Single Cell Mass Cytometry of Non-Small Cell Lung Cancer Cells Reveals Complexity of In Vivo and Three-Dimensional Models over the Petri-Dish
Abstract
1. Introduction
2. Materials and Methods
2.1. Two-Dimensional (2D) Cell Culture
2.2. T-75 Flask Surface Coating
2.3. Three-Dimensional Microcarrier Coating
2.4. Three-Dimensional (3D) Cell Culturing Using Bench-top Incubator System
2.5. Real Architecture for 3D Tissue (RAFT) Culturing
2.6. A549 Xenograft Tumor Model
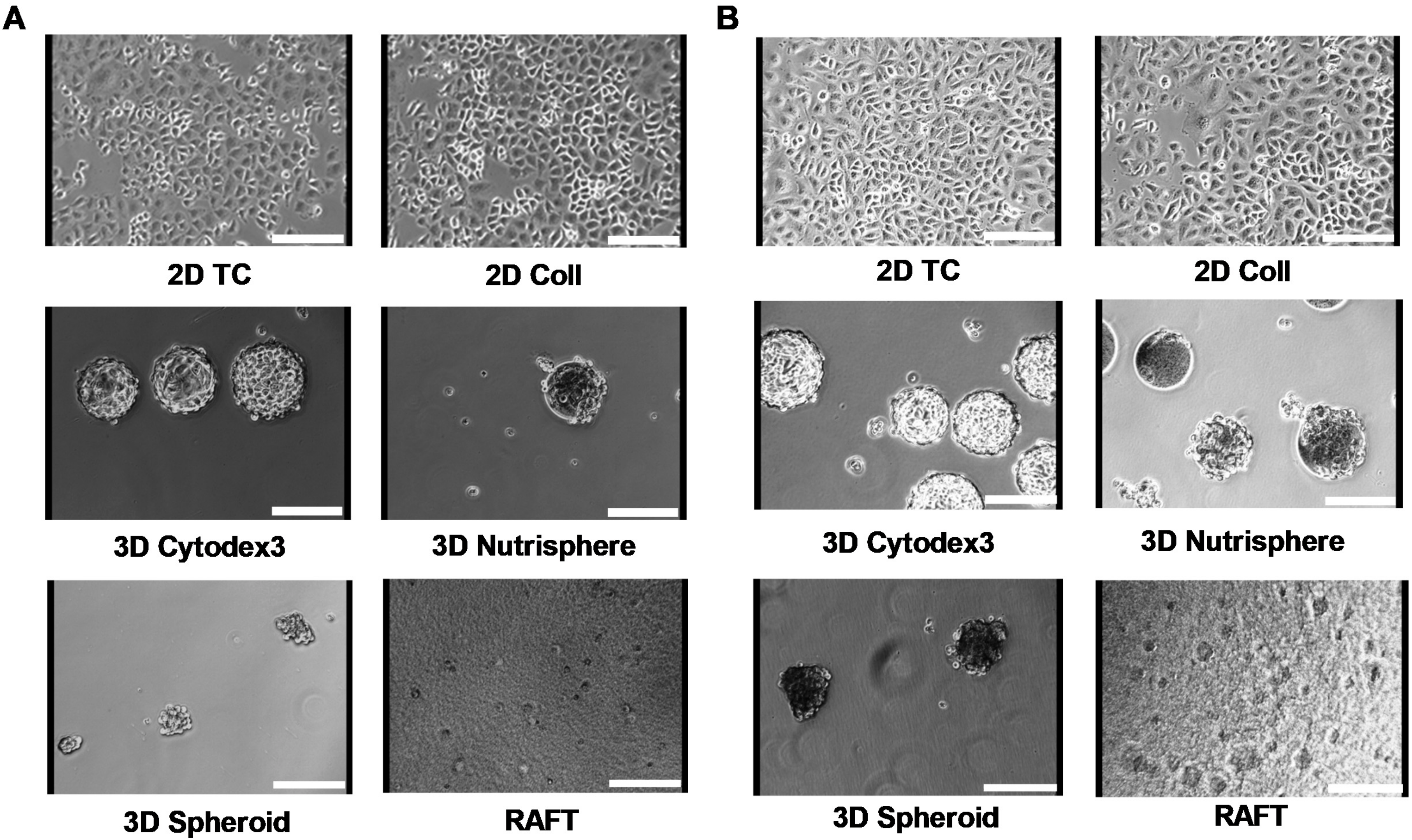
2.7. Imaging
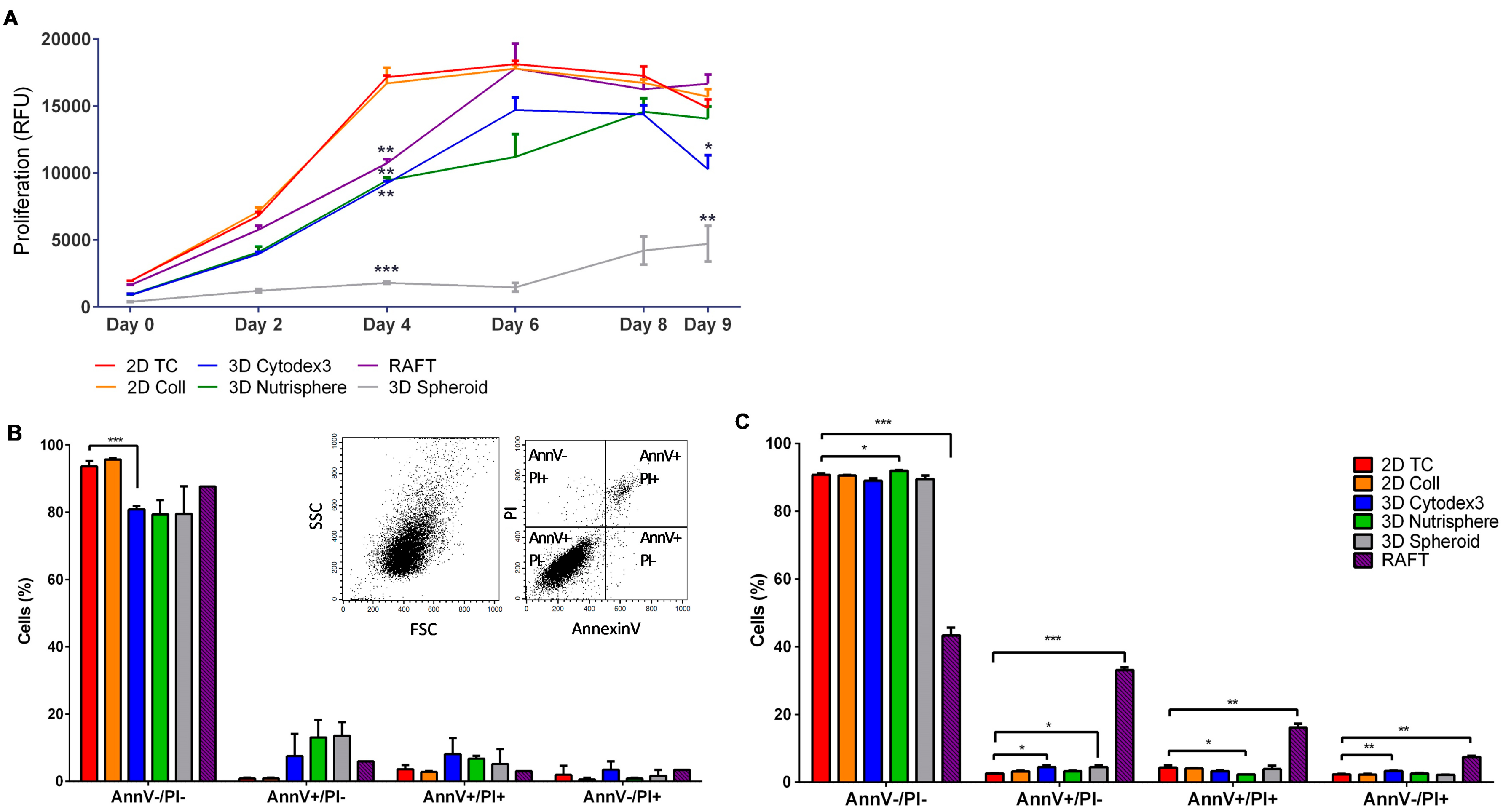
2.8. Cell Proliferation Assay
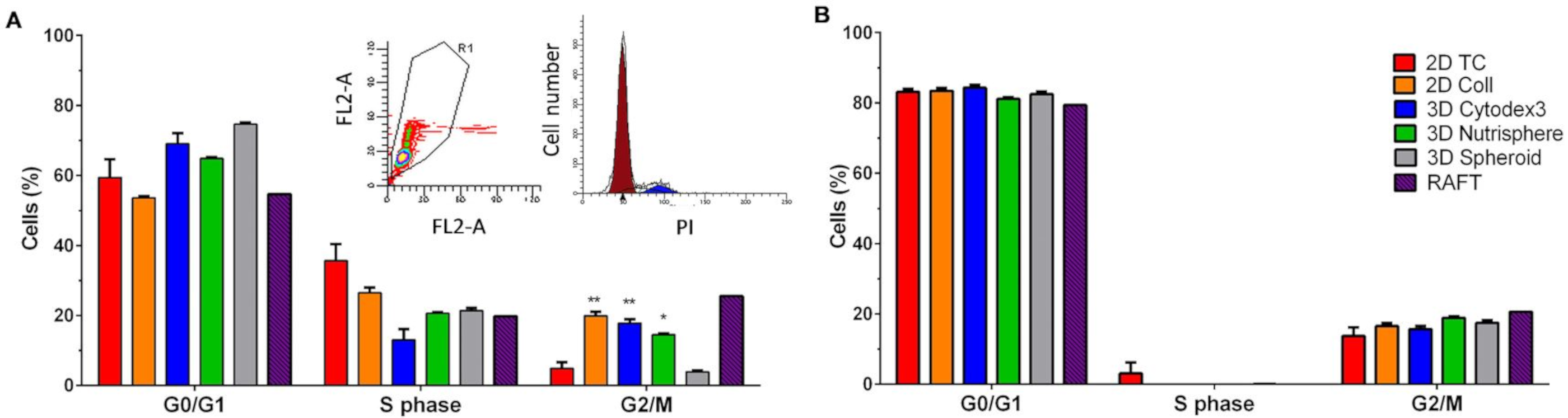
2.9. Cell Cycle Analysis
2.10. Apoptotic Assay
2.11. Profiling of RNAs with High-Throughput, Nanocapillary qRT-PCR
2.12. Gene Expression Analysis by High-Throughput qRT-PCR
2.13. Cluster Analysis
2.14. Single Cell Mass Cytometry
2.15. Statistical Analysis
3. Results
3.1. Long-term Growth Curve, Apoptosis, and Cell Cycle Phase Distribution of 3D Cultures
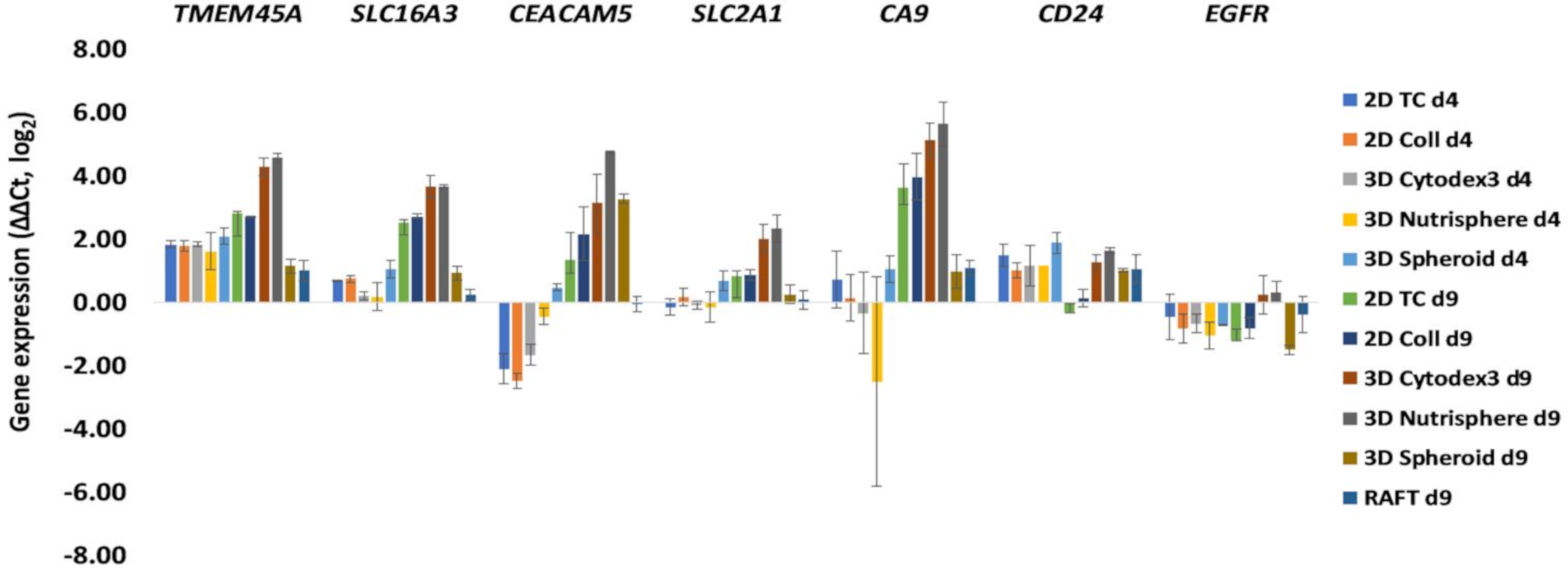
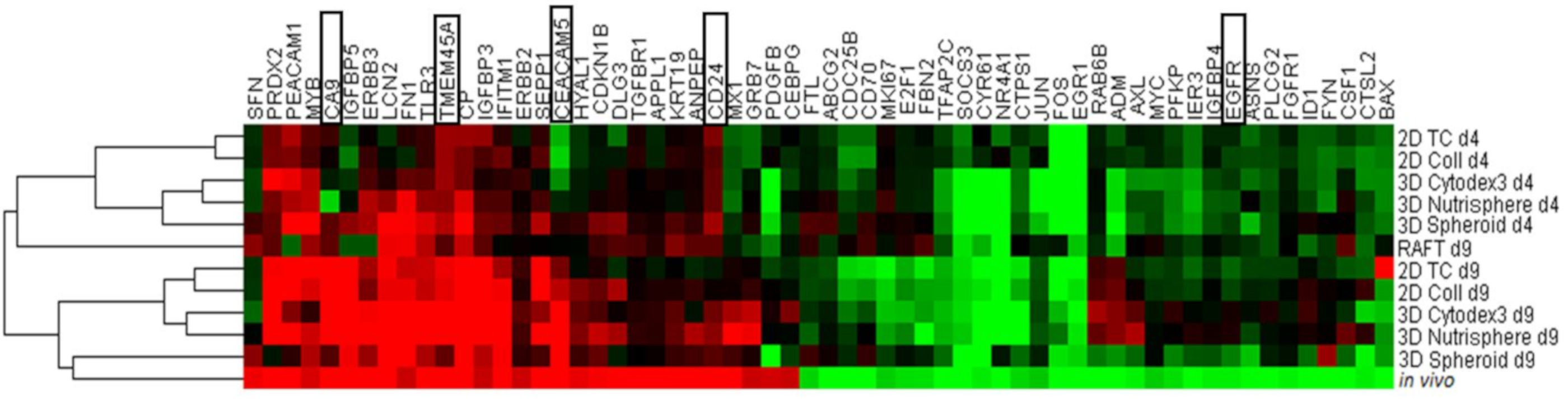
3.2. Selection of Genes with Differential Expression In Vivo and in 3D Models Compared to Monolayer Cultures
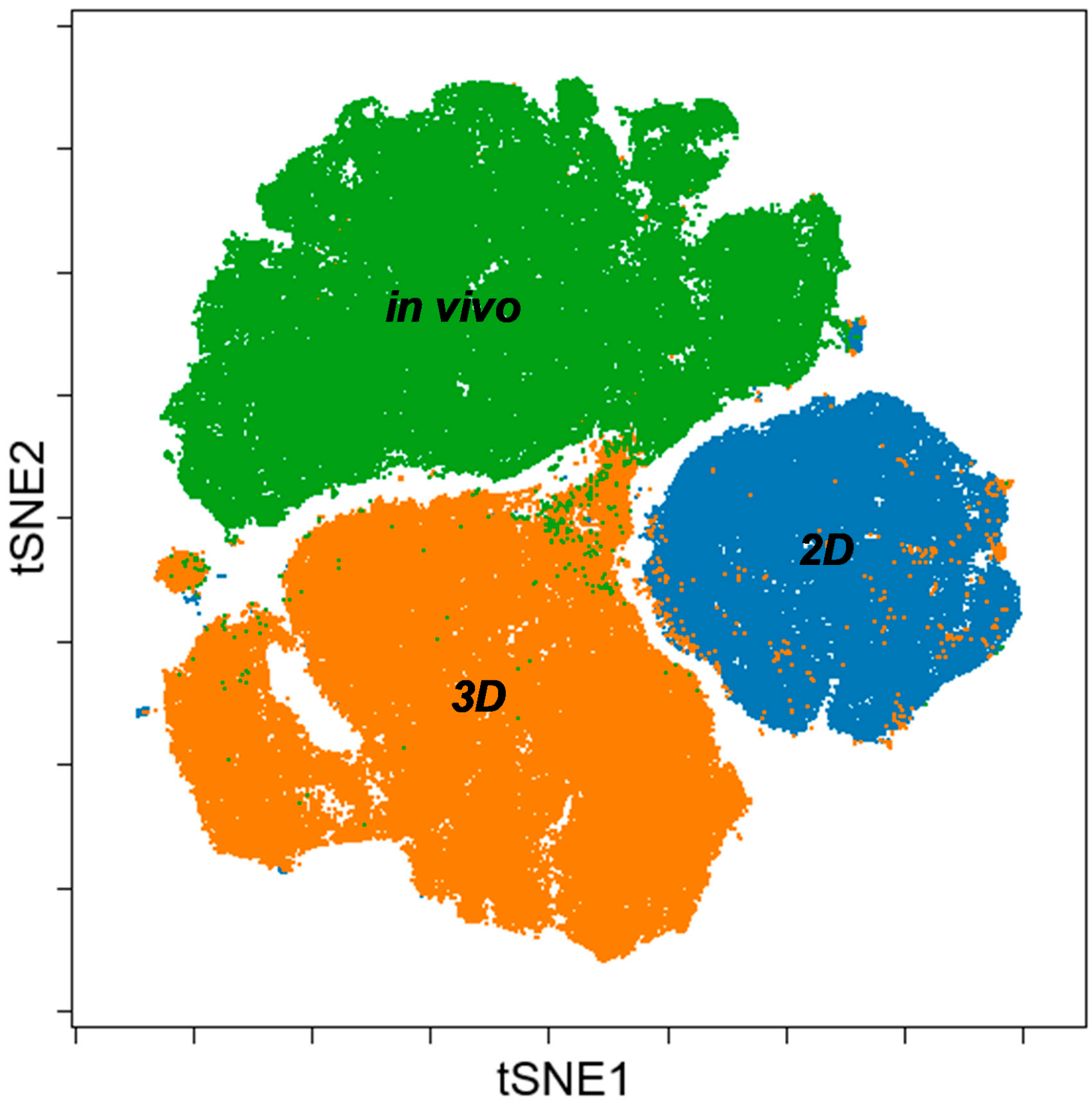
3.3. Single Cell-based Profiling Provides a Characteristic Map of Lung Cancer Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Doerr, A. Single-cell proteomics. Nat. Methods 2019, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Kim, J.K. Dissecting Cellular Heterogeneity Using Single-Cell RNA Sequencing. Mol. Cells 2019, 42, 189–199. [Google Scholar] [PubMed]
- Scurrah, C.R.; Simmons, A.J.; Lau, K.S. Single-Cell Mass Cytometry of Archived Human Epithelial Tissue for Decoding Cancer Signaling Pathways. Methods Mol. Biol. 2019, 1884, 215–229. [Google Scholar] [PubMed]
- Brasko, C.; Smith, K.; Molnar, C.; Farago, N.; Hegedus, L.; Balind, A.; Balassa, T.; Szkalisity, A.; Sukosd, F.; Kocsis, K.; et al. Intelligent image-based in situ single-cell isolation. Nat. Commun 2018, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Han, G.J.; Spitzer, M.H.; Bendall, S.C.; Fantl, W.J.; Nolan, G.P. Metal-isotope-tagged monoclonal antibodies for high-dimensional mass cytometry. Nat. Protoc. 2018, 13, 2121–2148. [Google Scholar] [CrossRef] [PubMed]
- Brainard, J.; Farver, C. The diagnosis of non-small cell lung cancer in the molecular era. Mod. Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kenfield, S.; Wei, E.; Colditz, G.; Stampfer, M.; Rosner, B. Comparison of aspects of smoking among four histologic types of lung cancer. Am. J. Epidemiol. 2006, 163, S108. [Google Scholar] [CrossRef][Green Version]
