miR-3938 hsa-miR-942-5p hsa-miR-3935 hsa-miR-646 hsa-miR-4753-3p |
| #13 | ASCRP3007511 | 0.005784275 | 0.960907885 | 1.8653692 | up | hsa_circRNA_100832 | FADS1 | hsa-miR-218-1-3p hsa-miR-193a-5p hsa-miR-764 hsa-miR-876-3p hsa-miR-138-5p |
| #14 | ASCRP3009116 | 0.007604484 | 0.960907885 | 1.8116207 | up | hsa_circRNA_405763 | TDRD12 | hsa-miR-4438 hsa-miR-182-5p hsa-miR-3188 hsa-miR-7-5p hsa-miR-1226-3p |
| #15 | ASCRP3002616 | 0.035838981 | 0.960907885 | 1.6842185 | up | hsa_circRNA_104864 | LPAR1 | hsa-miR-7-5p hsa-miR-588 hsa-miR-135a-3p hsa-miR-608 hsa-miR-140-3p |
| #16 | ASCRP3003960 | 0.009707576 | 0.960907885 | 1.679094 | up | hsa_circRNA_402206 | LTBP1 | hsa-miR-432-5p hsa-miR-6797-3p hsa-miR-3127-3p hsa-miR-6804-5p hsa-miR-18a-3p |
| #17 | ASCRP3003038 | 0.011416289 | 0.960907885 | 1.6747001 | up | hsa_circRNA_000782 | ZEB1 | hsa-miR-141-3p hsa-miR-200a-3p hsa-miR-148a-5p hsa-miR-136-5p hsa-miR-494-5p |
| #20 | ASCRP3001742 | 0.044814485 | 0.960907885 | 1.5884727 | up | hsa_circRNA_100582 | ZEB1 | hsa-miR-141-3p hsa-miR-200a-3p hsa-miR-148a-5p hsa-miR-452-3p hsa-miR-877-3p |
| #1 | ASCRP3007208 | 0.002787677 | 0.923030978 | 0.4292865 | down | hsa_circRNA_100512 | SMYD3 | hsa-miR-182-5p hsa-miR-181a-5p hsa-miR-532-3p hsa-miR-1224-3p hsa-miR-181b-5p |
| #2 | ASCRP3006013 | 0.007116011 | 0.960907885 | 0.4726294 | down | hsa_circRNA_100511 | SMYD3 | hsa-miR-532-3p hsa-miR-516a-3p hsa-miR-516b-3p hsa-miR-182-5p hsa-miR-181a-5p |
| #3 | ASCRP3004046 | 0.000140327 | 0.172024477 | 0.4904122 | down | hsa_circRNA_100835 | BSCL2 | hsa-miR-646 hsa-miR-199b-5p hsa-let-7g-5p hsa-let-7e-5p hsa-miR-658 |
| #6 | ASCRP3012578 | 7.30204E-05 | 0.108800441 | 0.5413913 | down | hsa_circRNA_103729 | PDE5A | hsa-miR-670-3p hsa-miR-583 hsa-miR-455-3p hsa-miR-510-3p hsa-miR-500a-5p |
| #7 | ASCRP3008231 | 0.009384685 | 0.960907885 | 0.5464973 | down | hsa_circRNA_100509 | SMYD3 | hsa-miR-516b-3p hsa-miR-516a-3p hsa-miR-671-5p hsa-miR-425-3p hsa-miR-584-5p |
| #8 | ASCRP3007154 | 0.004585789 | 0.960907885 | 0.5603480 | down | hsa_circRNA_100510 | SMYD3 | hsa-miR-516a-3p hsa-miR-516b-3p hsa-miR-182-5p hsa-miR-181a-5p hsa-miR-362-5p |
| #9 | ASCRP3001523 | 0.002025898 | 0.832713912 | 0.5671141 | down | hsa_circRNA_103415 | FOXP1 | hsa-miR-370-3p hsa-miR-558 hsa-miR-127-5p hsa-miR-93-3p hsa-miR-17-3p |
