Silencing Dicer-Like Genes Reduces Virulence and sRNA Generation in Penicillium italicum, the Cause of Citrus Blue Mold
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strain and Plasmid
2.2. Construction of Silencing Vectors
2.3. Preparation of Protoplasts, Transformation and Screening
2.4. Biomass Assay and Pathogenicity Testing
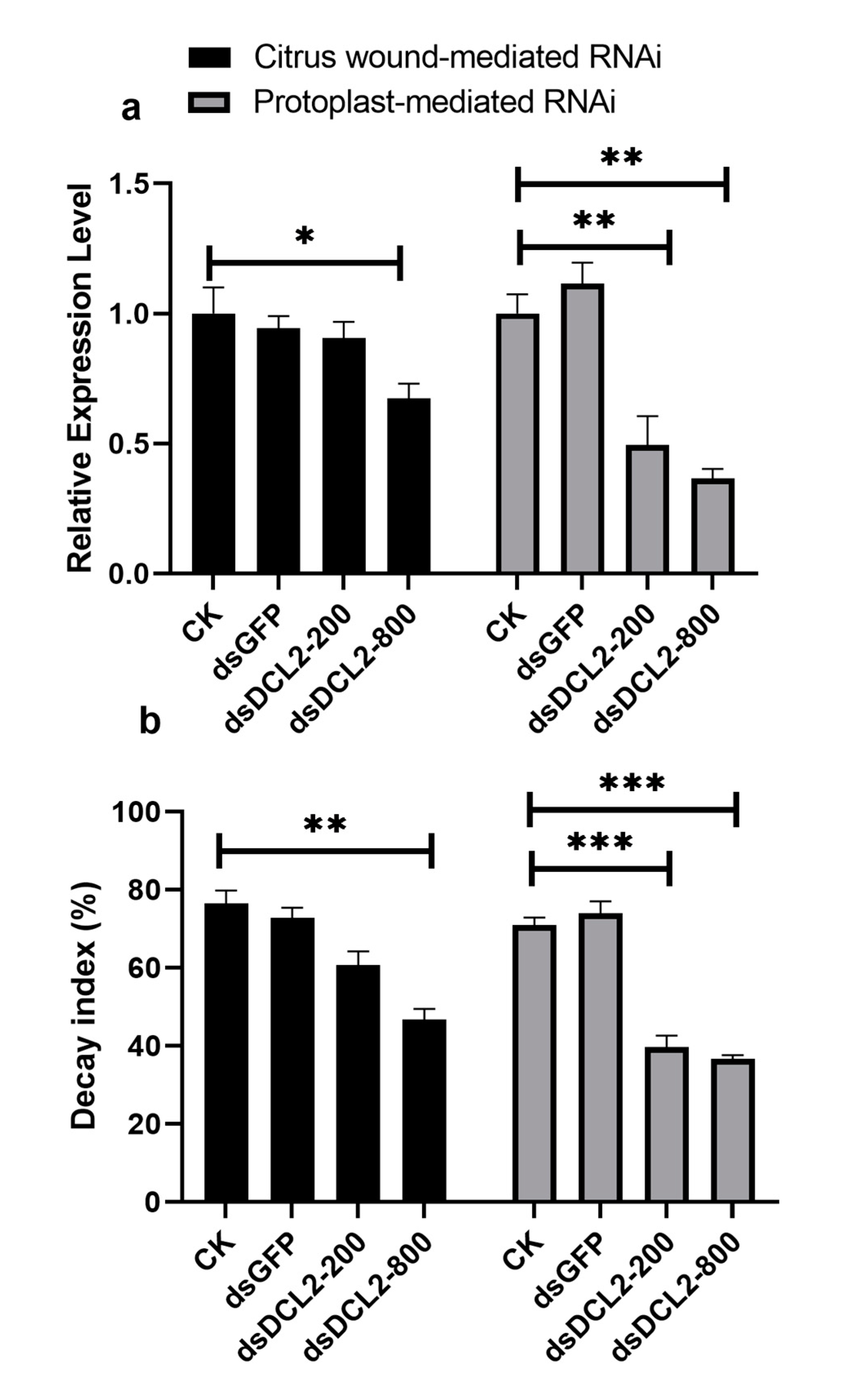
2.5. Preparation of Pit-DCL2 dsRNA and Application on the Citrus Fruit Surface
2.6. Small RNA Sequencing
2.7. qRT-PCR and Northern Blot Analysis
3. Results
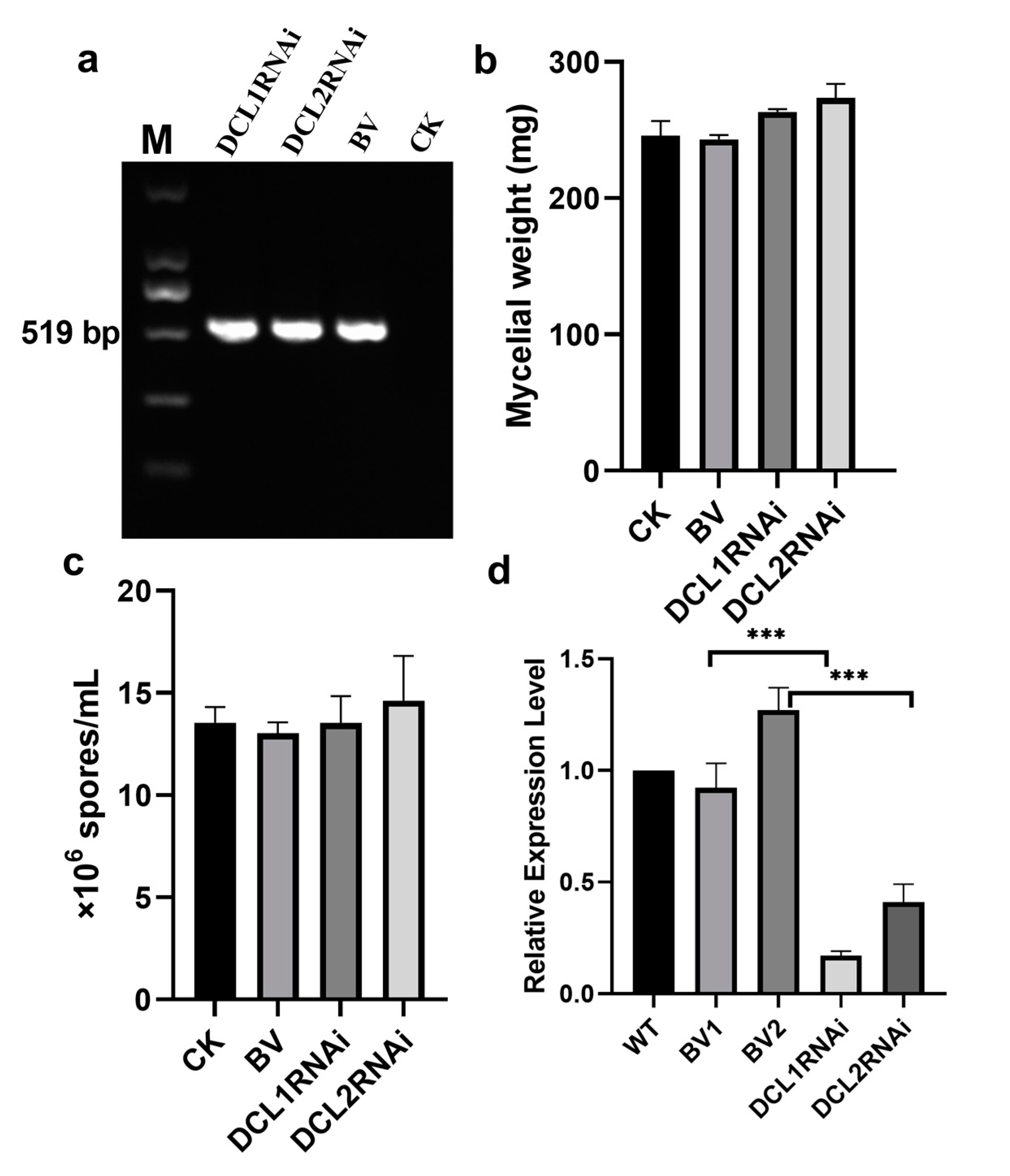
3.1. Generation of Silencing Transformants of Dicer-Like Genes in P. italicum
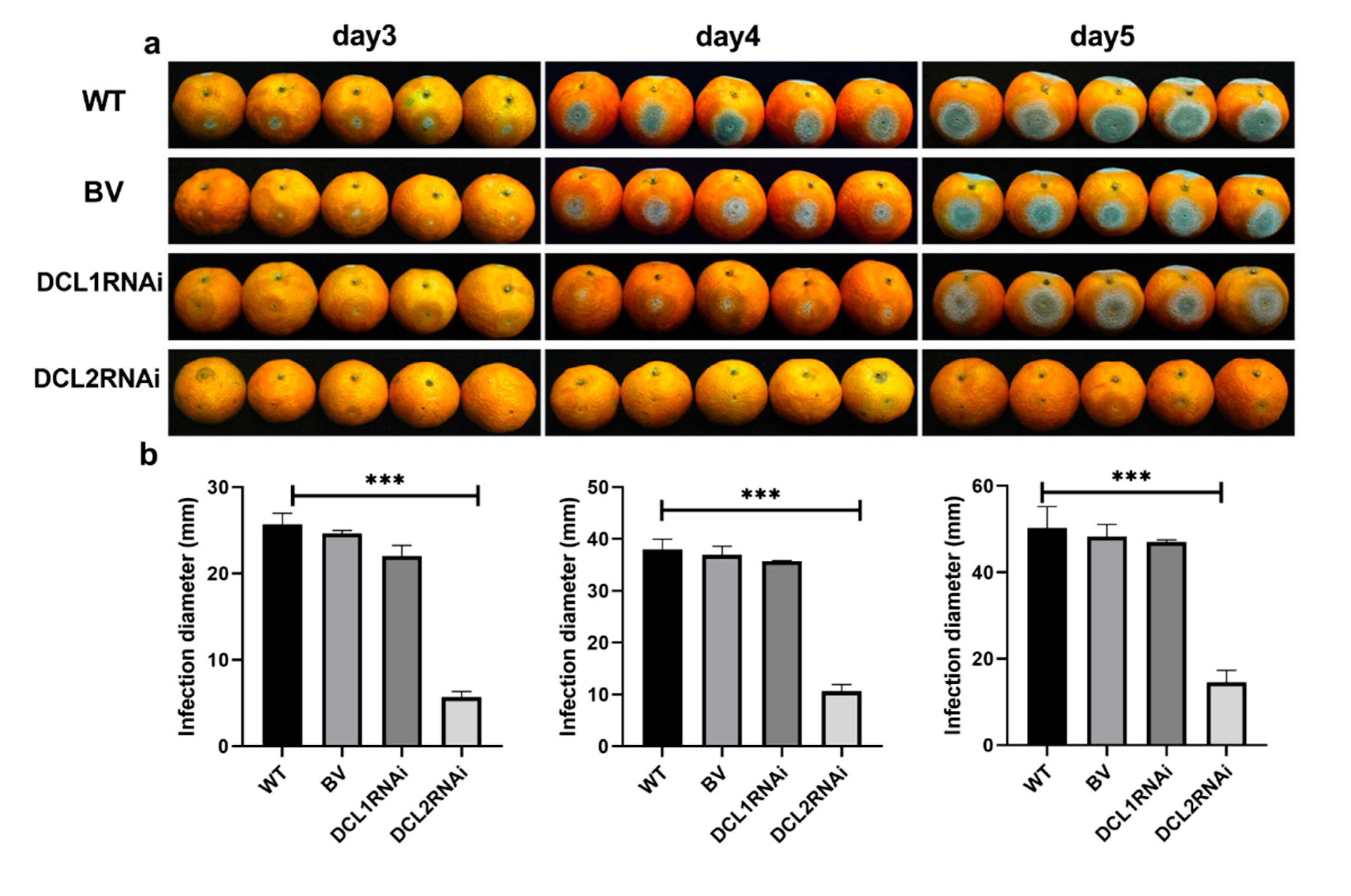
3.2. The Effect of Dicer-Like Gene Silencing on the Infection of Citrus Fruits by P. italicum
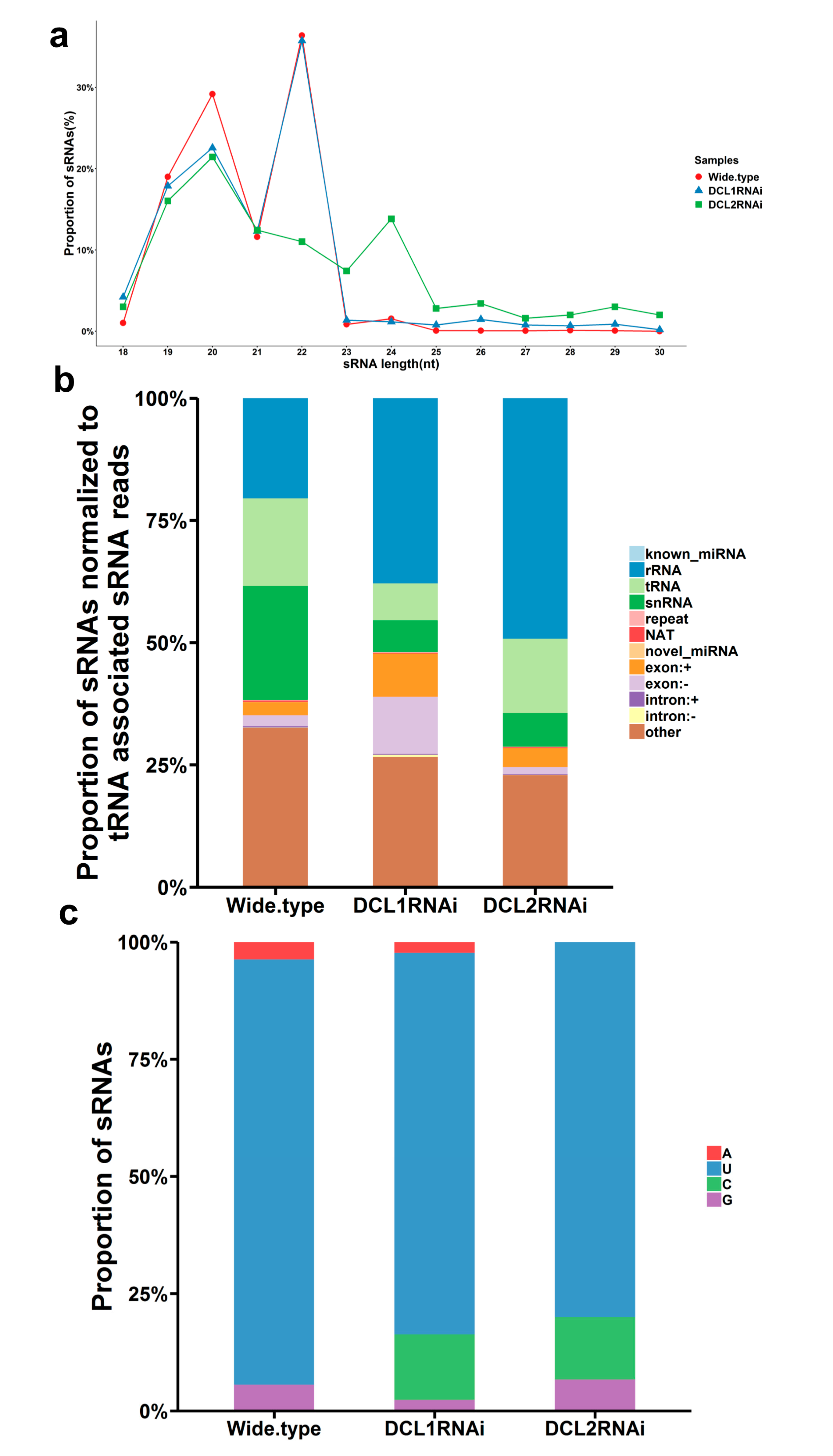
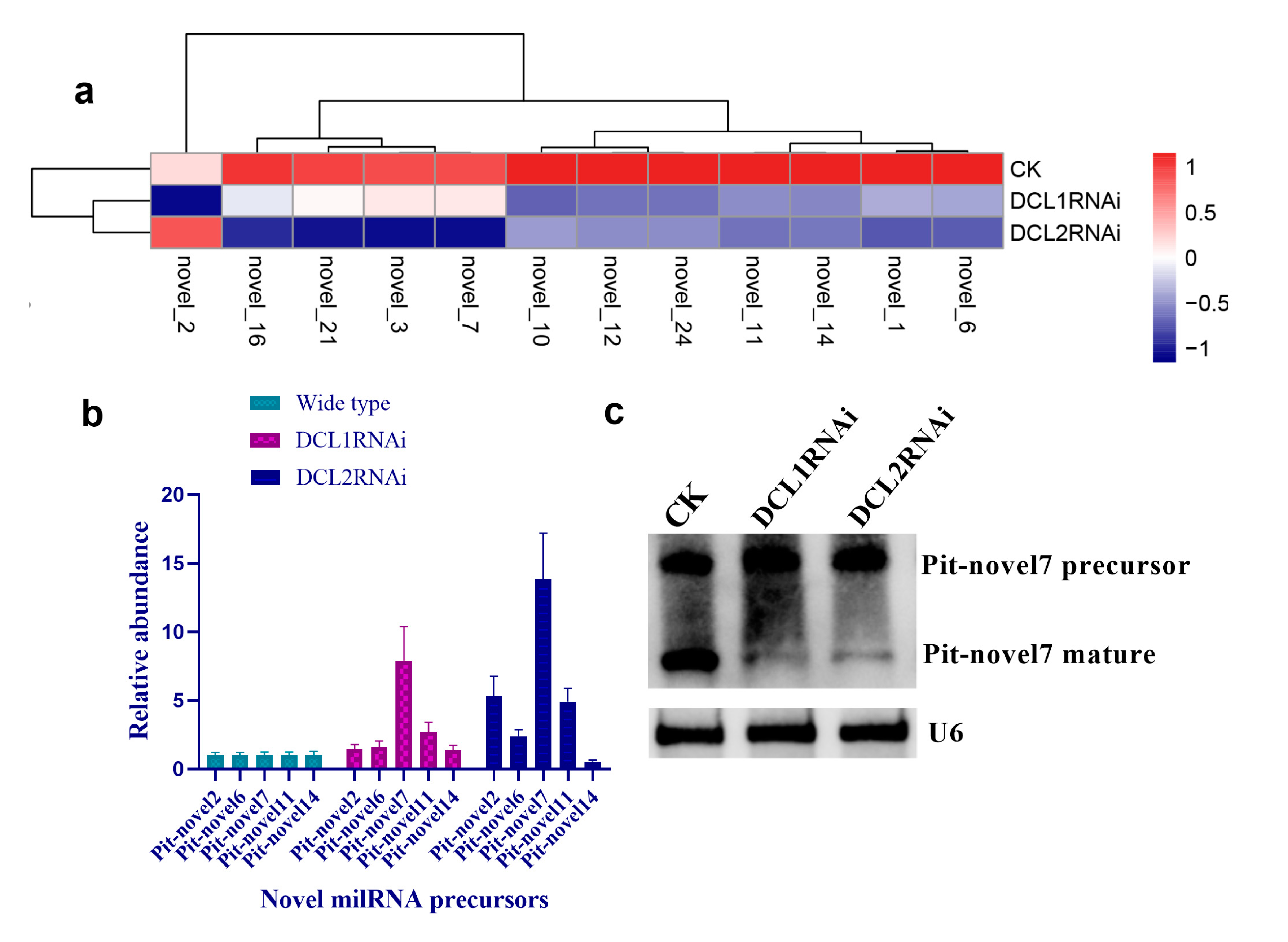
3.3. Characterization of Small RNAs in the RNAi Transformants
3.4. Structural Features, Expression Patterns and Predicted Targets of the Novel Pit-milRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- FAOSTAT Database. Available online: http://www.fao.org/faostat/en (accessed on 1 October 2017).
- Papoutsis, K.; Mathioudakis, M.M.; Hasperué, J.H.; Ziogas, V. Non-chemical treatments for preventing the postharvest fungal rotting of citrus caused by Penicillium digitatum (green mold) and Penicillium italicum (blue mold). Trends Food Sci. Technol. 2019, 86, 479–491. [Google Scholar] [CrossRef]