- Lavin, Y.; Kobayashi, S.; Leader, A.; Amir, E.D.; Elefant, N.; Bigenwald, C.; Remark, R.; Sweeney, R.; Becker, C.D.; Levine, J.H.; et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 2017, 169, 750–765. [Google Scholar] [CrossRef]
- Hickman, J.A.; Graeser, R.; de Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; van der Kuip, H.; Consortium, I.P. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef]
- Verjans, E.T.; Doijen, J.; Luyten, W.; Landuyt, B.; Schoofs, L. Three-dimensional cell culture models for anticancer drug screening: Worth the effort? J. Cell. Physiol. 2018, 233, 2993–3003. [Google Scholar] [CrossRef]
- Inch, W.R.; McCredie, J.A.; Sutherland, R.M. Growth of nodular carcinomas in rodents compared with multi-cell spheroids in tissue culture. Growth 1970, 34, 271–282. [Google Scholar] [PubMed]
- Sutherland, R.M.; McCredie, J.A.; Inch, W.R. Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J. Natl. Cancer Inst. 1971, 46, 113–120. [Google Scholar] [PubMed]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharm. 2018, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, G.J.; Tancos, Z.; Feher, L.Z.; Alfoldi, R.; Kobolak, J.; Dinnyes, A.; Puskas, L.G. Real architecture for 3D Tissue (RAFT) culture system improves viability and maintains insulin and glucagon production of mouse pancreatic islet cells. Cytotechnology 2017, 69, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Alhaque, S.; Themis, M.; Rashidi, H. Three-dimensional cell culture: From evolution to revolution. Philos. Trans. R Soc. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Ozsvari, B.; Puskas, L.G.; Nagy, L.I.; Kanizsai, I.; Gyuris, M.; Madacsi, R.; Feher, L.Z.; Gero, D.; Szabo, C. A cell-microelectronic sensing technique for the screening of cytoprotective compounds. Int. J. Mol. Med. 2010, 25, 525–530. [Google Scholar]
- Nagy, L.I.; Molnar, E.; Kanizsai, I.; Madacsi, R.; Ozsvari, B.; Feher, L.Z.; Fabian, G.; Marton, A.; Vizler, C.; Ayaydin, F.; et al. Lipid droplet binding thalidomide analogs activate endoplasmic reticulum stress and suppress hepatocellular carcinoma in a chemically induced transgenic mouse model. Lipids Health Dis. 2013, 12, 175. [Google Scholar] [CrossRef]
- Ernst, A.; Hofmann, S.; Ahmadi, R.; Becker, N.; Korshunov, A.; Engel, F.; Hartmann, C.; Felsberg, J.; Sabel, M.; Peterziel, H.; et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 6541–6550. [Google Scholar] [CrossRef]
