Granulins Regulate Aging Kinetics in the Adult Zebrafish Telencephalon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. DNA Extraction and Genotyping
2.3. Tissue Preparation and Immunohistochemistry
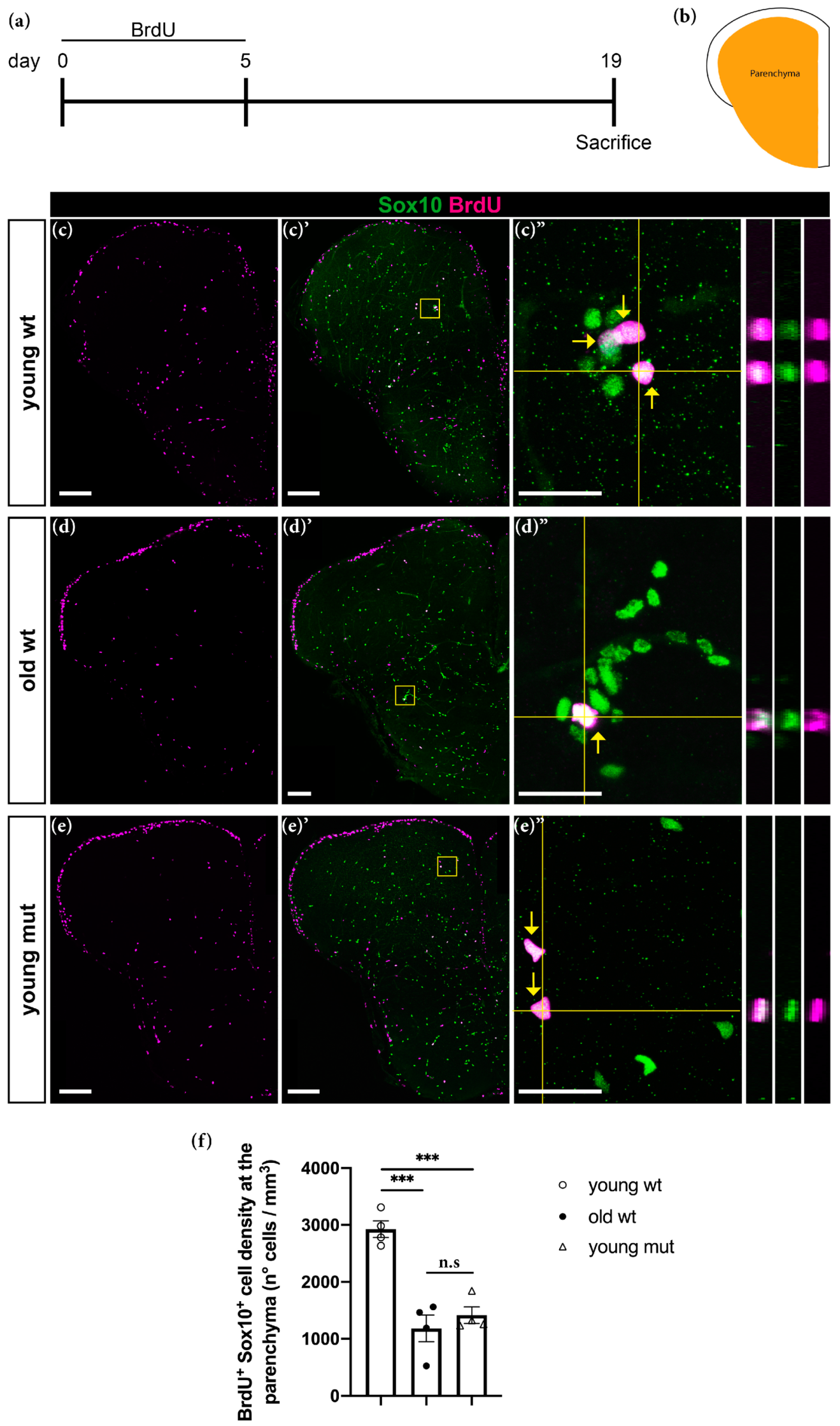
2.4. Bromodeoxyuridine (BrdU) Labelling Experiments
2.5. Image Acquisition and Processing
2.6. Quantitative Analysis and Statistical Tests
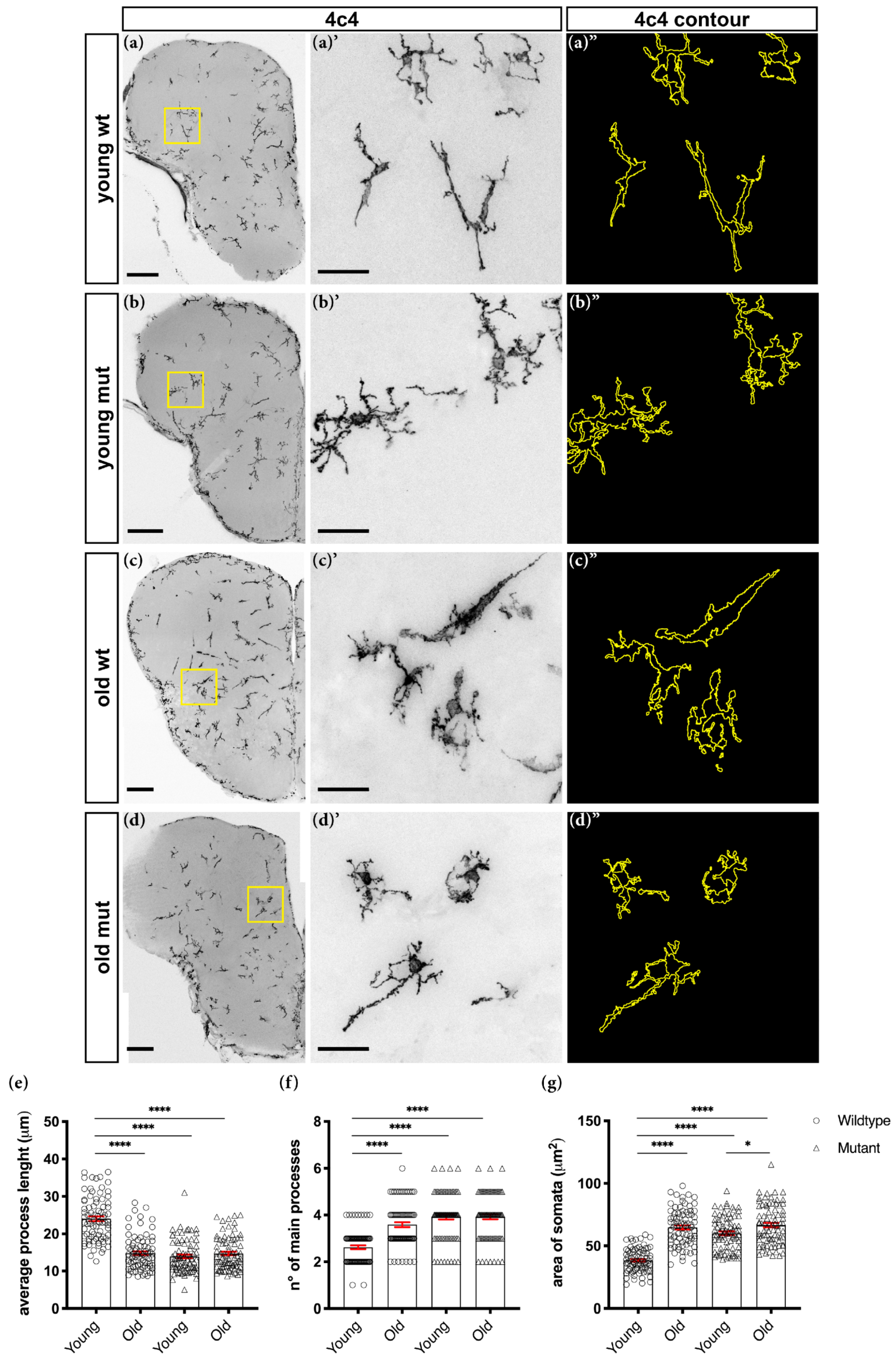
2.7. Morphological Analysis of Microglial Cells
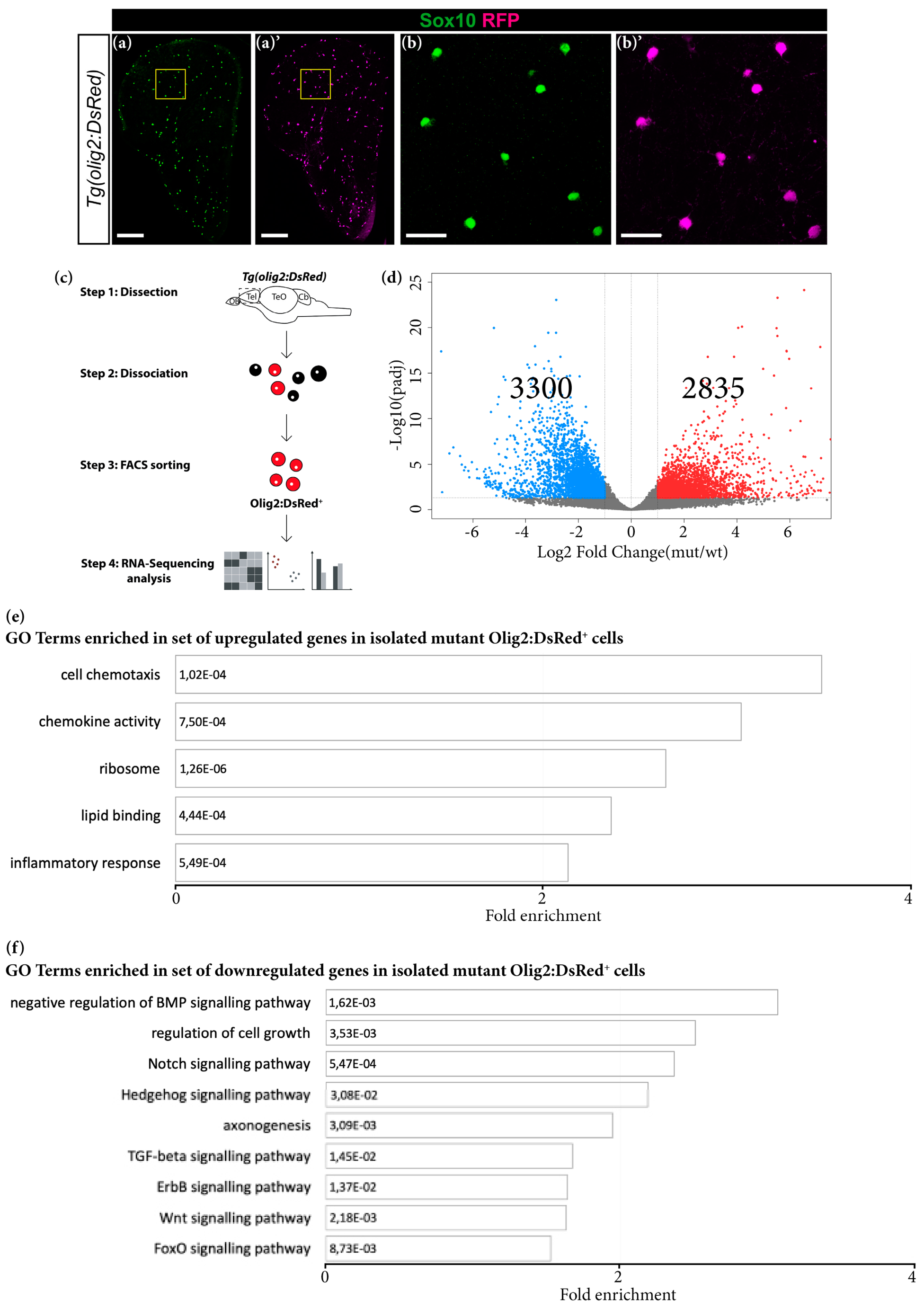
2.8. Fluorescence-Activated Cell Sorting (FACS) Isolation Method
2.9. Libraries for Deep Sequencing of Whole Telencephali and FACS Sorted Cells
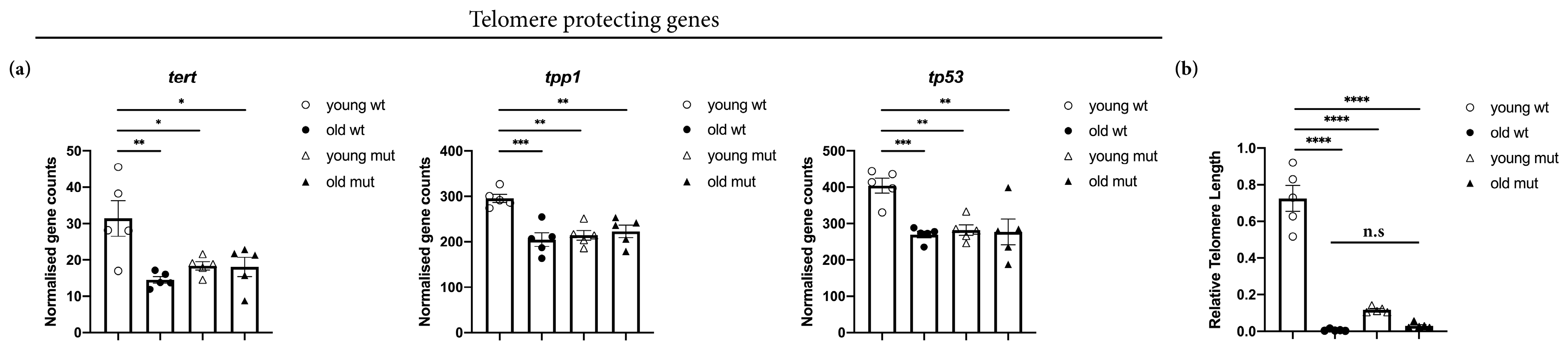
2.10. Quantification of Relative Telomere Length
3. Results
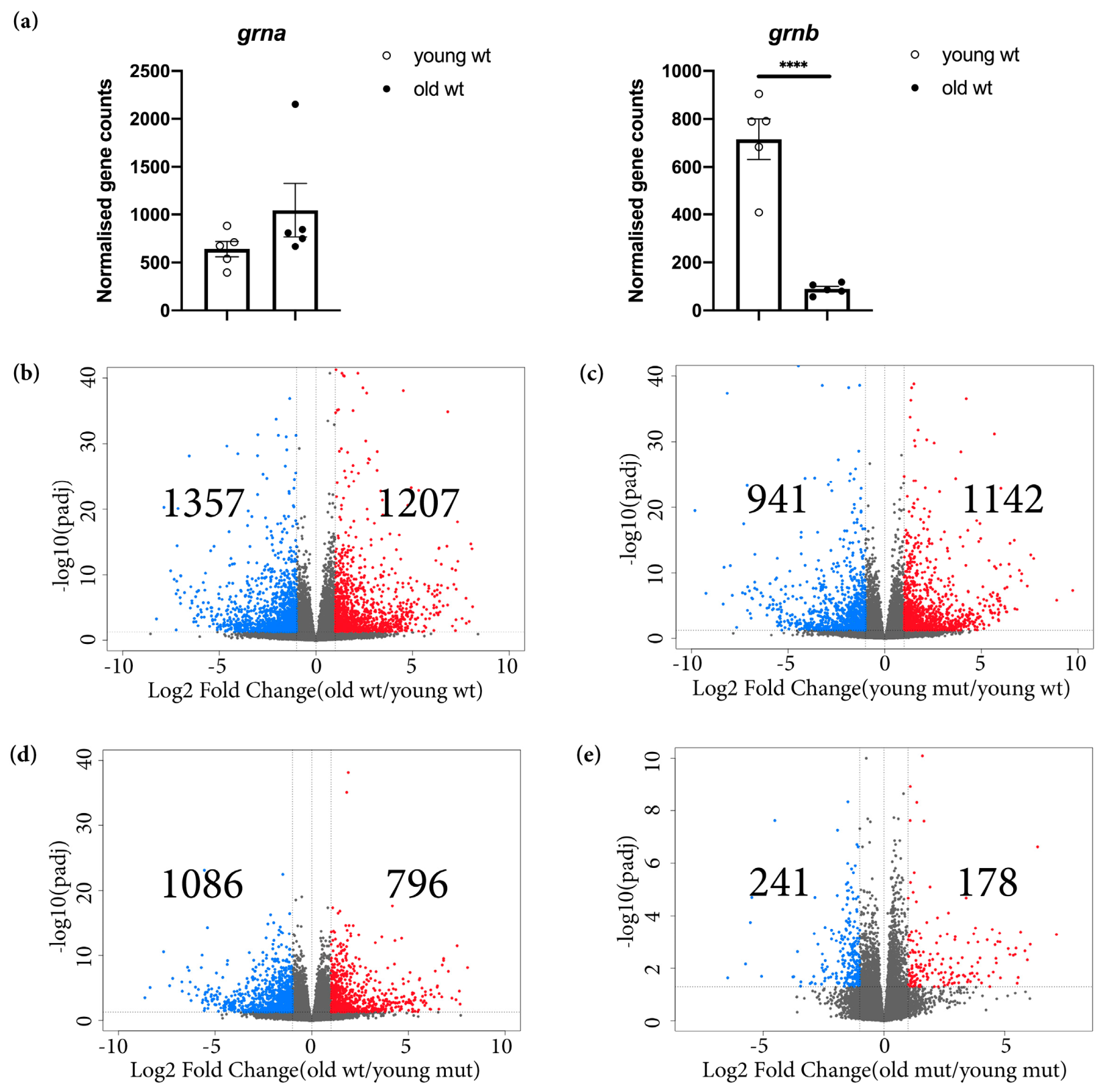
3.1. Grna and Grnb Deficiency Leads to a Pro-Inflammatory Transcriptional Signature in Microglial Cells in the Adult Zebrafish Telencephalon
3.2. Loss of Grna; Grnb Function Induces Premature Microglial Aging
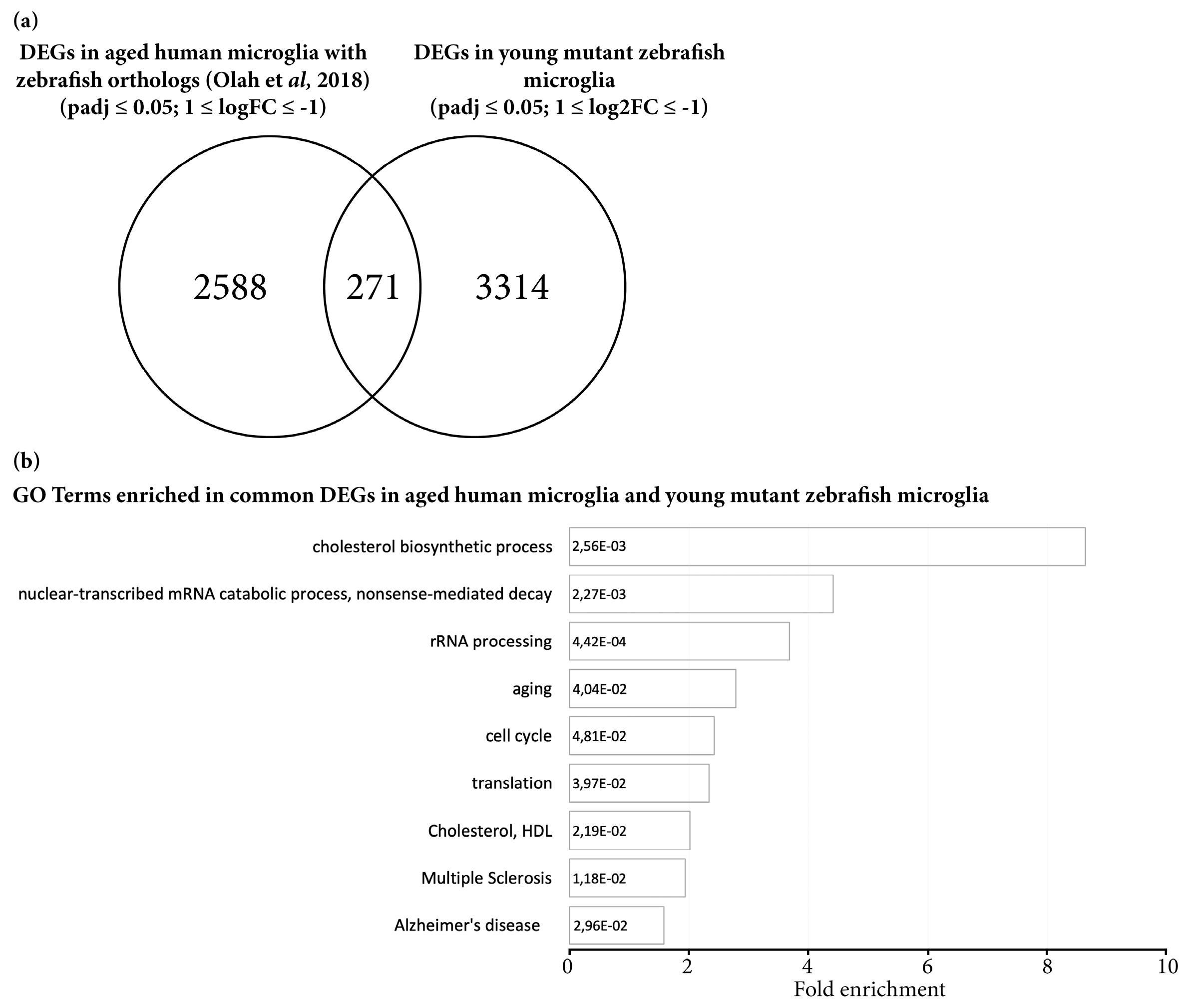
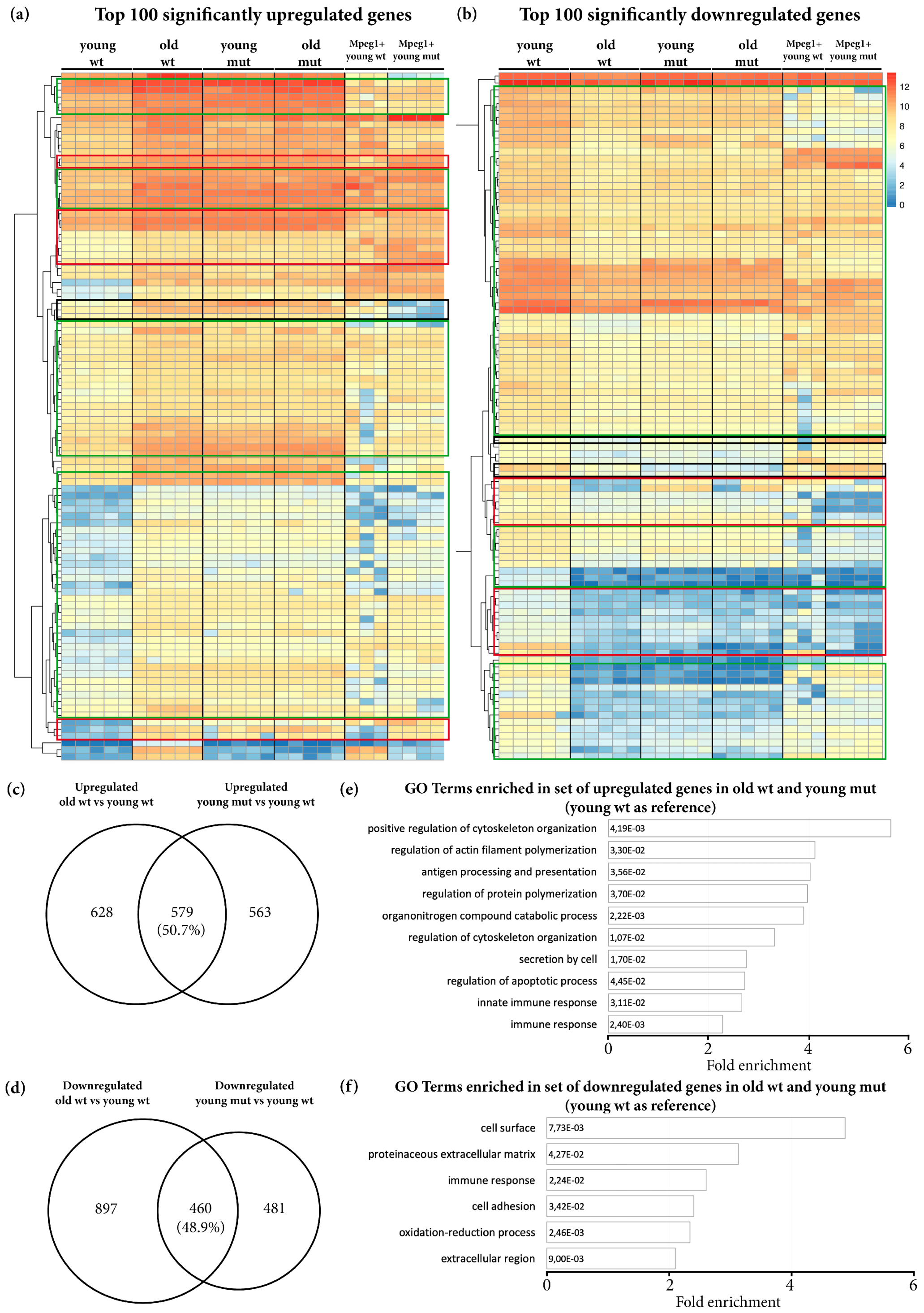
3.3. Transcriptome Analysis Reveals Premature Aging of Grna; Grnb-Deficient Brains
3.4. Grna and Grnb Deficiency Causes a Reduction in Neurogenesis in the Ventricular Zone of the Zebrafish Telencephalon, thus Mimicking the Phenotype of Reduction Observed during Aging
3.5. Grna and Grnb Deficiency Leads to Impaired Oligodendrocyte Precursor Cells Differentiation
3.6. Grna and Grnb Deficiency Decreases of Telomere Length
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NF-kB | Nuclear factor kappa B |
| wt | Wildtype |
| mut | Mutant |
| grna | Granulin a |
| grnb | Granulin b |
| mpeg1 | Macrophage expressed 1 |
| MAPK | Mitogen-activated protein kinase |
| JAK/STAT | Janus kinase/signal transducer and activator of transcription |
| PPAR | Peroxisome proliferator-activated receptor |
| il1b | Interleukin 1 beta |
| il6 | Interleukin 6 |
| tnfa | Tumor necrosis factor alpha |
| il12bβ | Interleukin 12b beta |
| Cxcl8a | Chemokine (C-X-C motif) ligand 8a |
| tgfb3 | Transforming growth factor beta 3 |
| igf1 | Insulin-like growth factor 1 |
| il10 | Interleukin 10 |
| il4 | Interleukin 4 |
| stat6 | Signal transducer and activator of transcription 6 |
| VZ | Ventricular zone |
| SVZ | Subventricular zone |
| sox10 | SRY-box transcription Factor 10 |
| olig2 | Oligodendrocyte lineage transcription factor 2 |
| BMP | Bone morphogenetic protein |
| TGF-b | Transforming growth factor-beta |
| ErbB | EGFR family signalling pathway |
| tert | Telomerase reverse transcriptase |
| tp53 | Tumor protein p53 |
| tpp1 | Tripeptidyl peptidase 1 |
References
- Mattson, M.P.; Arumugam, T.V. Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, R.; Koch, H.; Doll, B.A.; Tegtmeier, F.; Einhorn, T.A.; Hollinger, J.O. Fracture healing in the elderly patient. Exp. Gerontol. 2006, 41, 1080–1093. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.L.; Kohlmaier, A.; Polesello, C.; Veelken, C.; Edgar, B.A.; Tapon, N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 2010, 137, 4147–4158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conboy, I.M.; Rando, T.A. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle 2012, 11, 2260–2267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molofsky, A.V.; Slutsky, S.G.; Joseph, N.M.; He, S.; Pardal, R.; Krishnamurthy, J.; Sharpless, N.E.; Morrison, S.J. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 2006, 443, 448–452. [Google Scholar] [CrossRef] [Green Version]