- Prusky, D.; McEvoy, J.L.; Saftner, R.; Conway, W.S.; Jones, R. Relationship Between Host Acidification and Virulence of Penicillium spp. on Apple and Citrus Fruit. Phytopathology 2004, 94, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Macarisin, D.; Cohen, L.; Eick, A.; Rafael, G.; Belausov, E.; Wisniewski, M.; Droby, S. Penicillium digitatum Suppresses Production of Hydrogen Peroxide in Host Tissue During Infection of Citrus Fruit. Phytopathology 2007, 97, 1491–1500. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zong, Y.; Du, Z.; Chen, Y.; Zhang, Z.; Qin, G.; Zhao, W.; Tian, S. Genomic Characterization Reveals Insights Into Patulin Biosynthesis and Pathogenicity in Penicillium Species. Mol. Plant Microbe Interact. 2015, 28, 635–647. [Google Scholar] [CrossRef]
- Ballester, A.R.; Marcet-Houben, M.; Levin, E.; Sela, N.; Selma-Lázaro, C.; Carmona, L.; Wisniewski, M.; Droby, S.; González-Candelas, L.; Gabaldón, T. Genome, Transcriptome, and Functional Analyses of Penicillium expansum Provide New Insights Into Secondary Metabolism and Pathogenicity. Mol. Plant Microbe Interact. 2015, 28, 232–248. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Kogel, K.H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Nakayashiki, H.; Kadotani, N.; Mayama, S. Evolution and diversification of RNA silencing proteins in fungi. J. Mol. Evol. 2006, 63, 127–135. [Google Scholar] [CrossRef]
- Carmell, M.A.; Xuan, Z.; Zhang, M.Q.; Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002, 16, 2733–2742. [Google Scholar] [CrossRef]
- Dang, Y.; Yang, Q.; Xue, Z.; Liu, Y. RNA interference in fungi: Pathways, functions, and applications. Eukaryot. Cell 2011, 10, 1148–1155. [Google Scholar] [CrossRef]
- Meng, H.; Wang, Z.; Wang, Y.; Zhu, H.; Huang, B. Dicer and Argonaute Genes Involved in RNA Interference in the Entomopathogenic Fungus Metarhizium robertsii. Appl. Environ. Microbiol. 2017, 83, e03230. [Google Scholar] [CrossRef]