- LaBarbera, D.V.; Reid, B.G.; Yoo, B.H. The multicellular tumor spheroid model for high-throughput cancer drug discovery. Expert Opin. Drug Discov. 2012, 7, 819–830. [Google Scholar] [CrossRef]
- Magdeldin, T.; Lopez-Davila, V.; Villemant, C.; Cameron, G.; Drake, R.; Cheema, U.; Loizidou, M. The efficacy of cetuximab in a tissue-engineered three-dimensional in vitro model of colorectal cancer. J. Tissue Eng. 2014. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, G.J.; Balog, J.A.; Demjen, A.; Alfoldi, R.; Vegi, V.L.; Feher, L.Z.; Man, I.; Kotogany, E.; Guban, B.; Batar, P.; et al. Imidazo[1,2-b]pyrazole-7-carboxamides Induce Apoptosis in Human Leukemia Cells at Nanomolar Concentrations. Molecules 2018, 23, 2845. [Google Scholar] [CrossRef] [PubMed]
- Demjen, A.; Alfoldi, R.; Angyal, A.; Gyuris, M.; Hackler, L., Jr.; Szebeni, G.J.; Wolfling, J.; Puskas, L.G.; Kanizsai, I. Synthesis, cytotoxic characterization, and SAR study of imidazo[1,2-b]pyrazole-7-carboxamides. Arch. Pharm. 2018. [Google Scholar] [CrossRef] [PubMed]
- Abbott, A. Cell culture: biology’s new dimension. Nature 2003, 424, 870–872. [Google Scholar] [CrossRef]
- Weigelt, B.; Lo, A.T.; Park, C.C.; Gray, J.W.; Bissell, M.J. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 2010, 122, 35–43. [Google Scholar] [CrossRef]
- Horning, J.L.; Sahoo, S.K.; Vijayaraghavalu, S.; Dimitrijevic, S.; Vasir, J.K.; Jain, T.K.; Panda, A.K.; Labhasetwar, V. 3-D tumor model for in vitro evaluation of anticancer drugs. Mol. Pharm. 2008, 5, 849–862. [Google Scholar] [CrossRef]
- Szebeni, G.J.; Balazs, A.; Madarasz, I.; Pocz, G.; Ayaydin, F.; Kanizsai, I.; Fajka-Boja, R.; Alfoldi, R.; Hackler, L., Jr.; Puskas, L.G. Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells. Int. J. Mol. Sci. 2017, 18, 2105. [Google Scholar] [CrossRef]
- Molnar, J.; Szebeni, G.J.; Csupor-Loffler, B.; Hajdu, Z.; Szekeres, T.; Saiko, P.; Ocsovszki, I.; Puskas, L.G.; Hohmann, J.; Zupko, I. Investigation of the Antiproliferative Properties of Natural Sesquiterpenes from Artemisia asiatica and Onopordum acanthium on HL-60 Cells in Vitro. Int. J. Mol. Sci. 2016, 17, 83. [Google Scholar] [CrossRef]
- Man, I.; Szebeni, G.J.; Plangar, I.; Szabo, E.R.; Tokes, T.; Szabo, Z.; Nagy, Z.; Fekete, G.; Fajka-Boja, R.; Puskas, L.G.; et al. Novel real-time cell analysis platform for the dynamic monitoring of ionizing radiation effects on human tumor cell lines and primary fibroblasts. Mol. Med. Rep. 2015, 12, 4610–4619. [Google Scholar] [CrossRef]