| #10 | ASCRP3009267 | 0.000205329 | 0.180247633 | 0.5766464 | down | hsa_circRNA_102854 | PDK1 | hsa-miR-362-5p hsa-miR-500a-5p hsa-miR-30a-5p hsa-miR-30e-5p hsa-miR-21-3p |
| #14 | ASCRP3003982 | 2.29715E-05 | 0.045636674 | 0.6169628 | down | hsa_circRNA_406933 | PDE1C | hsa-miR-138-5p hsa-miR-1248 hsa-miR-3653-5p hsa-miR-6755-3p hsa-miR-26b-3p |
| #16 | ASCRP3007600 | 0.009304848 | 0.960907885 | 0.6298330 | down | hsa_circRNA_103414 | FOXP1 | hsa-miR-370-3p hsa-miR-558 hsa-miR-127-5p hsa-miR-93-3p hsa-miR-17-3p |
| #19 | ASCRP3010569 | 0.000767043 | 0.481218738 | 0.6493059 | down | hsa_circRNA_102856 | PDK1 | hsa-miR-637 hsa-miR-362-5p hsa-miR-500a-5p hsa-miR-30a-5p hsa-miR-511-5p |
| #21 | ASCRP3000965 | 0.001161463 | 0.604800463 | 0.6630723 | down | hsa_circRNA_102855 | PDK1 | hsa-miR-637 hsa-miR-362-5p hsa-miR-500a-5p hsa-miR-30a-5p hsa-miR-511-5p |
| Rank | probeID | P-value | FDR | FC (abs) | Reg | circRNA | Gene Symbol | miRNA Response Elements |
|---|---|---|---|---|---|---|---|---|
| #1 | ASCRP3005562 | 1.63024E-06 | 0.003238743 | 5.3480122 | up | hsa_circRNA_104101 | FKBP5 | hsa-miR-1468-5p hsa-miR-642a-5p hsa-miR-561-5p hsa-miR-29b-1-5p hsa-miR-708-5p |
| #2 | ASCRP3000434 | 0.000396713 | 0.055633194 | 4.6683484 | up | hsa_circRNA_022382 | FADS2 | hsa-miR-5586-5p hsa-miR-4726-5p hsa-miR-4640-5p hsa-miR-3138 hsa-miR-8080 |
| #3 | ASCRP3000836 | 0.000102233 | 0.031215885 | 4.496583 | up | hsa_circRNA_002161 | FKBP5 | hsa-miR-619-5p hsa-miR-20b-3p hsa-miR-367-3p hsa-miR-153-5p hsa-miR-665 |
| #4 | ASCRP3009795 | 1.8396E-07 | 0.001096402 | 3.9770883 | up | hsa_circRNA_081069 | COL1A2 | hsa-miR-4733-3p hsa-miR-665 hsa-miR-5096 hsa-miR-412-3p hsa-miR-4459 |
| #6 | ASCRP3006708 | 7.97805E-05 | 0.030676897 | 3.7163892 | up | hsa_circRNA_102121 | COL1A1 | hsa-miR-412-3p hsa-miR-214-3p hsa-miR-194-3p hsa-miR-761 hsa-miR-362-5p |
| #7 | ASCRP3003129 | 0.000497582 | 0.061309307 | 3.5929669 | up | hsa_circRNA_100833 | FADS2 | hsa-miR-765 hsa-miR-495-3p hsa-miR-665 hsa-miR-193b-5p hsa-miR-124-5p |
| #8 | ASCRP3000835 | 0.000417696 | 0.057894568 | 3.1908719 | up | hsa_circRNA_076155 | FKBP5 | hsa-miR-6845-3p hsa-miR-1468-5p hsa-miR-642a-5p hsa-miR-6834-3p hsa-miR-561-5p |
| #15 | ASCRP3002750 | 8.236E-07 | 0.001963462 | 2.7030084 | up | hsa_circRNA_400670 | FGFR2 | hsa-miR-449c-5p hsa-miR-885-3p hsa-miR-216b-3p hsa-miR-3147 hsa-miR-4691-3p |
| #16 | ASCRP3009380 | 0.000395625 | 0.055633194 | 2.6823979 | up | hsa_circRNA_400294 | COL11A1 | hsa-miR-4668-3p hsa-miR-4659a-3p hsa-miR-4659b-3p hsa-miR-2113 hsa-miR-548aq-3p |
