Single-Nucleus Sequencing of an Entire Mammalian Heart: Cell Type Composition and Velocity
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Nuclei
2.2. Computational Data Analysis
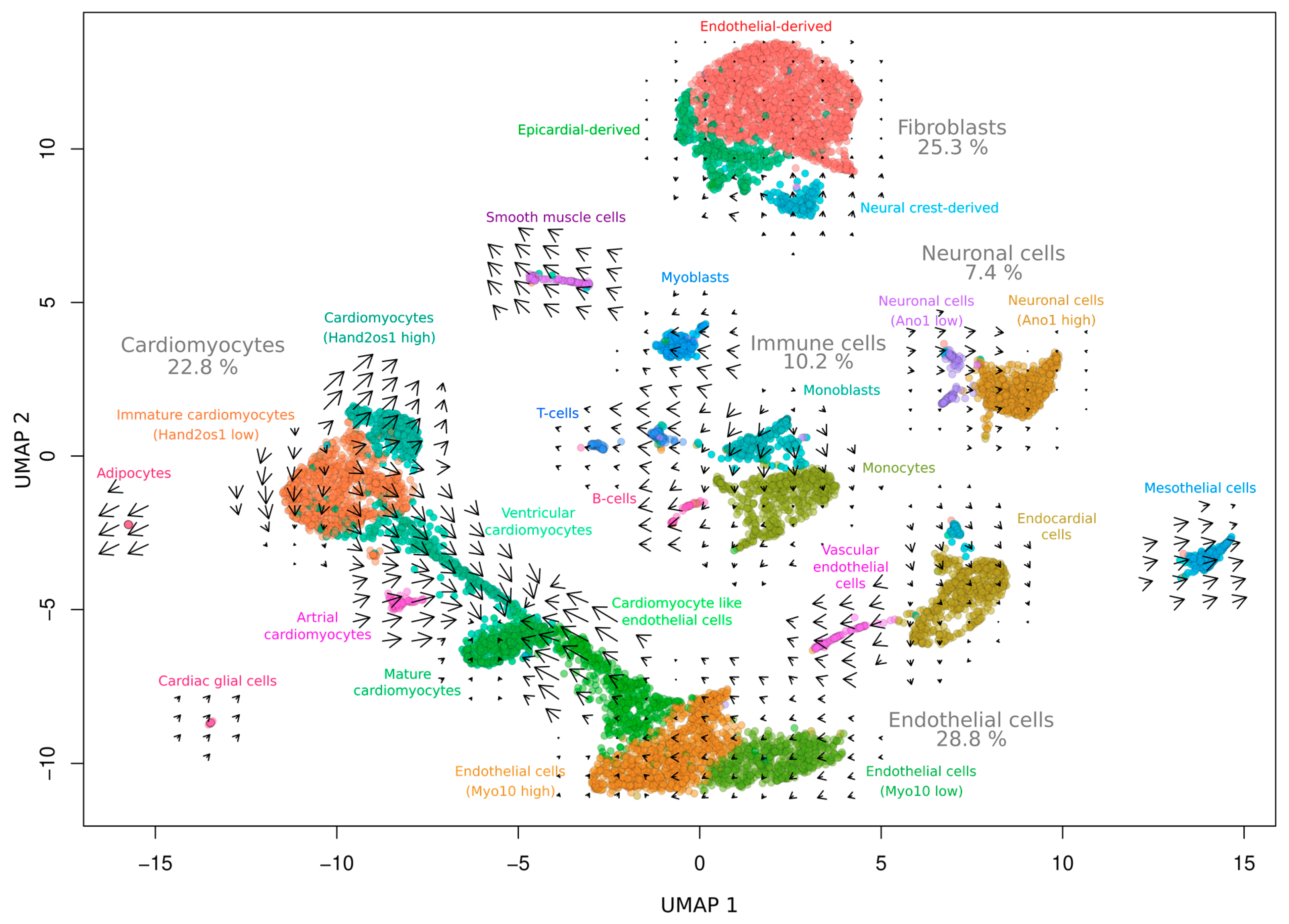
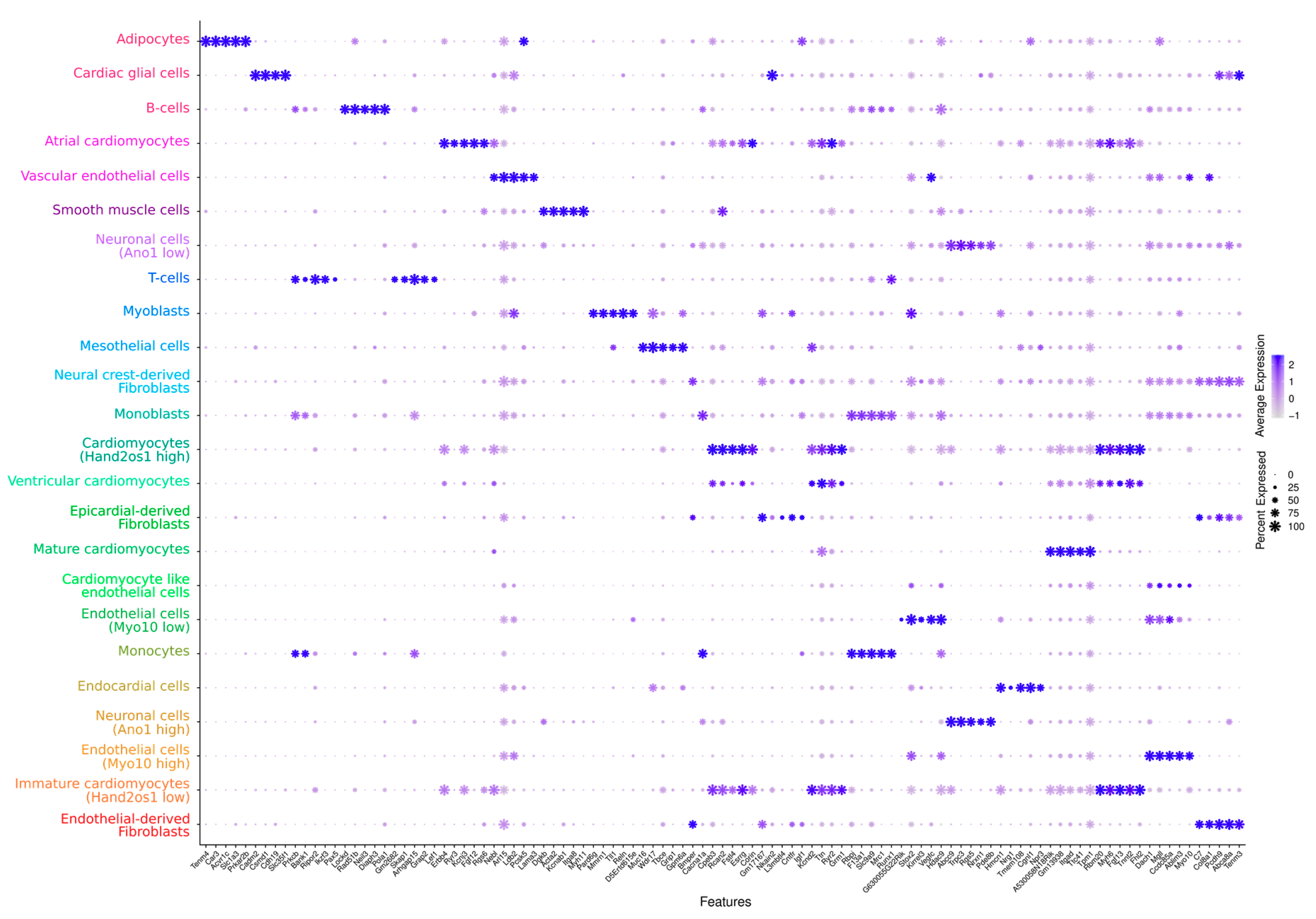
3. Results and Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Schaum, N.; Karkanias, J.; Neff, N.F.; May, A.P.; Quake, S.R.; Wyss-Coray, T.; Darmanis, S.; Batson, J.; Botvinnik, O.; Chen, M.B.; et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nat. Nat. Publ. Group 2018, 562, 367–372. [Google Scholar]
- Ackers-Johnson, M.; Tan, W.L.W.; Foo, R.S.-Y. Following hearts, one cell at a time: Recent applications of single-cell RNA sequencing to the understanding of heart disease. Nat. Commun. 2018, 9, 4434. [Google Scholar] [CrossRef]
- Bakken, T.E.; Hodge, R.D.; Miller, J.A.; Yao, Z.; Nguyen, T.N.; Aevermann, B.; Barkan, E.; Bertagnolli, D.; Casper, T.; Dee, N.; et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS ONE 2018, 13, e0209648. [Google Scholar] [CrossRef] [PubMed]
- Linscheid, N.; Logantha, S.J.R.J.; Poulsen, P.C.; Zhang, S.; Schrölkamp, M.; Egerod, K.L.; Thompson, J.J.; Kitmitto, A.; Galli, G.; Humphries, M.J.; et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat. Commun. 2019, 10, 2889. [Google Scholar] [CrossRef]