- Bast, L.; Calzolari, F.; Strasser, M.K.; Hasenauer, J.; Theis, F.J.; Ninkovic, J.; Marr, C. Increasing Neural Stem Cell Division Asymmetry and Quiescence Are Predicted to Contribute to the Age-Related Decline in Neurogenesis. Cell Rep. 2018, 25, 3231–3240.e8. [Google Scholar] [CrossRef] [Green Version]
- Lupo, G.; Gioia, R.; Nisi, P.S.; Biagioni, S.; Cacci, E. Molecular Mechanisms of Neurogenic Aging in the Adult Mouse Subventricular Zone. J. Exp. Neurosci. 2019, 13, 1179069519829040. [Google Scholar] [CrossRef]
- Enwere, E.; Shingo, T.; Gregg, C.; Fujikawa, H.; Ohta, S.; Weiss, S. Aging results in reduced epidermal growth factor receptor signaling, diminished olfactory neurogenesis, and deficits in fine olfactory discrimination. J. Neurosci. 2004, 24, 8354–8365. [Google Scholar] [CrossRef] [Green Version]
- Daynac, M.; Morizur, L.; Chicheportiche, A.; Mouthon, M.A.; Boussin, F.D. Age-related neurogenesis decline in the subventricular zone is associated with specific cell cycle regulation changes in activated neural stem cells. Sci. Rep. 2016, 6, 21505. [Google Scholar] [CrossRef] [Green Version]
- Silva-Vargas, V.; Maldonado-Soto, A.R.; Mizrak, D.; Codega, P.; Doetsch, F. Age-Dependent Niche Signals from the Choroid Plexus Regulate Adult Neural Stem Cells. Cell Stem Cell 2016, 19, 643–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baruch, K.; Deczkowska, A.; David, E.; Castellano, J.M.; Miller, O.; Kertser, A.; Berkutzki, T.; Barnett-Itzhaki, Z.; Bezalel, D.; Wyss-Coray, T.; et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 2014, 346, 89–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepko, T.; Pusch, M.; Muller, T.; Schulte, D.; Ehses, J.; Kiebler, M.; Hasler, J.; Huttner, H.B.; Vandenbroucke, R.E.; Vandendriessche, C.; et al. Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain. EMBO J. 2019, 38, e100481. [Google Scholar] [CrossRef] [PubMed]
- Lupo, G.; Nisi, P.S.; Esteve, P.; Paul, Y.L.; Novo, C.L.; Sidders, B.; Khan, M.A.; Biagioni, S.; Liu, H.K.; Bovolenta, P.; et al. Molecular profiling of aged neural progenitors identifies Dbx2 as a candidate regulator of age-associated neurogenic decline. Aging Cell 2018, 17, e12745. [Google Scholar] [CrossRef]
- Apostolopoulou, M.; Kiehl, T.R.; Winter, M.; Cardenas De La Hoz, E.; Boles, N.C.; Bjornsson, C.S.; Zuloaga, K.L.; Goderie, S.K.; Wang, Y.; Cohen, A.R.; et al. Non-monotonic Changes in Progenitor Cell Behavior and Gene Expression during Aging of the Adult V-SVZ Neural Stem Cell Niche. Stem Cell Rep. 2017, 9, 1931–1947. [Google Scholar] [CrossRef] [Green Version]
- Leeman, D.S.; Hebestreit, K.; Ruetz, T.; Webb, A.E.; McKay, A.; Pollina, E.A.; Dulken, B.W.; Zhao, X.; Yeo, R.W.; Ho, T.T.; et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 2018, 359, 1277–1283. [Google Scholar] [CrossRef] [Green Version]