| #18 | ASCRP3004582 | 6.12173E-05 | 0.026061058 | 2.544377 | up | hsa_circRNA_103987 | SPARC | hsa-miR-194-3p hsa-miR-22-5p hsa-miR-342-5p hsa-miR-328-3p hsa-miR-30d-3p |
| #22 | ASCRP3013118 | 0.0007731 | 0.075807131 | 2.2866119 | up | hsa_circRNA_100834 | FADS2 | hsa-miR-873-5p hsa-miR-23b-5p hsa-miR-181a-2-3p hsa-miR-93-3p hsa-miR-299-3p |
| #23 | ASCRP3001707 | 0.006322882 | 0.219733971 | 2.2821711 | up | hsa_circRNA_406763 | FKBP5 | hsa-miR-1273g-3p hsa-miR-1285-3p hsa-miR-619-5p hsa-miR-1183 hsa-miR-512-5p |
| #26 | ASCRP3004478 | 0.001383209 | 0.110656729 | 2.1118736 | up | hsa_circRNA_102558 | TGFB1 | hsa-miR-219a-2-3p hsa-miR-514a-5p hsa-miR-602 hsa-miR-1301-3p hsa-miR-150-3p |
| #33 | ASCRP3002980 | 0.000198815 | 0.036800403 | 2.0231353 | up | hsa_circRNA_007482 | COL5A1 | hsa-miR-6746-5p hsa-miR-639 hsa-miR-608 hsa-miR-8082 hsa-miR-4534 |
| #34 | ASCRP3013535 | 0.000013325 | 0.015778114 | 2.0144106 | up | hsa_circRNA_047037 | LDLRAD4 | hsa-miR-4767 hsa-miR-2277-5p hsa-miR-6737-3p hsa-miR-5008-3p hsa-miR-6089 |
| #41 | ASCRP3000681 | 1.45603E-05 | 0.015778114 | 1.9222447 | up | hsa_circRNA_035152 | FBN1 | hsa-miR-181b-5p hsa-miR-181d-5p hsa-miR-181a-5p hsa-miR-181c-5p hsa-miR-6841-3p |
| #42 | ASCRP3003323 | 0.000124468 | 0.032253525 | 1.9115005 | up | hsa_circRNA_008421 | FBN1 | hsa-miR-519c-3p hsa-miR-519a-3p hsa-miR-519b-3p hsa-miR-6762-5p hsa-miR-136-5p |
| #46 | ASCRP3012377 | 0.000184393 | 0.036347001 | 1.8787574 | up | hsa_circRNA_001552 | ZEB1 | hsa-miR-548a-5p hsa-miR-153-5p hsa-miR-548d-5p hsa-miR-136-5p hsa-miR-568 |
| #52 | ASCRP3012658 | 8.81337E-05 | 0.031215885 | 1.8418985 | up | hsa_circRNA_404737 | ZEB1 | hsa-miR-1277-5p hsa-miR-3171 hsa-miR-2054 hsa-miR-548ab hsa-miR-3149 |
| #55 | ASCRP3000740 | 0.000138804 | 0.033621593 | 1.822582 | up | hsa_circRNA_002765 | ZEB1 | hsa-miR-6728-3p hsa-miR-1909-5p hsa-miR-1277-5p hsa-miR-6877-3p hsa-miR-8089 |
| #63 | ASCRP3010383 | 0.004955326 | 0.20113116 | 1.7629155 | up | hsa_circRNA_102231 | P4HB | hsa-miR-1301-3p hsa-miR-103a-2-5p hsa-miR-541-5p hsa-miR-133a-5p hsa-miR-598-5p |
| #71 | ASCRP3008500 | 2.9687E-07 | 0.001179563 | 1.6937109 | up | hsa_circRNA_405335 | FBN1 | hsa-miR-181b-5p hsa-miR-181d-5p hsa-miR-181a-5p hsa-miR-4801 hsa-miR-181c-5p |