- Hu, P.; Liu, J.; Zhao, J.; Wilkins, B.J.; Lupino, K.; Wu, H.; Pei, L. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018, 32, 1344–1357. [Google Scholar] [CrossRef] [PubMed]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Dietl, G.; Langhammer, M.; Renne, U. Model simulations for genetic random drift in the outbred strain Fzt:DU. Arch. Anim. Breed. 2004, 47, 595–604. [Google Scholar] [CrossRef]
- Melsted, P.; Booeshaghi, A.S.; Gao, F.; Beltrame, E.D.V.; Lu, L.; Hjorleifsson, K.E.; Gehring, J.; Pachter, L. Modular and Efficient Pre-Processing of Single-Cell RNA-Seq; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2019; p. 673285. [Google Scholar]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.A.; et al. Revisiting cardiac cellular composition. Circ. Res. Lippincott Williams Wilkins 2016, 118, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146, dev176198. [Google Scholar] [CrossRef] [PubMed]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Müller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657.e8. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.M.; Anderson, U.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef] [PubMed]
- De Soysa, T.Y.; Ranade, S.S.; Okawa, S.; Ravichandran, S.; Huang, Y.; Salunga, H.T.; Schricker, A.; Del Sol, A.; Gifford, C.A.; Srivastava, D. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 2019, 572, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Condorelli, G.; Borello, U.; De Angelis, L.; Latronico, M.; Sirabella, D.; Coletta, M.; Galli, R.; Balconi, G.; Follenzi, A.; Frati, G.; et