- Feng, H.; Xu, M.; Liu, Y.; Dong, R.; Gao, X.; Huang, L. Dicer-Like Genes Are Required for H2O2 and KCl Stress Responses, Pathogenicity and Small RNA Generation in Valsa mali. Front. Microbiol. 2017, 8, 1166. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; An, B.; Hou, X.; Guo, Y.; Luo, H.; He, C. Dicer-like Proteins Regulate the Growth, Conidiation, and Pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Front. Microbiol. 2018, 8, 2621. [Google Scholar] [CrossRef] [PubMed]
- Romano, N.; Macino, G. Quelling: Transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992, 6, 3343–3353. [Google Scholar] [CrossRef]
- Wang, L.; Xu, X.; Yang, J.; Chen, L.; Liu, B.; Liu, T.; Jin, Q. Integrated microRNA and mRNA analysis in the pathogenic filamentous fungus Trichophyton rubrum. Bmc Genom. 2018, 19, 933. [Google Scholar] [CrossRef]
- Derbyshire, M.; Mbengue, M.; Barascud, M.; Navaud, O.; Raffaele, S. Small RNAs from the plant pathogenic fungus Sclerotinia sclerotiorum highlight host candidate genes associated with quantitative disease resistance. Mol. Plant Pathol. 2019, 20, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Ji, H.M.; Gao, Y.; Cao, X.X.; Mao, H.Y.; Ouyang, S.Q.; Liu, P. An integrated analysis of mRNA and sRNA transcriptional profiles in tomato root: Insights on tomato wilt disease. PLoS ONE 2018, 13, e0206765. [Google Scholar] [CrossRef] [PubMed]
- Hunt, M.; Banerjee, S.; Surana, P.; Liu, M.; Fuerst, G.; Mathioni, S.; Meyers, B.C.; Nettleton, D.; Wise, R.P. Small RNA discovery in the interaction between barley and the powdery mildew pathogen. BMC Genom. 2019, 20, 610. [Google Scholar]