| #78 | ASCRP3005846 | 0.005602103 | 0.215854454 | 1.673856 | up | hsa_circRNA_100094 | HSPG2 | hsa-miR-185-3p hsa-miR-671-3p hsa-miR-638 hsa-miR-675-5p hsa-miR-552-3p |
| #80 | ASCRP3005992 | 0.000153855 | 0.034452544 | 1.6644833 | up | hsa_circRNA_104118 | RUNX2 | hsa-miR-335-3p hsa-miR-216a-5p hsa-miR-450a-2-3p hsa-miR-125a-3p hsa-miR-502-5p |
| #91 | ASCRP3003055 | 0.011456522 | 0.286111338 | 1.6111455 | up | hsa_circRNA_402986 | PLOD2 | hsa-miR-6884-3p hsa-miR-7151-5p hsa-miR-6887-3p hsa-miR-6873-3p hsa-miR-1285-5p |
| #92 | ASCRP3012598 | 0.008597966 | 0.246958447 | 1.6044765 | up | hsa_circRNA_046265 | P4HB | hsa-miR-4763-3p hsa-miR-6808-5p hsa-miR-5047 hsa-miR-4692 hsa-miR-1207-5p |
| #94 | ASCRP3010648 | 0.001722361 | 0.12220558 | 1.5990799 | up | hsa_circRNA_073237 | VCAN | hsa-miR-4778-3p hsa-miR-122-5p hsa-miR-2115-5p hsa-miR-6854-5p hsa-miR-335-3p |
| #97 | ASCRP3000965 | 0.004929519 | 0.20113116 | 1.5916847 | up | hsa_circRNA_102855 | PDK1 | hsa-miR-637 hsa-miR-362-5p hsa-miR-500a-5p hsa-miR-30a-5p hsa-miR-511-5p |
| #99 | ASCRP3003038 | 0.001851474 | 0.125885791 | 1.5899916 | up | hsa_circRNA_000782 | ZEB1 | hsa-miR-141-3p hsa-miR-200a-3p hsa-miR-148a-5p hsa-miR-136-5p hsa-miR-494-5p |
| #103 | ASCRP3001742 | 0.001136042 | 0.096473031 | 1.5793354 | up | hsa_circRNA_100582 | ZEB1 | hsa-miR-141-3p hsa-miR-200a-3p hsa-miR-148a-5p hsa-miR-452-3p hsa-miR-877-3p |
| #105 | ASCRP3009267 | 0.036604648 | 0.45977598 | 1.5758009 | up | hsa_circRNA_102854 | PDK1 | hsa-miR-362-5p hsa-miR-500a-5p hsa-miR-30a-5p hsa-miR-30e-5p hsa-miR-21-3p |
| #124 | ASCRP3005963 | 0.001148645 | 0.096473031 | 1.5235693 | up | hsa_circRNA_104119 | RUNX2 | hsa-miR-7-5p hsa-miR-663a hsa-miR-149-3p hsa-miR-885-3p hsa-miR-661 |
| #56 | ASCRP3006502 | 0.002984365 | 0.160966674 | 0.5802392 | down | hsa_circRNA_062035 | COL6A1 | hsa-miR-5001-5p hsa-miR-4685-5p hsa-miR-661 hsa-miR-4739 hsa-miR-6089 |
| #60 | ASCRP3007208 | 0.00578642 | 0.217584002 | 0.6086586 | down | hsa_circRNA_100512 | SMYD3 | hsa-miR-182-5p hsa-miR-181a-5p hsa-miR-532-3p hsa-miR-1224-3p hsa-miR-181b-5p |
| #77 | ASCRP3010338 | 0.005350258 | 0.210791919 | 0.6319221 | down | hsa_circRNA_102171 | SMURF2 | hsa-miR-539-5p hsa-let-7f-2-3p hsa-miR-216a-3p hsa-miR-578 hsa-miR-587 |
| #79 | ASCRP3008231 | 0.011029053 | 0.282723243 | 0.6357313 | down | hsa_circRNA_100509 | SMYD3 | hsa-miR-516b-3p hsa-miR-516a-3p hsa-miR-671-5p hsa-miR-425-3p hsa-miR-584-5p |