- Piccin, D.; Tufford, A.; Morshead, C.M. Neural stem and progenitor cells in the aged subependyma are activated by the young niche. Neurobiol. Aging 2014, 35, 1669–1679. [Google Scholar] [CrossRef]
- Villeda, S.A.; Luo, J.; Mosher, K.I.; Zou, B.; Britschgi, M.; Bieri, G.; Stan, T.M.; Fainberg, N.; Ding, Z.; Eggel, A.; et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 2011, 477, 90–94. [Google Scholar] [CrossRef] [Green Version]
- Butovsky, O.; Ziv, Y.; Schwartz, A.; Landa, G.; Talpalar, A.E.; Pluchino, S.; Martino, G.; Schwartz, M. Microglia activated by IL-4 or IFN-gamma differentially induce neurogenesis and oligodendrogenesis from adult stem/progenitor cells. Mol. Cell. Neurosci. 2006, 31, 149–160. [Google Scholar] [CrossRef]
- Wong, W.T. Microglial aging in the healthy CNS: Phenotypes, drivers, and rejuvenation. Front. Cell. Neurosci. 2013, 7, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziv, Y.; Ron, N.; Butovsky, O.; Landa, G.; Sudai, E.; Greenberg, N.; Cohen, H.; Kipnis, J.; Schwartz, M. Immune cells contribute to the maintenance of neurogenesis and spatial learning abilities in adulthood. Nat. Neurosci. 2006, 9, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Ekdahl, C.T.; Kokaia, Z.; Lindvall, O. Brain inflammation and adult neurogenesis: The dual role of microglia. Neuroscience 2009, 158, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Inflammaging: Disturbed interplay between autophagy and inflammasomes. Aging 2012, 4, 166–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Li, J.; Purkayastha, S.; Tang, Y.; Zhang, H.; Yin, Y.; Li, B.; Liu, G.; Cai, D. Hypothalamic programming of systemic ageing involving IKK-beta, NF-kappaB and GnRH. Nature 2013, 497, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Pont, A.R.; Sadri, N.; Hsiao, S.J.; Smith, S.; Schneider, R.J. mRNA decay factor AUF1 maintains normal aging, telomere maintenance, and suppression of senescence by activation of telomerase transcription. Mol. Cell 2012, 47, 5–15. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Bateman, A. Progranulin (granulin-epithelin precursor, PC-cell-derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J. Mol. Med. (Berl.) 2003, 81, 600–612. [Google Scholar] [CrossRef]
- Petkau, T.L.; Neal, S.J.; Orban, P.C.; MacDonald, J.L.; Hill, A.M.; Lu, G.; Feldman, H.H.; Mackenzie, I.R.; Leavitt, B.R. Progranulin expression in the developing and adult murine brain. J. Comp. Neurol. 2010, 518, 3931–3947. [Google Scholar] [CrossRef]
- Cenik, B.; Sephton, C.F.; Kutluk Cenik, B.; Herz, J.; Yu, G. Progranulin: A proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J. Biol. Chem. 2012, 287, 32298–32306. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Nathan, C.; Jin, W.; Sim, D.; Ashcroft, G.S.; Wahl, S.M.; Lacomis, L.; Erdjument-Bromage, H.; Tempst, P.; Wright, C.D.; et al. Conversion of proepithelin to epithelins: Roles of SLPI and elastase in host defense and wound repair. Cell 2002, 111, 867–878. [Google Scholar] [CrossRef]