- Vass, L.; Kelemen, J.Z.; Feher, L.Z.; Lorincz, Z.; Kulin, S.; Cseh, S.; Dorman, G.; Puskas, L.G. Toxicogenomics screening of small molecules using high-density, nanocapillary real-time PCR. Int. J. Mol. Med. 2009, 23, 65–74. [Google Scholar] [PubMed]
- Farago, N.; Kocsis, A.K.; Lovas, S.; Molnar, G.; Boldog, E.; Rozsa, M.; Szemenyei, V.; Vamos, E.; Nagy, L.I.; Tamas, G.; et al. Digital PCR to determine the number of transcripts from single neurons after patch-clamp recording. Biotechniques 2013, 54, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.I.; Feher, L.Z.; Szebeni, G.J.; Gyuris, M.; Sipos, P.; Alfoldi, R.; Ozsvari, B.; Hackler, L., Jr.; Balazs, A.; Batar, P.; et al. Curcumin and its analogue induce apoptosis in leukemia cells and have additive effects with bortezomib in cellular and xenograft models. Biomed. Res. Int. 2015, 2015, 968981. [Google Scholar] [CrossRef] [PubMed]
- Amir, E.D.; Davis, K.L.; Tadmor, M.D.; Simonds, E.F.; Levine, J.H.; Bendall, S.C.; Shenfeld, D.K.; Krishnaswamy, S.; Nolan, G.P.; Pe’er, D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013, 31, 545. [Google Scholar] [CrossRef] [PubMed]
- Fabian, G.; Farago, N.; Feher, L.Z.; Nagy, L.I.; Kulin, S.; Kitajka, K.; Bito, T.; Tubak, V.; Katona, R.L.; Tiszlavicz, L.; et al. High-density real-time PCR-based in vivo toxicogenomic screen to predict organ-specific toxicity. Int. J. Mol. Sci. 2011, 12, 6116–6134. [Google Scholar] [CrossRef] [PubMed]
- Meijer, T.W.; Schuurbiers, O.C.; Kaanders, J.H.; Looijen-Salamon, M.G.; de Geus-Oei, L.F.; Verhagen, A.F.; Lok, J.; van der Heijden, H.F.; Rademakers, S.E.; Span, P.N.; et al. Differences in metabolism between adeno- and squamous cell non-small cell lung carcinomas: Spatial distribution and prognostic value of GLUT1 and MCT4. Lung Cancer 2012, 76, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Dhup, S.; Dadhich, R.K.; Copetti, T.; Sonveaux, P. Anticancer targets in the glycolytic metabolism of tumors: A comprehensive review. Front. Pharm. 2011, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.; Shao, J.; Picon, H.M.; Bristow, C.; Ge, Z.; Peoples, M.; Robinson, F.; Jeter-Jones, S.L.; Schlosberg, C.; Grzeskowiak, C.L.; et al. A functional genomic screen in vivo identifies CEACAM5 as a clinically relevant driver of breast cancer metastasis. NPJ Breast Cancer 2018, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Li, Y.; Zhang, X. Stemness-Related Markers in Cancer. Cancer Trans. Med. 2017, 3, 87–95. [Google Scholar]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharm. 2018, 9, 1345. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 52. [Google Scholar] [CrossRef]