| #89 | ASCRP3006013 | 0.003248887 | 0.167106571 | 0.6532234 | down | hsa_circRNA_100511 | SMYD3 | hsa-miR-532-3p hsa-miR-516a-3p hsa-miR-516b-3p hsa-miR-182-5p hsa-miR-181a-5p |
| #91 | ASCRP3013164 | 0.000112608 | 0.031215885 | 0.6539596 | down | hsa_circRNA_083999 | FGFR1 | hsa-miR-6769a-5p hsa-miR-6769b-5p hsa-miR-3692-3p hsa-miR-130a-5p hsa-miR-1226-5p |
| #97 | ASCRP3005052 | 0.002076144 | 0.135233004 | 0.6664074 | down | hsa_circRNA_084010 | FGFR1 | hsa-miR-759 hsa-miR-298 hsa-miR-4435 hsa-miR-92b-5p hsa-miR-490-5p |
| CircRNA ID | Best Transcript | CircRNA Regulation in OSTEO vs. CRL Monolayer (Fold Change) | Binding Site For | miRNA Regulation in OSTEO vs. CRL Monolayer (Fold change) |
| hsa_circRNA_103249 | FBLN1 | Upregulated (2.28) | hsa-let-7i-5p | Downregulated (0.56) |
| hsa_circRNA_405468 | MT2A | Upregulated (2.00) | hsa-miR-450a-1-3p | Downregulated (0.62) |
| hsa_circRNA_406308 | COL8A1 | Upregulated (2.04) | hsa-miR-548az-5p | Upregulated (1.86) |
| hsa_circRNA_405763 | TDRD12 | Upregulated (1.81) | hsa-miR-7-5p | Upregulated (1.64) |
| hsa_circRNA_104864 | LPAR1 | Upregulated (1.68) | hsa-miR-7-5p | Upregulated (1.64) |
| hsa_circRNA_057186 | AGPS | Upregulated (1.91) | hsa-miR-942-5p | Upregulated (1.59) |
| hsa_circRNA_100835 | BSCL2 | Downregulated (0.49) | hsa-miR-199b-5p | Upregulated (2.65) |
| CircRNA ID | Best Transcript | CircRNA Regulation in CHONDRO vs. CRL Pellet (Fold Change) | Binding Site For | miRNA Regulation in CHONDRO vs. CRL Pellet (Fold Change) |
| hsa_circRNA_102121 | COL1A1 | Upregulated (3.72) | hsa-miR-214-3p | Upregulated (1.85) |
| hsa_circRNA_081069 | COL1A2 | Upregulated (3.98) | hsa-miR-665 | Downregulated (0.37) |
| hsa_circRNA_100833 | FADS2 | Upregulated (3.59) | hsa-miR-665 hsa-miR-495-3p | Downregulated (0.37) Downregulated (0.54) |
| hsa_circRNA_100834 | FADS2 | Upregulated (2.29) | hsa-miR-23b-5p hsa-miR-299-3p | Upregulated (4.13) Downregulated (0.61) |
| hsa_circRNA_035152 | FBN1 | Upregulated (1.92) | hsa-miR-181a-5p hsa-miR-181b-5p hsa-miR-181d-5p | Upregulated (3.89) Upregulated (3.70) Upregulated (1.82) |
| hsa_circRNA_405335 | FBN1 | Upregulated (1.69) | hsa-miR-181a-5p hsa-miR-181b-5p hsa-miR-181d-5p | Upregulated (3.89) Upregulated (3.70) Upregulated (1.82) |
| hsa_circRNA_400670 | FGFR2 | Upregulated (2.70) | hsa-miR-449c-5p | Upregulated (4.81) |
| hsa_circRNA_002161 | FKBP5 | Upregulated (4.50) | hsa-miR-665 | Downregulated (0.37) |