- Jian, J.; Konopka, J.; Liu, C. Insights into the role of progranulin in immunity, infection, and inflammation. J. Leukoc. Biol. 2013, 93, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Wils, H.; Kleinberger, G.; Pereson, S.; Janssens, J.; Capell, A.; Van Dam, D.; Cuijt, I.; Joris, G.; De Deyn, P.P.; Haass, C.; et al. Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J. Pathol. 2012, 228, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Valenzano, D.R.; Benayoun, B.A.; Singh, P.P.; Zhang, E.; Etter, P.D.; Hu, C.K.; Clément-Ziza, M.; Willemsen, D.; Cui, R.; Harel, I.; et al. The African Turquoise Killifish Genome Provides Insights into Evolution and Genetic Architecture of Lifespan. Cell 2015, 163, 1539–1554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhinn, H.; Abeliovich, A. Differential Aging Analysis in Human Cerebral Cortex Identifies Variants in TMEM106B and GRN that Regulate Aging Phenotypes. Cell Syst. 2017, 4, 404.e5–415.e5. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Mackenzie, I.R.; Pickering-Brown, S.M.; Gass, J.; Rademakers, R.; Lindholm, C.; Snowden, J.; Adamson, J.; Sadovnick, A.D.; Rollinson, S.; et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006, 442, 916–919. [Google Scholar] [CrossRef]
- Cruts, M.; Gijselinck, I.; van der Zee, J.; Engelborghs, S.; Wils, H.; Pirici, D.; Rademakers, R.; Vandenberghe, R.; Dermaut, B.; Martin, J.-J.; et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006, 442, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R.; Damiano, J.; Franceschetti, S.; Carpenter, S.; Canafoglia, L.; Morbin, M.; Rossi, G.; Pareyson, D.; Mole, S.E.; Staropoli, J.F.; et al. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am. J. Hum. Genet. 2012, 90, 1102–1107. [Google Scholar] [CrossRef] [Green Version]
- Almeida, M.R.; Macario, M.C.; Ramos, L.; Baldeiras, I.; Ribeiro, M.H.; Santana, I. Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol. Aging 2016, 41, 200.e1–200.e5. [Google Scholar] [CrossRef]
- Lui, H.; Zhang, J.; Makinson, S.R.; Cahill, M.K.; Kelley, K.W.; Huang, H.Y.; Shang, Y.; Oldham, M.C.; Martens, L.H.; Gao, F.; et al. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell 2016, 165, 921–935. [Google Scholar] [CrossRef] [Green Version]
- Götzl, J.K.; Brendel, M.; Werner, G.; Parhizkar, S.; Sebastian Monasor, L.; Kleinberger, G.; Colombo, A.V.; Deussing, M.; Wagner, M.; Winkelmann, J.; et al. Opposite microglial activation stages upon loss of PGRN or TREM2 result in reduced cerebral glucose metabolism. EMBO Mol. Med. 2019, 11. [Google Scholar] [CrossRef]
- Yin, F.; Banerjee, R.; Thomas, B.; Zhou, P.; Qian, L.; Jia, T.; Ma, X.; Ma, Y.; Iadecola, C.; Beal, M.F.; et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 2010, 207, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Martens, L.H.; Zhang, J.; Barmada, S.J.; Zhou, P.; Kamiya, S.; Sun, B.; Min, S.W.; Gan, L.; Finkbeiner, S.; Huang, E.J.; et al. Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J. Clin. Investig. 2012, 122, 3955–3959. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Matsuwaki, T.; Yamanouchi, K.; Nishihara, M. Exacerbated inflammatory responses related to activated microglia after traumatic brain injury in progranulin-deficient mice. Neuroscience 2013, 231, 49–60. [Google Scholar] [CrossRef]