- Levina, V.; Marrangoni, A.M.; DeMarco, R.; Gorelik, E.; Lokshin, A.E. Drug-selected human lung cancer stem cells: Cytokine network, tumorigenic and metastatic properties. PLoS ONE 2008. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, N.; Yusoff, N.M.; Zakaria, Z.; Lim, M.N.; Baharuddin, P.J.; Fakiruddin, K.S.; Yahaya, B. Human non-small cell lung cancer expresses putative cancer stem cell markers and exhibits the transcriptomic profile of multipotent cells. BMC Cancer 2015, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Nangia-Makker, P.; Hogan, V.; Raz, A. Galectin-3 and cancer stemness. Glycobiology 2018, 28, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Pawelczyk, K.; Piotrowska, A.; Ciesielska, U.; Jablonska, K.; Gletzel-Plucinska, N.; Grzegrzolka, J.; Podhorska-Okolow, M.; Dziegiel, P.; Nowinska, K. Role of PD-L1 Expression in Non-Small Cell Lung Cancer and Their Prognostic Significance according to Clinicopathological Factors and Diagnostic Markers. Int. J. Mol. Sci. 2019, 20, 824. [Google Scholar] [CrossRef] [PubMed]
- Karantza, V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 2011, 30, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chen, Z.; Wang, J.; Shao, X.; Cui, Z.; Yang, C.; Zhu, Z.; Xiong, D. Overexpression of cell surface cytokeratin 8 in multidrug-resistant MCF-7/MX cells enhances cell adhesion to the extracellular matrix. Neoplasia 2008, 10, 1275–1284. [Google Scholar] [CrossRef]
- Godfroid, E.; Geuskens, M.; Dupressoir, T.; Parent, I.; Szpirer, C. Cytokeratins are exposed on the outer surface of established human mammary carcinoma cells. J. Cell Sci. 1991, 99, 595–607. [Google Scholar]
- Kunz-Schughart, L.A.; Freyer, J.P.; Hofstaedter, F.; Ebner, R. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J. Biomol. Screen. 2004, 9, 273–285. [Google Scholar] [CrossRef]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef]
- Amoedo, N.D.; Valencia, J.P.; Rodrigues, M.F.; Galina, A.; Rumjanek, F.D. How does the metabolism of tumour cells differ from that of normal cells. Biosci. Rep. 2013. [Google Scholar] [CrossRef] [PubMed]