- Feng, H.; Xu, M.; Zheng, X.; Zhu, T.; Gao, X.; Huang, L. microRNAs and Their Targets in Apple (Malus domestica cv. “Fuji”) Involved in Response to Infection of Pathogen Valsa mali. Front. Plant Sci. 2017, 8, 2081. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Ma, X.; Tang, Z.; Qin, J.; Meng, Y. The use of high-throughput sequencing methods for plant microRNA research. RNA Biol. 2015, 12, 709–719. [Google Scholar] [CrossRef]
- Gong, L.; Tan, H.; Chen, F.; Li, T.; Zhu, J.; Jian, Q.; Yuan, D.; Xu, L.; Hu, W.; Jiang, Y.; et al. Novel synthesized 2, 4-DAPG analogues: Antifungal activity, mechanism and toxicology. Sci. Rep. 2016, 6, 32266. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, L.; Hu, Q.; Zhang, B.; Wu, W.; Jin, F.; Jiang, J. Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci. BMC Biotechnol. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Visser, M.; Gordon, T.R.; Wingfield, B.D.; Wingfield, M.J.; Viljoen, A. Transformation of Fusarium oxysporum f. sp cubense, causal agent of Fusarium wilt of banana, with the green fluorescent protein (GFP) gene. Australas Plant Pathol 2004, 33, 69–75. [Google Scholar] [CrossRef]
- Proctor, R.H.; Desjardins, A.E.; Plattner, R.D.; Hohn, T.M. A polyketide synthase gene required for biosynthesis of fumonisin mycotoxins in Gibberella fujikuroi slating population A. Fungal Genet. Biol. 1999, 27, 100–112. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An Integrative microRNA Evolutionary Analysis Platform for Next-generation Sequencing Experiments. BMC Bioinform. 2010, 13, 140. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef]
- Denman, R.B. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques 1993, 15, 1090–1095. [Google Scholar]