- Altmann, C.; Vasic, V.; Hardt, S.; Heidler, J.; Häussler, A.; Wittig, I.; Schmidt, M.H.H.; Tegeder, I. Progranulin promotes peripheral nerve regeneration and reinnervation: Role of notch signaling. Mol. Neurodegener. 2016, 11, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toh, H.; Chitramuthu, B.P.; Bennett, H.P.J.; Bateman, A. Structure, function, and mechanism of progranulin; the brain and beyond. J. Mol. Neurosci. 2011, 45, 538–548. [Google Scholar] [CrossRef]
- Ryan, C.L.; Baranowski, D.C.; Chitramuthu, B.P.; Malik, S.; Li, Z.; Cao, M.; Minotti, S.; Durham, H.D.; Kay, D.G.; Shaw, C.A.; et al. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci. 2009, 10, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Damme, P.; Van Hoecke, A.; Lambrechts, D.; Vanacker, P.; Bogaert, E.; van Swieten, J.; Carmeliet, P.; Van Den Bosch, L.; Robberecht, W. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J. Cell Biol. 2008, 181, 37–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadieux, B.; Chitramuthu, B.P.; Baranowski, D.; Bennett, H.P.J. The zebrafish progranulin gene family and antisense transcripts. BMC Genomics 2005, 6, 156. [Google Scholar] [CrossRef] [Green Version]
- Carneiro, M.C.; de Castro, I.P.; Ferreira, M.G. Telomeres in aging and disease: Lessons from zebrafish. Dis. Model. Mech. 2016, 9, 737–748. [Google Scholar] [CrossRef] [Green Version]
- Van Houcke, J.; De Groef, L.; Dekeyster, E.; Moons, L. The zebrafish as a gerontology model in nervous system aging, disease, and repair. Ageing Res. Rev. 2015, 24, 358–368. [Google Scholar] [CrossRef]
- Cayuela, M.L.; Claes, K.B.M.; Ferreira, M.G.; Henriques, C.M.; van Eeden, F.; Varga, M.; Vierstraete, J.; Mione, M.C. The Zebrafish as an Emerging Model to Study DNA Damage in Aging, Cancer and Other Diseases. Front. Cell Dev. Biol. 2018, 6, 178. [Google Scholar] [CrossRef] [Green Version]
- Solchenberger, B.; Russell, C.; Kremmer, E.; Haass, C.; Schmid, B. Granulin knock out zebrafish lack frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis pathology. PLoS ONE 2015, 10, e0118956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Park, H.C.; Topczewska, J.M.; Mawdsley, D.J.; Appel, B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003, 25, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Bernut, A.; Herrmann, J.L.; Kissa, K.; Dubremetz, J.F.; Gaillard, J.L.; Lutfalla, G.; Kremer, L. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc. Natl. Acad. Sci. USA 2014, 111, E943–E952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Baumgart, E.V.; Barbosa, J.S.; Bally-Cuif, L.; Götz, M.; Ninkovic, J. Stab wound injury of the zebrafish telencephalon: A model for comparative analysis of reactive gliosis. Glia 2012, 60, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.; Gonzalez-Sanchez, R.; Di Giaimo, R.; Baumgardt Violette, E.; Theis, F.; Götz, M.; Ninkovic, J. Live imaging of adult neural stem cell behavior in the intact and injured zebrafish brain. Science 2015, 348, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Adolf, B.; Chapouton, P.; Lam, C.S.; Topp, S.; Tannhauser, B.; Strahle, U.; Gotz, M.; Bally-Cuif, L. Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev. Biol. 2006, 295, 278–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, J.; Beckervordersandforth, R.; Tripathi, P.; Steiner-Mezzadri, A.; Ninkovic, J.; Gotz, M. Prospective isolation of adult neural stem cells from the mouse subependymal zone. Nat. Protoc. 2011, 6, 1981–1989. [Google Scholar] [CrossRef]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.11–11.14.19. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arslan-Ergul, A.; Erbaba, B.; Karoglu, E.T.; Halim, D.O.; Adams, M.M. Short-term dietary restriction in old zebrafish changes cell senescence mechanisms. Neuroscience 2016, 334, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Cawthon, R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002, 30, e47. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Lim, M.X.; Png, C.W.; Tay, C.Y.; Teo, J.D.; Jiao, H.; Lehming, N.; Tan, K.S.; Zhang, Y. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect. Immun. 2014, 82, 4789–4801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perner, F.; Perner, C.; Ernst, T.; Heidel, F.H. Roles of JAK2 in Aging, Inflammation, Hematopoiesis and Malignant Transformation. Cells 2019, 8, 854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heming, M.; Gran, S.; Jauch, S.L.; Fischer-Riepe, L.; Russo, A.; Klotz, L.; Hermann, S.; Schafers, M.; Roth, J.; Barczyk-Kahlert, K. Peroxisome Proliferator-Activated Receptor-gamma Modulates the Response of Macrophages to Lipopolysaccharide and Glucocorticoids. Front. Immunol. 2018, 9, 893. [Google Scholar] [CrossRef]
- Becker, T.; Becker, C.G. Regenerating descending axons preferentially reroute to the gray matter in the presence of a general macrophage/microglial reaction caudal to a spinal transection in adult zebrafish. J. Comp. Neurol. 2001, 433, 131–147. [Google Scholar] [CrossRef]
- Olah, M.; Patrick, E.; Villani, A.C.; Xu, J.; White, C.C.; Ryan, K.J.; Piehowski, P.; Kapasi, A.; Nejad, P.; Cimpean, M.; et al. A transcriptomic atlas of aged human microglia. Nat. Commun. 2018, 9, 539. [Google Scholar] [CrossRef] [Green Version]
- Hart, A.D.; Wyttenbach, A.; Perry, V.H.; Teeling, J.L. Age related changes in microglial phenotype vary between CNS regions: Grey versus white matter differences. Brain Behav. Immun. 2012, 26, 754–765. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, M.; Groth, M.; Priebe, S.; Savino, A.; Testa, G.; Dix, A.; Ripa, R.; Spallotta, F.; Gaetano, C.; Ori, M.; et al. RNA-seq of the aging brain in the short-lived fish N. furzeri - conserved pathways and novel genes associated with neurogenesis. Aging Cell 2014, 13, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Evers, B.M.; Rodriguez-Navas, C.; Tesla, R.J.; Prange-Kiel, J.; Wasser, C.R.; Yoo, K.S.; McDonald, J.; Cenik, B.; Ravenscroft, T.A.; Plattner, F.; et al. Lipidomic and Transcriptomic Basis of Lysosomal Dysfunction in Progranulin Deficiency. Cell Rep. 2017, 20, 2565–2574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotzl, J.K.; Colombo, A.V.; Fellerer, K.; Reifschneider, A.; Werner, G.; Tahirovic, S.; Haass, C.; Capell, A. Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol. Neurodegener. 2018, 13, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattarai, P.; Thomas, A.K.; Cosacak, M.I.; Papadimitriou, C.; Mashkaryan, V.; Froc, C.; Reinhardt, S.; Kurth, T.; Dahl, A.; Zhang, Y.; et al. IL4/STAT6 Signaling Activates Neural Stem Cell Proliferation and Neurogenesis upon Amyloid-beta42 Aggregation in Adult Zebrafish Brain. Cell Rep. 2016, 17, 941–948. [Google Scholar] [CrossRef] [Green Version]