- Voss, M.J.; Niggemann, B.; Zanker, K.S.; Entschladen, F. Tumour reactions to hypoxia. Curr. Mol. Med. 2010, 10, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Airley, R.E.; Mobasheri, A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: Novel pathways and targets for anticancer therapeutics. Chemotherapy 2007, 53, 233–256. [Google Scholar] [CrossRef]
- Gonzalez, C.D.; Alvarez, S.; Ropolo, A.; Rosenzvit, C.; Bagnes, M.F.; Vaccaro, M.I. Autophagy, Warburg, and Warburg reverse effects in human cancer. Biomed. Res. Int. 2014, 2014, 926729. [Google Scholar] [CrossRef] [PubMed]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef] [PubMed]
- Pickl, M.; Ries, C.H. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene 2009, 28, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Svirshchevskaya, E.; Doronina, E.; Grechikhina, M.; Matushevskaya, E.; Kotsareva, O.; Fattakhova, G.; Sapozhnikov, A.; Felix, K. Characteristics of multicellular tumor spheroids formed by pancreatic cells expressing different adhesion molecules. Life Sci. 2019, 219, 343–352. [Google Scholar] [CrossRef]
- Stadler, M.; Scherzer, M.; Walter, S.; Holzner, S.; Pudelko, K.; Riedl, A.; Unger, C.; Kramer, N.; Weil, B.; Neesen, J.; et al. Exclusion from spheroid formation identifies loss of essential cell-cell adhesion molecules in colon cancer cells. Sci. Rep. 2018, 8, 1151. [Google Scholar] [CrossRef]
- Jia, W.; Jiang, X.; Liu, W.; Wang, L.; Zhu, B.; Zhu, H.; Liu, X.; Zhong, M.; Xie, D.; Huang, W.; et al. Effects of three-dimensional collagen scaffolds on the expression profiles and biological functions of glioma cells. Int. J. Oncol. 2018, 52, 1787–1800. [Google Scholar] [CrossRef]
- Pacheco-Marin, R.; Melendez-Zajgla, J.; Castillo-Rojas, G.; Mandujano-Tinoco, E.; Garcia-Venzor, A.; Uribe-Carvajal, S.; Cabrera-Orefice, A.; Gonzalez-Torres, C.; Gaytan-Cervantes, J.; Mitre-Aguilar, I.B.; et al. Transcriptome profile of the early stages of breast cancer tumoral spheroids. Sci. Rep. 2016, 6, 23373. [Google Scholar] [CrossRef]
- Joosse, S.A.; Hannemann, J.; Spotter, J.; Bauche, A.; Andreas, A.; Muller, V.; Pantel, K. Changes in keratin expression during metastatic progression of breast cancer: Impact on the detection of circulating tumor cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 993–1003. [Google Scholar] [CrossRef] [PubMed]
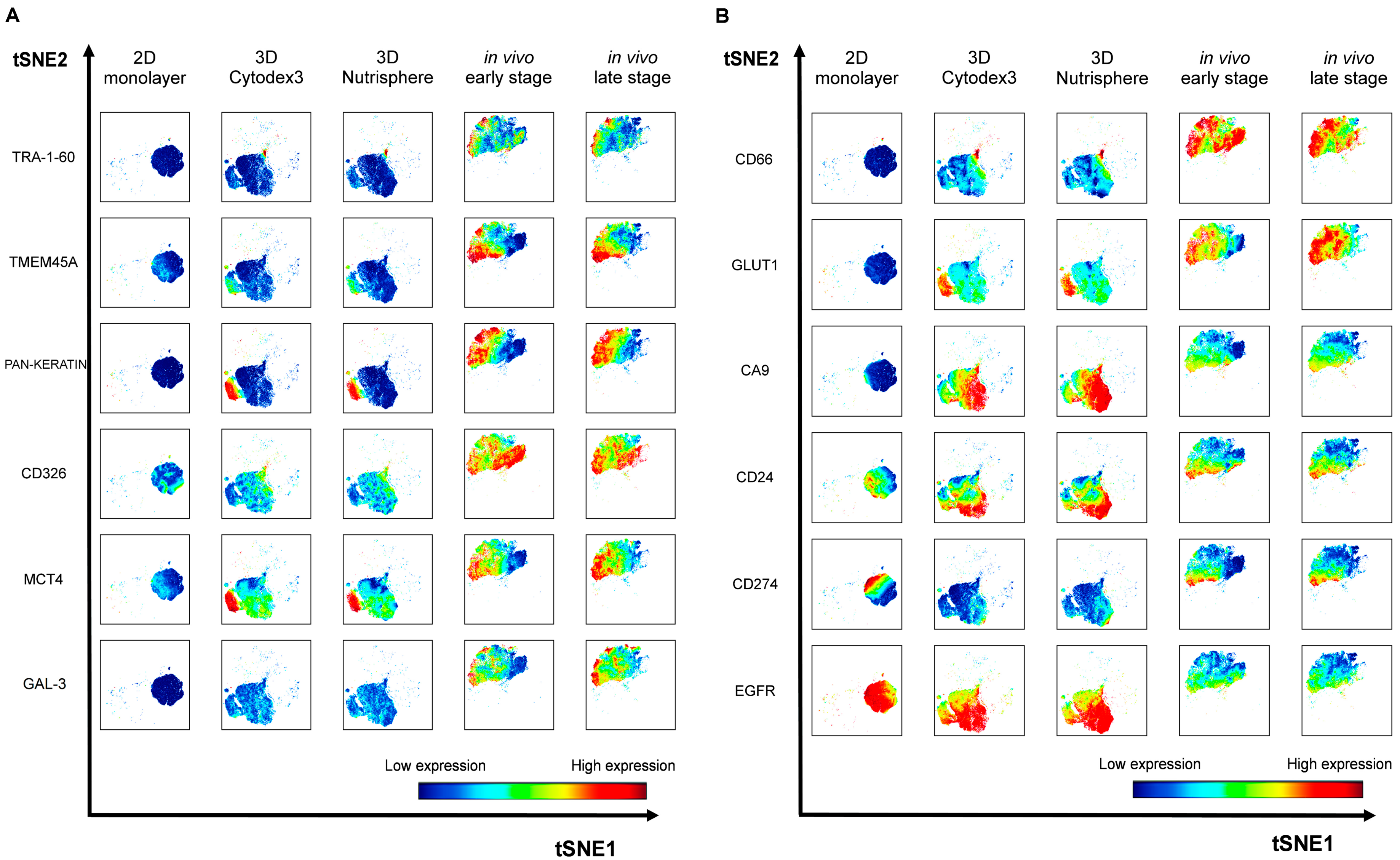
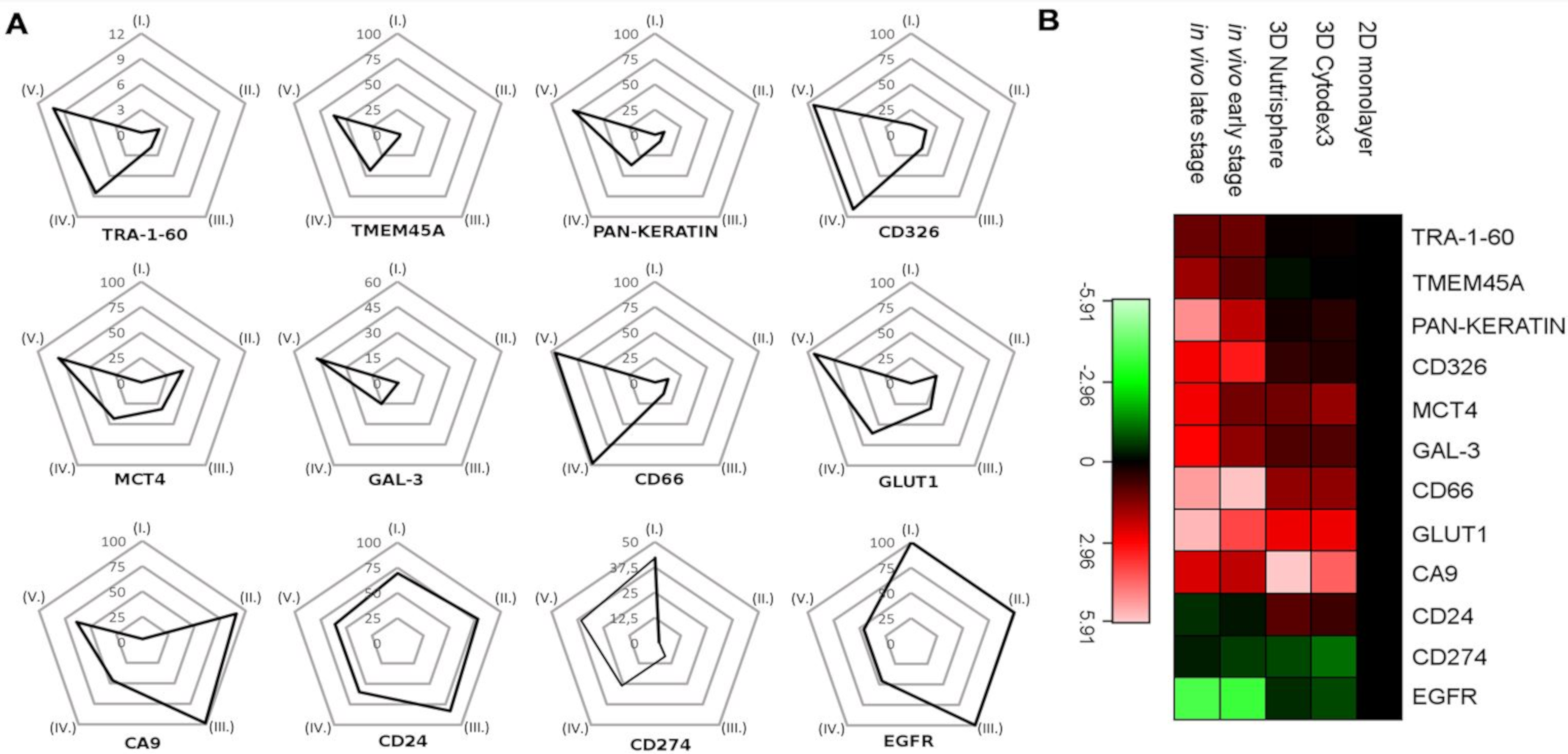
| Catalogue Number | Supplier | Target | Metal Tag |
|---|---|---|---|
| 3144017B | Fluidigm | HLA-A,B,C | 144_Nd |
| 3141006B | Fluidigm | CD326 (EpCam) | 141_Pr |
| 3148012B | Fluidigm | TRA-1-60 | 148_Nd |