- Dai, X.B.; Zhuang, Z.H.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2–ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef]
- Machado, A.K.; Brown, N.A.; Urban, M.; Kanyuka, K.; Hammond-Kosack, K.E. RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag. Sci. 2018, 74, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ye, T.; Gao, Z. Cloning and functional analysis of succinate dehydrogenase gene PsSDHA in Phytophthora sojae. Microb. Pathog. 2017, 108, 40–48. [Google Scholar] [CrossRef]
- Enayati, S.; Azizi, M.; Aminollahi, E.; Ranjvar Shahrivar, M.; Khalaj, V. T7-RNA polymerase dependent RNAi system in Aspergillus fumigatus: A proof of concept study. FEMS Microbiol. Lett. 2016, 363, fnw029. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef]
- Wang, Q.; Zhuang, X.; Mu, J.; Deng, Z.B.; Jiang, H.; Zhang, L.; Xiang, X.; Wang, B.; Yan, J.; Miller, D.; et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013, 4, 1867. [Google Scholar] [CrossRef]
- Zhang, C.; Montgomery, T.A.; Fischer, S.E.; Garcia, S.M.; Riedel, C.G.; Fahlgren, N.; Sullivan, C.M.; Carrington, J.C.; Ruvkun, G. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr. Biol. 2012, 22, 881–890. [Google Scholar] [CrossRef]
- Song, X.S.; Gu, K.X.; Duan, X.X.; Xiao, X.M.; Hou, Y.P.; Duan, Y.B.; Wang, J.X.; Yu, N.; Zhou, M.G. Secondary amplification of siRNA machinery limits the application of spray-induced gene silencing. Mol. Plant Pathol. 2018, 19, 2543–2560. [Google Scholar] [CrossRef]
- Wang, M.; Thomas, N.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Curr. Opin. Plant Biol. 2017, 38, 133–141. [Google Scholar] [CrossRef]