- Nolan, Y.; Maher, F.O.; Martin, D.S.; Clarke, R.M.; Brady, M.T.; Bolton, A.E.; Mills, K.H.; Lynch, M.A. Role of interleukin-4 in regulation of age-related inflammatory changes in the hippocampus. J. Biol. Chem. 2005, 280, 9354–9362. [Google Scholar] [CrossRef] [Green Version]
- Edelmann, K.; Glashauser, L.; Sprungala, S.; Hesl, B.; Fritschle, M.; Ninkovic, J.; Godinho, L.; Chapouton, P. Increased radial glia quiescence, decreased reactivation upon injury and unaltered neuroblast behavior underlie decreased neurogenesis in the aging zebrafish telencephalon. J. Comp. Neurol. 2013, 521, 3099–3115. [Google Scholar] [CrossRef]
- Barbosa, J.S.; Ninkovic, J. Adult neural stem cell behavior underlying constitutive and restorative neurogenesis in zebrafish. Neurogenesis (Austin) 2016, 3, e1148101. [Google Scholar] [CrossRef] [Green Version]
- März, M.; Schmidt, R.; Rastegar, S.; Strähle, U. Expression of the transcription factor Olig2 in proliferating cells in the adult zebrafish telencephalon. Dev. Dyn. 2010, 239, 3336–3349. [Google Scholar] [CrossRef]
- Wang, L.C.; Almazan, G. Role of Sonic Hedgehog Signaling in Oligodendrocyte Differentiation. Neurochem. Res. 2016, 41, 3289–3299. [Google Scholar] [CrossRef]
- Bartus, K.; Burnside, E.R.; Galino, J.; James, N.D.; Bennett, D.L.H.; Bradbury, E.J. ErbB receptor signaling directly controls oligodendrocyte progenitor cell transformation and spontaneous remyelination after spinal cord injury. Glia 2019, 67, 1036–1046. [Google Scholar] [CrossRef] [Green Version]
- Palazuelos, J.; Klingener, M.; Aguirre, A. TGFβ signaling regulates the timing of CNS myelination by modulating oligodendrocyte progenitor cell cycle exit through SMAD3/4/FoxO1/Sp1. J. Neurosci. 2014, 34, 7917–7930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Juárez, A.; Titus, H.E.; Silbak, S.H.; Pressler, J.W.; Rizvi, T.A.; Bogard, M.; Bennett, M.R.; Ciraolo, G.; Williams, M.T.; Vorhees, C.V.; et al. Oligodendrocyte Nf1 Controls Aberrant Notch Activation and Regulates Myelin Structure and Behavior. Cell Rep. 2017, 19, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Grinspan, J.B. Bone Morphogenetic Proteins: Inhibitors of Myelination in Development and Disease. Vitam. Horm. 2015, 99, 195–222. [Google Scholar]
- Whittemore, K.; Vera, E.; Martínez-Nevado, E.; Sanpera, C.; Blasco, M.A. Telomere shortening rate predicts species life span. Proc. Natl. Acad. Sci. USA 2019, 116, 15122–15127. [Google Scholar] [CrossRef] [Green Version]
- Kao, A.W.; McKay, A.; Singh, P.P.; Brunet, A.; Huang, E.J. Progranulin, lysosomal regulation and neurodegenerative disease. Nat. Rev. Neurosci. 2017, 18, 325–333. [Google Scholar] [CrossRef]
- Chitramuthu, B.P.; Bennett, H.P.J.; Bateman, A. Progranulin: A new avenue towards the understanding and treatment of neurodegenerative disease. Brain 2017, 140, 3081–3104. [Google Scholar] [CrossRef]