| 3149018B | Fluidigm | CD66-a,c,e | 149_Sm |
| 3156026B | Fluidigm | CD274 (PD-L1) | 156_Gd |
| 3162027A | Fluidigm | Pan-Keratin | 162_Dy |
| 3166007B | Fluidigm | CD24 | 166_Er |
| 3170009B | Fluidigm | EGFR | 170_Er |
| 3153026B | Fluidigm | Galectin-3 (Gal-3) | 153_Eu |
| MAB2188-100 | R&D Systems | CA9 | 158_Gd |
| MAB1418 | R&D Systems | GLUT1 | 154_Sm |
| sc-376140 | Santa Cruz Biotech. | MCT4 | 171_Yb |
| orb357227 | Biorbyt | TMEM45A | 169_TM |
| Gene Symbol | Assay ID | ΔΔCt (log2) | SD | Significance (p) | Gene Symbol | Assay ID | ΔΔCt (log2) | SD | Significance (p) |
|---|---|---|---|---|---|---|---|---|---|
| CEACAM5 | Hs00944025_m1 | 9.11 | 1.77 | 0.0220 | JUN | Hs00277190_s1 | −2.34 | 1.97 | 0.2740 |
| APPL1 | Hs00179382_m1 | 8.17 | 0.12 | 0.0001 | MYC | Hs99999003_m1 | −2.42 | 1.96 | 0.2306 |
| LCN2 | Hs01008571_m1 | 5.58 | 0.68 | 0.0082 | MKI67 | Hs01032443_m1 | −2.43 | 0.45 | 0.0249 |
| SEPP1 | Hs01032845_m1 | 5.26 | 1.10 | 0.0293 | FTL | Hs00830226_gH | −2.51 | 1.21 | 0.1293 |
| PRDX2 | Hs03044902_g1 | 4.71 | 1.45 | 0.0590 | CTPS | Hs00157163_m1 | −2.53 | 1.41 | 0.1831 |
| TGFBR1 | Hs00610318_m1 | 4.38 | 0.02 | 0.0001 | E2F1 | Hs00153451_m1 | −2.63 | 1.58 | 0.1555 |
| CP | Hs00236810_m1 | 4.36 | 0.08 | 0.1173 | PFKP | Hs00242993_m1 | −2.64 | 1.70 | 0.1746 |
| ANPEP | Hs00952642_m1 | 3.79 | 2.19 | 0.2148 | FBN2 | Hs00266592_m1 | −2.65 | 0.17 | 0.2083 |
| DLG3 | Hs00221664_m1 | 3.72 | 0.22 | 0.0213 | CYR61 | Hs00155479_m1 | −2.79 | 2.20 | 0.2390 |
| CA9 | Hs00154208_m1 | 3.71 | 1.42 | 0.0886 | CTSL2 | Hs00822401_m1 | −2.81 | 1.03 | 0.0769 |
| CD24 | Hs00273561_s1 | 3.66 | 0.54 | 0.0182 | EGFR | Hs01076078_m1 | −2.82 | 0.38 | 0.0345 |
| IFITM1 | Hs00705137_s1 | 3.60 | 0.25 | 0.0057 | IGFBP4 | Hs00181767_m1 | −2.83 | 1.73 | 0.1601 |
| PECAM1 | Hs00169777_m1 | 3.54 | 0.00 | 0.0217 | FGFR1 | Hs00241111_m1 | −2.87 | 1.74 | 0.2655 |
| MX1 | Hs00895608_m1 | 3.53 | 0.92 | 0.0702 | AXL | Hs01064444_m1 | −2.87 | 1.81 | 0.1685 |
| TMEM45A | Hs01046616_m1 | 3.49 | 1.42 | 0.2538 | ASNS | Hs00370265_m1 | −2.98 | 1.02 | 0.0890 |
| KRT19 | Hs00761767_s1 | 3.38 | 1.35 | 0.0793 | SOCS3 | Hs02330328_s1 | −3.04 | 1.01 | 0.1962 |
| TLR3 | Hs00152933_m1 | 3.37 | 0.48 | 0.0330 | IER3 | Hs00174674_m1 | −3.18 | 1.78 | 0.2419 |
| ERBB3 | Hs00176538_m1 | 3.27 | 0.95 | 0.0524 | CDC25B | Hs01550934_m1 | −3.20 | 1.51 | 0.1116 |
| IGFBP5 | Hs01052296_m1 | 3.18 | 1.03 | 0.0563 | BAX | Hs00180269_m1 | −3.32 | 1.22 | 0.0804 |