- Zeng, J.; Gupta, V.K.; Jiang, Y.; Yang, B.; Gong, L.; Zhu, H. Cross-Kingdom Small RNAs Among Animals, Plants and Microbes. Cells 2019, 8, 371. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Dellota, E.J.; Yamane, D.; Jin, H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017, 14, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.; Song, N.; Zhao, M.; Liu, R.; Feng, H.; Wang, X.; Kang, Z. Puccinia striiformis f. sp tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol 2017, 215, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Kettles, G.J.; Hofinger, B.J.; Hu, P.; Bayon, C.; Rudd, J.J.; Balmer, D.; Courbot, M.; Hammond-Kosack, K.E.; Scalliet, G.; Kanyuka, K. sRNA Profiling Combined With Gene Function Analysis Reveals a Lack of Evidence for Cross-Kingdom RNAi in the Wheat - Zymoseptoria tritici Pathosystem. Front. Plant Sci. 2019, 10, 892. [Google Scholar] [CrossRef] [PubMed]
- Raman, V.; Simon, S.A.; Demirci, F.; Nakano, M.; Meyers, B.C.; Donofrio, N.M. Small RNA Functions Are Required for Growth and Development of Magnaporthe oryzae. Mol. Plant Microbe Interact. 2017, 30, 517–530. [Google Scholar] [CrossRef]
- Lau, S.K.; Chow, W.N.; Wong, A.Y.; Yeung, J.M.; Bao, J.; Zhang, N.; Lok, S.; Woo, P.C.; Yuen, K.Y. Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl. Trop. Dis. 2013, 7, e2398. [Google Scholar] [CrossRef]
- Dahlmann, T.A.; Kück, U. Dicer-Dependent Biogenesis of Small RNAs and Evidence for MicroRNA-Like RNAs in the Penicillin Producing Fungus Penicillium chrysogenum. PLoS ONE 2015, 10, e0125989. [Google Scholar] [CrossRef]
- Dubey, H.; Kiran, K.; Jaswal, R.; Jain, P.; Kayastha, A.M.; Bhardwaj, S.C.; Mondal, T.K.; Sharma, T.R. Discovery and profiling of small RNAs from Puccinia triticina by deep sequencing and identification of their potential targets in wheat. Funct. Integr. Genom. 2019, 19, 391–407. [Google Scholar] [CrossRef]