| hsa_circRNA_406763 | FKBP5 | Upregulated (2.28) | hsa-miR-1285-3p | Upregulated (1.82) |
| hsa_circRNA_104101 | FKBP5 | Upregulated (5.35) | hsa-miR-29b-1-5p hsa-miR-708-5p | Downregulated (0.46) Downregulated (0.48) |
| hsa_circRNA_100094 | HSPG2 | Upregulated (1.67) | hsa-miR-675-5p | Upregulated (24.65) |
| hsa_circRNA_047037 | LDLRAD4 | Upregulated (2.01) | hsa-miR-2277-5p | Upregulated (1.52) |
| hsa_circRNA_102854 | PDK1 | Upregulated (1.58) | hsa-miR-30e-5p hsa-miR-21-3p | Upregulated (1.56) Upregulated (2.29) |
| hsa_circRNA_104119 | RUNX2 | Upregulated (1.52) | hsa-miR-7-5p | Upregulated (1.42) |
| hsa_circRNA_104118 | RUNX2 | Upregulated (1.66) | hsa-miR-335-3p hsa-miR-450a-2-3p | Downregulated (0.21) Downregulated (0.60) |
| hsa_circRNA_073237 | VCAN | Upregulated (1.60) | hsa-miR-335-3p | Downregulated (0.21) |
| hsa_circRNA_100582 | ZEB1 | Upregulated (1.58) | hsa-miR-148a-5p | Upregulated (2.57) |
| hsa_circRNA_000782 | ZEB1 | Upregulated (1.59) | hsa-miR-148a-5p | Upregulated (2.57) |
| hsa_circRNA_100511 | SMYD3 | Downregulated (0.65) | hsa-miR-181a-5p | Upregulated (3.89) |
| hsa_circRNA_100512 | SMYD3 | Downregulated (0.61) | hsa-miR-181a-5p hsa-miR-181b-5p | Upregulated (3.89) Upregulated (3.70) |
| hsa_circRNA_100509 | SMYD3 | Downregulated (0.64) | hsa-miR-671-5p | Upregulated (1.80) |
| hsa_circRNA_102171 | SMURF2 | Downregulated (0.63) | hsa-let-7f-2-3p | Upregulated (1.72) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Della Bella, E.; Menzel, U.; Basoli, V.; Tourbier, C.; Alini, M.; Stoddart, M.J. Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells 2020, 9, 398. https://doi.org/10.3390/cells9020398
Della Bella E, Menzel U, Basoli V, Tourbier C, Alini M, Stoddart MJ. Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells. 2020; 9(2):398. https://doi.org/10.3390/cells9020398
Chicago/Turabian StyleDella Bella, Elena, Ursula Menzel, Valentina Basoli, Céline Tourbier, Mauro Alini, and Martin J. Stoddart. 2020. "Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells" Cells 9, no. 2: 398. https://doi.org/10.3390/cells9020398
APA StyleDella Bella, E., Menzel, U., Basoli, V., Tourbier, C., Alini, M., & Stoddart, M. J. (2020). Differential Regulation of circRNA, miRNA, and piRNA during Early Osteogenic and Chondrogenic Differentiation of Human Mesenchymal Stromal Cells. Cells, 9(2), 398. https://doi.org/10.3390/cells9020398