| CDKN1B | Hs00153277_m1 | 2.91 | 0.22 | 0.0217 | TFAP2C | Hs00231476_m1 | −3.60 | 1.58 | 0.0921 |
| SFN | Hs00968567_s1 | 2.83 | 0.61 | 0.0329 | ABCG2 | Hs01053790_m1 | −3.66 | 0.85 | 0.0368 |
| HYAL1 | Hs00201046_m1 | 2.74 | 0.38 | 0.0096 | NR4A1 | Hs00374226_m1 | −3.66 | 0.83 | 0.0250 |
| MYB | Hs00920554_m1 | 2.54 | 1.61 | 0.2887 | RAB6B | Hs00981572_m1 | −3.66 | 0.89 | 0.0446 |
| PDGFB | Hs00966522_m1 | 2.47 | 0.47 | 0.0853 | CD70 | Hs00174297_m1 | −3.94 | 0.00 | 0.1385 |
| ERBB2 | Hs01001580_m1 | 2.40 | 1.25 | 0.1180 | CSF1 | Hs00174164_m1 | −4.03 | 0.19 | 0.0107 |
| GRB7 | Hs00918009_g1 | 2.39 | 0.67 | 0.3032 | FYN | Hs00176628_m1 | −4.09 | 1.86 | 0.0988 |
| FN1 | Hs01549976_m1 | 2.38 | 1.25 | 0.1188 | PLCG2 | Hs00182192_m1 | −4.13 | 2.19 | 0.1465 |
| CEBPG | Hs00156454_m1 | 2.37 | 1.99 | 0.5677 | EGR1 | Hs00152928_m1 | −4.19 | 2.60 | 0.1582 |
| IGFBP3 | Hs00181211_m1 | 2.34 | 1.03 | 0.1114 | ID1 | Hs03676575_s1 | −4.38 | 0.22 | 0.0155 |
| ADM | Hs00181605_m1 | −4.51 | 0.10 | 0.0013 | |||||
| FOS | Hs00170630_m1 | −4.61 | 2.40 | 0.1254 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alföldi, R.; Balog, J.Á.; Faragó, N.; Halmai, M.; Kotogány, E.; Neuperger, P.; Nagy, L.I.; Fehér, L.Z.; Szebeni, G.J.; Puskás, L.G. Single Cell Mass Cytometry of Non-Small Cell Lung Cancer Cells Reveals Complexity of In Vivo and Three-Dimensional Models over the Petri-Dish. Cells 2019, 8, 1093. https://doi.org/10.3390/cells8091093
Alföldi R, Balog JÁ, Faragó N, Halmai M, Kotogány E, Neuperger P, Nagy LI, Fehér LZ, Szebeni GJ, Puskás LG. Single Cell Mass Cytometry of Non-Small Cell Lung Cancer Cells Reveals Complexity of In Vivo and Three-Dimensional Models over the Petri-Dish. Cells. 2019; 8(9):1093. https://doi.org/10.3390/cells8091093
Chicago/Turabian StyleAlföldi, Róbert, József Á. Balog, Nóra Faragó, Miklós Halmai, Edit Kotogány, Patrícia Neuperger, Lajos I. Nagy, Liliána Z. Fehér, Gábor J. Szebeni, and László G. Puskás. 2019. "Single Cell Mass Cytometry of Non-Small Cell Lung Cancer Cells Reveals Complexity of In Vivo and Three-Dimensional Models over the Petri-Dish" Cells 8, no. 9: 1093. https://doi.org/10.3390/cells8091093
APA StyleAlföldi, R., Balog, J. Á., Faragó, N., Halmai, M., Kotogány, E., Neuperger, P., Nagy, L. I., Fehér, L. Z., Szebeni, G. J., & Puskás, L. G. (2019). Single Cell Mass Cytometry of Non-Small Cell Lung Cancer Cells Reveals Complexity of In Vivo and Three-Dimensional Models over the Petri-Dish. Cells, 8(9), 1093. https://doi.org/10.3390/cells8091093