- Kang, K.; Zhong, J.; Jiang, L.; Liu, G.; Gou, C.Y.; Wu, Q.; Wang, Y.; Luo, J.; Gou, D. Identification of microRNA-Like RNAs in the filamentous fungus Trichoderma reesei by solexa sequencing. PLoS ONE 2013, 8, e76288. [Google Scholar] [CrossRef]
- Halter, T.; Imkampe, J.; Mazzotta, S.; Wierzba, M.; Postel, S.; Bücherl, C.; Kiefer, C.; Stahl, M.; Chinchilla, D.; Wang, X.; et al. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 2014, 24, 134–143. [Google Scholar] [CrossRef]
- Figueiredo, A.; Monteiro, F.; Sebastiana, M. Subtilisin-like proteases in plant-pathogen recognition and immune priming: A perspective. Front. Plant Sci. 2014, 5, 739. [Google Scholar] [CrossRef]

| Mature_ID | Mature_Seq | Precursor Position | MFE | Target Accession | Target Annotation | Alignment |
|---|---|---|---|---|---|---|
| Pit-novel1 | ugccaaaguaguuggacucgcu | contig333:14656..14796:+ | −50.4 | Cs7g21420 Cs7g21400 | Leucine-rich repeat (LRR) receptor kinase MDIS1-interacting receptor like kinase 2 |  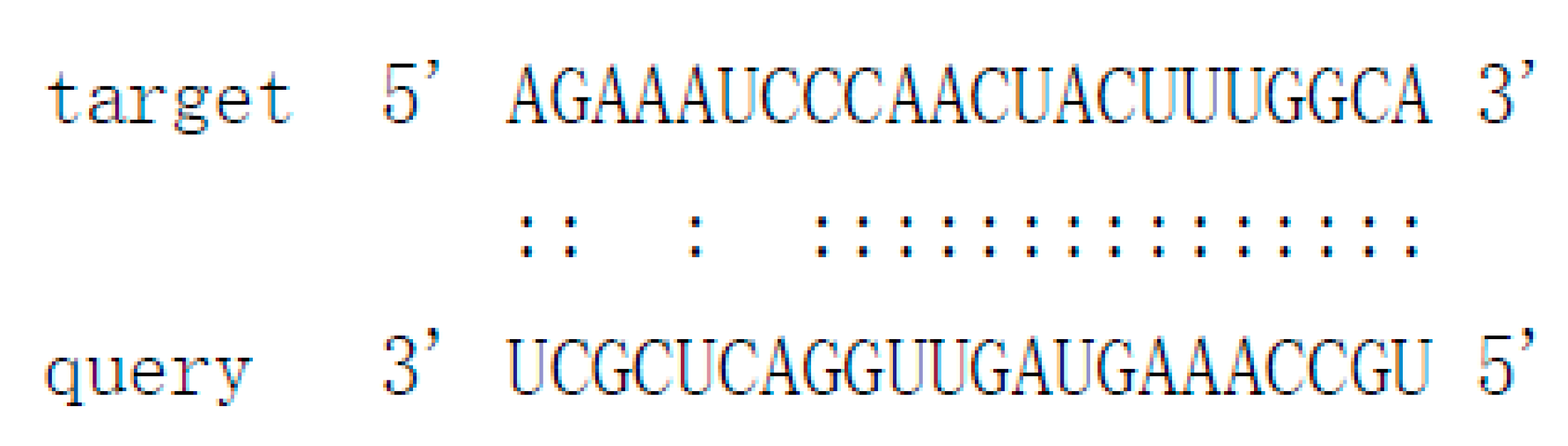 |
| Pit-novel2 | uaauugaucccgcuugcucccacg | contig2630:133598..133733:+ | −75.9 | N/A | N/A | |
| Pit-novel3 | uauggugacaaaaggcuucauu | contig1137:40522..40624:- | −49.8 | N/A | N/A | |
| Pit-novel6 | uacguagcagcgauccucuagc | contig837:270..404:+ | −43.7 | N/A | N/A | |
| Pit-novel7 | uggcuggagcaugcgcuugauu | contig1283:115664..115928:- | −90.6 | Cs7g19510 | AP2/B3-like transcriptional factor | 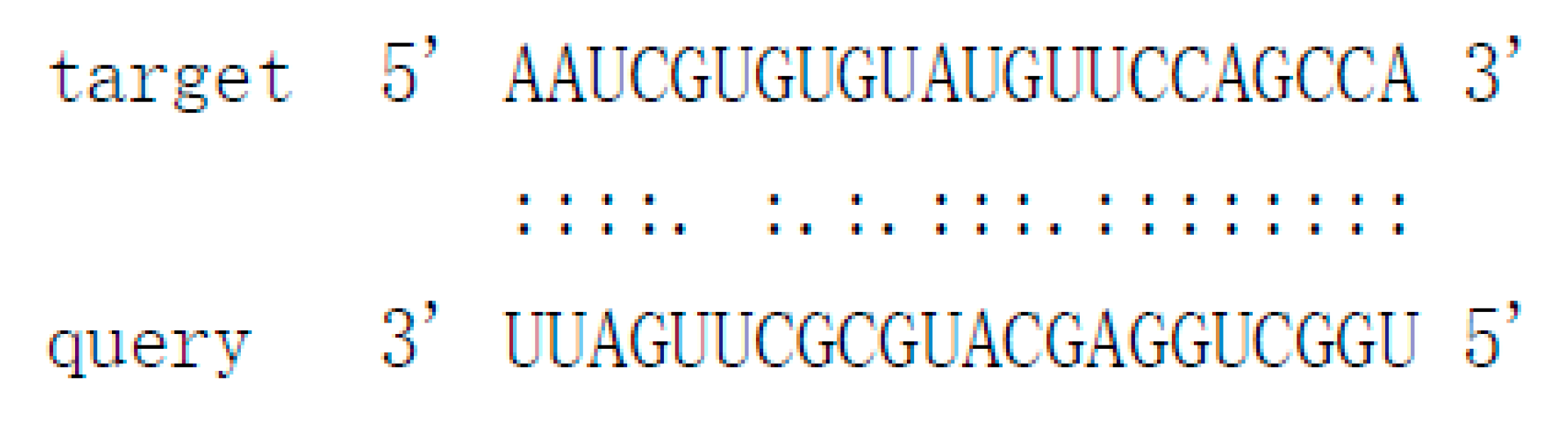 |
| Pit-novel10 | ugggaccuccgaaguauucgg | contig80:152579..152678:+ | −61.14 | N/A | N/A | |