- Spittau, B. Aging Microglia-Phenotypes, Functions and Implications for Age-Related Neurodegenerative Diseases. Front. Aging Neurosci. 2017, 9, 194. [Google Scholar] [CrossRef]
- Von Bernhardi, R.; Eugenín-von Bernhardi, L.; Eugenín, J. Microglial cell dysregulation in brain aging and neurodegeneration. Front. Aging Neurosci. 2015, 7, 124. [Google Scholar] [CrossRef] [Green Version]
- Gabuzda, D.; Yankner, B.A. Physiology: Inflammation links ageing to the brain. Nature 2013, 497, 197–198. [Google Scholar] [CrossRef] [Green Version]
- Kizil, C.; Dudczig, S.; Kyritsis, N.; Machate, A.; Blaesche, J.; Kroehne, V.; Brand, M. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012, 7, 27. [Google Scholar] [CrossRef] [Green Version]
- Kizil, C.; Kyritsis, N.; Dudczig, S.; Kroehne, V.; Freudenreich, D.; Kaslin, J.; Brand, M. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev. Cell 2012, 23, 1230–1237. [Google Scholar] [CrossRef] [Green Version]
- Kyritsis, N.; Kizil, C.; Zocher, S.; Kroehne, V.; Kaslin, J.; Freudenreich, D.; Iltzsche, A.; Brand, M. Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 2012, 338, 1353–1356. [Google Scholar] [CrossRef]
- Miyamoto, N.; Pham, L.D.; Hayakawa, K.; Matsuzaki, T.; Seo, J.H.; Magnain, C.; Ayata, C.; Kim, K.W.; Boas, D.; Lo, E.H.; et al. Age-related decline in oligodendrogenesis retards white matter repair in mice. Stroke 2013, 44, 2573–2578. [Google Scholar] [CrossRef] [Green Version]
- El Waly, B.; Macchi, M.; Cayre, M.; Durbec, P. Oligodendrogenesis in the normal and pathological central nervous system. Front. Neurosci. 2014, 8, 145. [Google Scholar] [CrossRef] [Green Version]
- Petersen, M.A.; Ryu, J.K.; Chang, K.J.; Etxeberria, A.; Bardehle, S.; Mendiola, A.S.; Kamau-Devers, W.; Fancy, S.P.J.; Thor, A.; Bushong, E.A.; et al. Fibrinogen Activates BMP Signaling in Oligodendrocyte Progenitor Cells and Inhibits Remyelination after Vascular Damage. Neuron 2017, 96, 1003.e1007–1012.e1007. [Google Scholar] [CrossRef] [Green Version]
- Ju, Z.; Lenhard Rudolph, K. Telomere dysfunction and stem cell ageing. Biochimie 2008, 90, 24–32. [Google Scholar] [CrossRef]
- Ahmed, Z.; Sheng, H.; Xu, Y.F.; Lin, W.L.; Innes, A.E.; Gass, J.; Yu, X.; Wuertzer, C.A.; Hou, H.; Chiba, S.; et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 2010, 177, 311–324. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zambusi, A.; Pelin Burhan, Ö.; Di Giaimo, R.; Schmid, B.; Ninkovic, J. Granulins Regulate Aging Kinetics in the Adult Zebrafish Telencephalon. Cells 2020, 9, 350. https://doi.org/10.3390/cells9020350
Zambusi A, Pelin Burhan Ö, Di Giaimo R, Schmid B, Ninkovic J. Granulins Regulate Aging Kinetics in the Adult Zebrafish Telencephalon. Cells. 2020; 9(2):350. https://doi.org/10.3390/cells9020350
Chicago/Turabian StyleZambusi, Alessandro, Özge Pelin Burhan, Rossella Di Giaimo, Bettina Schmid, and Jovica Ninkovic. 2020. "Granulins Regulate Aging Kinetics in the Adult Zebrafish Telencephalon" Cells 9, no. 2: 350. https://doi.org/10.3390/cells9020350
APA StyleZambusi, A., Pelin Burhan, Ö., Di Giaimo, R., Schmid, B., & Ninkovic, J. (2020). Granulins Regulate Aging Kinetics in the Adult Zebrafish Telencephalon. Cells, 9(2), 350. https://doi.org/10.3390/cells9020350