al. Cardiomyocytes induce endothelial cells to trans-differentiate into cardiac muscle: Implications for myocardium regeneration. Proc. Natl. Acad. Sci. USA 2001, 98, 10733–10738. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolfien, M.; Galow, A.-M.; Müller, P.; Bartsch, M.; Brunner, R.M.; Goldammer, T.; Wolkenhauer, O.; Hoeflich, A.; David, R. Single-Nucleus Sequencing of an Entire Mammalian Heart: Cell Type Composition and Velocity. Cells 2020, 9, 318. https://doi.org/10.3390/cells9020318
Wolfien M, Galow A-M, Müller P, Bartsch M, Brunner RM, Goldammer T, Wolkenhauer O, Hoeflich A, David R. Single-Nucleus Sequencing of an Entire Mammalian Heart: Cell Type Composition and Velocity. Cells. 2020; 9(2):318. https://doi.org/10.3390/cells9020318
Chicago/Turabian StyleWolfien, Markus, Anne-Marie Galow, Paula Müller, Madeleine Bartsch, Ronald M. Brunner, Tom Goldammer, Olaf Wolkenhauer, Andreas Hoeflich, and Robert David. 2020. "Single-Nucleus Sequencing of an Entire Mammalian Heart: Cell Type Composition and Velocity" Cells 9, no. 2: 318. https://doi.org/10.3390/cells9020318
APA StyleWolfien, M., Galow, A.-M., Müller, P., Bartsch, M., Brunner, R. M., Goldammer, T., Wolkenhauer, O., Hoeflich, A., & David, R. (2020). Single-Nucleus Sequencing of an Entire Mammalian Heart: Cell Type Composition and Velocity. Cells, 9(2), 318. https://doi.org/10.3390/cells9020318