| Pit-novel11 | uuugggugauuuucaggcuc | contig988:35989..36189:+ | −77.72 | Cs4g07930 | Subtilisin-like protease | 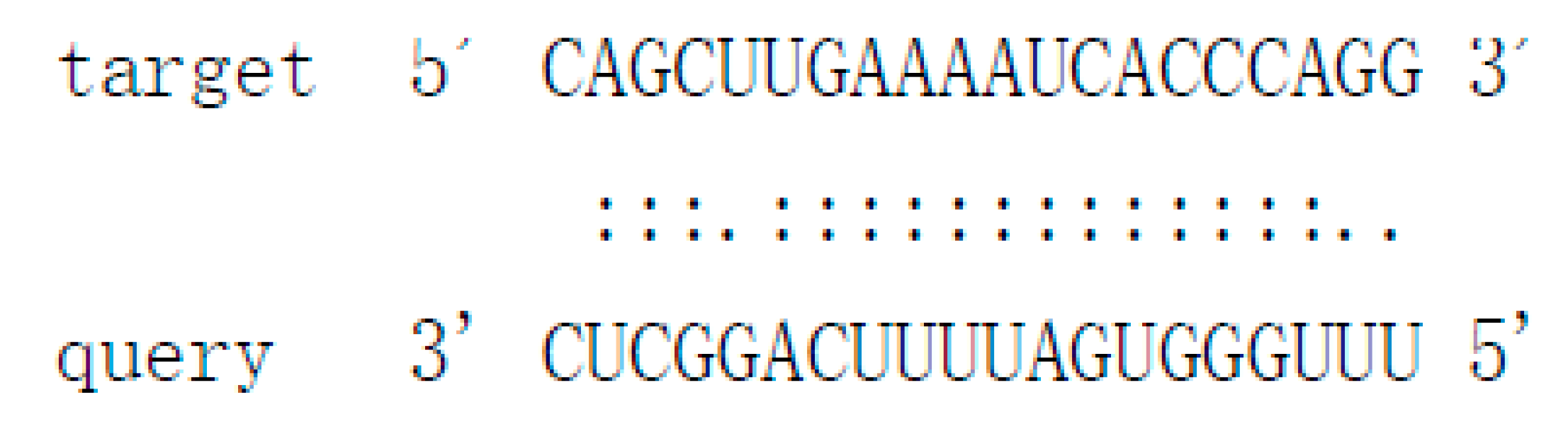 |
| Pit-novel12 | uacuucggaggucccacugu | contig2212:6256..6357:- | −64.1 | N/A | N/A | |
| Pit-novel14 | ugaguaggagagucauuugcu | contig769:62932..63030:+ | −51.3 | Cs7g09590 | Protein-tyrosine-phosphatase MKP1-like isoform X1 | 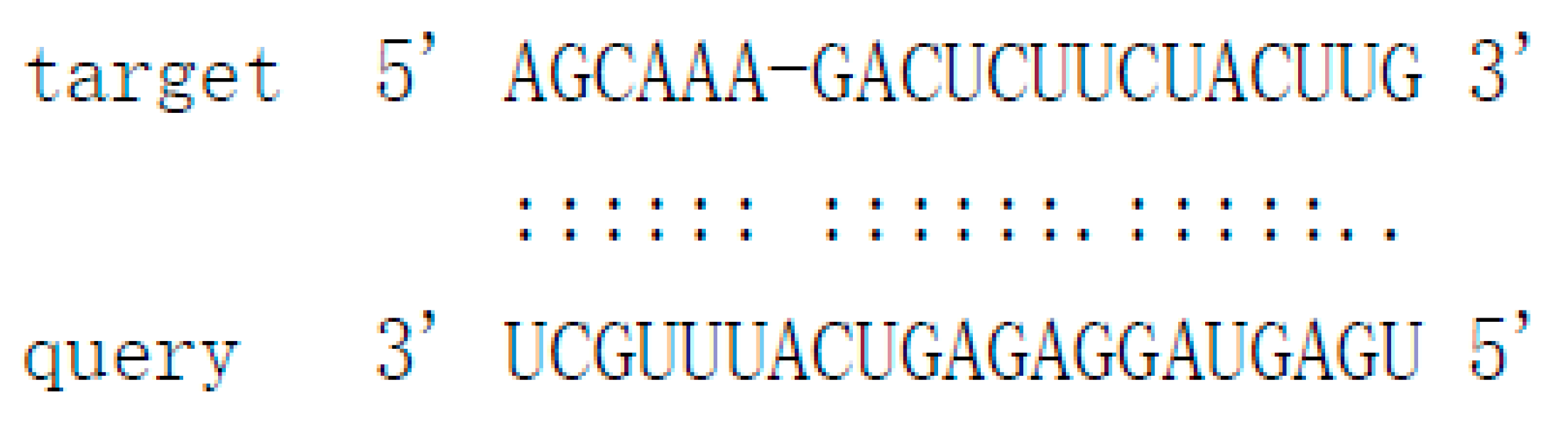 |
| Pit-novel16 | uaugacgauacugcuccauac | contig1182:333..382:+ | −17.1 | N/A | N/A | |
| Pit-novel21 | uggugacaaaaggcuucauuuu | contig50:167259..167366:- | −44.9 | N/A | N/A | |
| Pit-novel24 | uacuucggaggucccacuguacga | contig840:52392..52496:+ | −65.4 | N/A | N/A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Zhu, H.; Jiang, Y.; Shan, Y.; Gong, L. Silencing Dicer-Like Genes Reduces Virulence and sRNA Generation in Penicillium italicum, the Cause of Citrus Blue Mold. Cells 2020, 9, 363. https://doi.org/10.3390/cells9020363
Yin C, Zhu H, Jiang Y, Shan Y, Gong L. Silencing Dicer-Like Genes Reduces Virulence and sRNA Generation in Penicillium italicum, the Cause of Citrus Blue Mold. Cells. 2020; 9(2):363. https://doi.org/10.3390/cells9020363
Chicago/Turabian StyleYin, Chunxiao, Hong Zhu, Yueming Jiang, Yang Shan, and Liang Gong. 2020. "Silencing Dicer-Like Genes Reduces Virulence and sRNA Generation in Penicillium italicum, the Cause of Citrus Blue Mold" Cells 9, no. 2: 363. https://doi.org/10.3390/cells9020363
APA StyleYin, C., Zhu, H., Jiang, Y., Shan, Y., & Gong, L. (2020). Silencing Dicer-Like Genes Reduces Virulence and sRNA Generation in Penicillium italicum, the Cause of Citrus Blue Mold. Cells, 9(2), 363. https://doi.org/10.3390/cells9020363







