Extracellular Matrix Structure and Composition in the Early Four-Chambered Embryonic Heart
Abstract
1. Introduction
2. Materials and Methods
2.1. Chick Embryonic Heart Dissection
2.2. Fixation, Immunofluorescent Staining and Clearing
2.3. High-Resolution Confocal Microscopy
2.4. Quantitative Analysis of 3D Image Z-Stacks of Laminin and Fibronectin
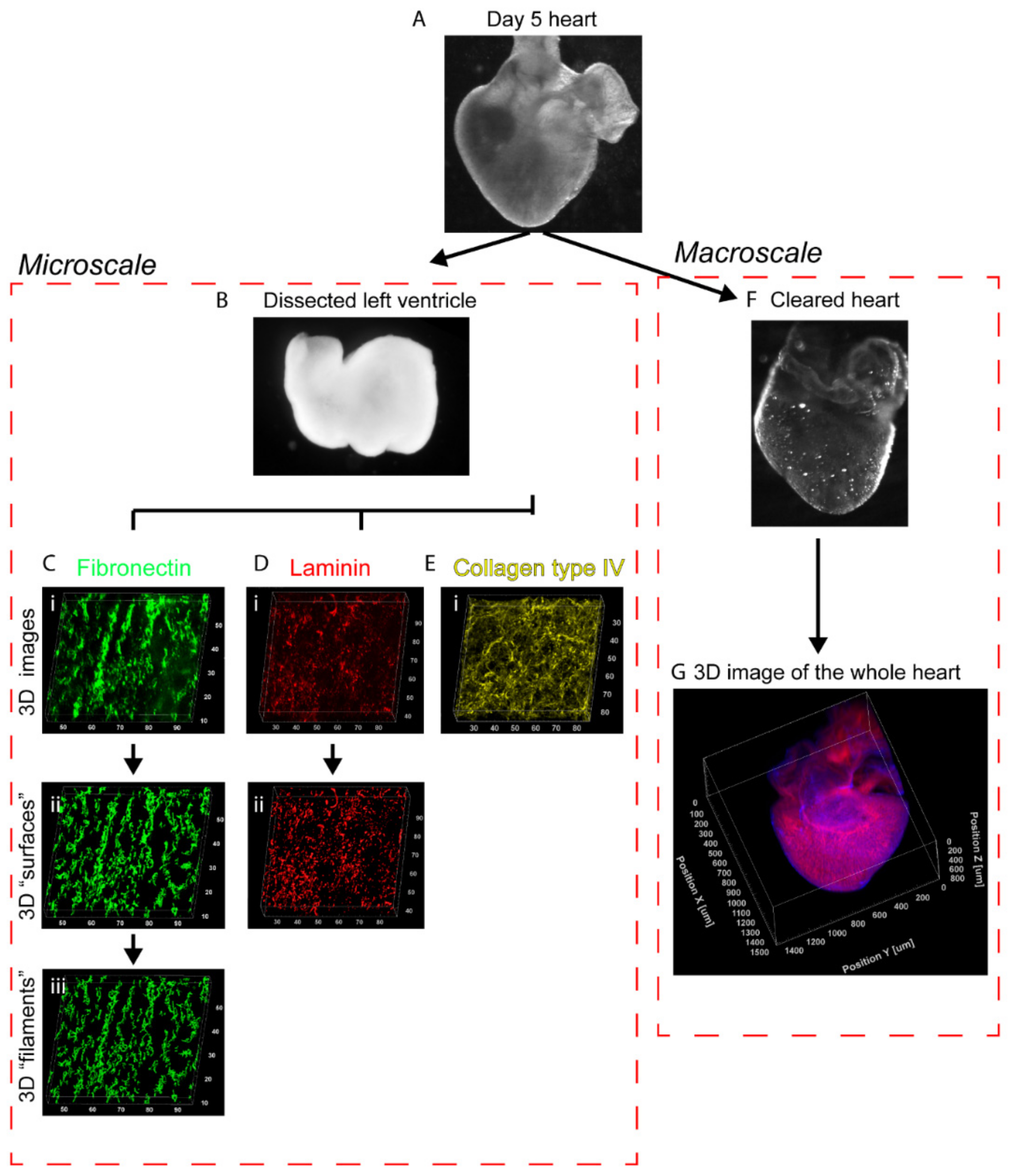
3. Results
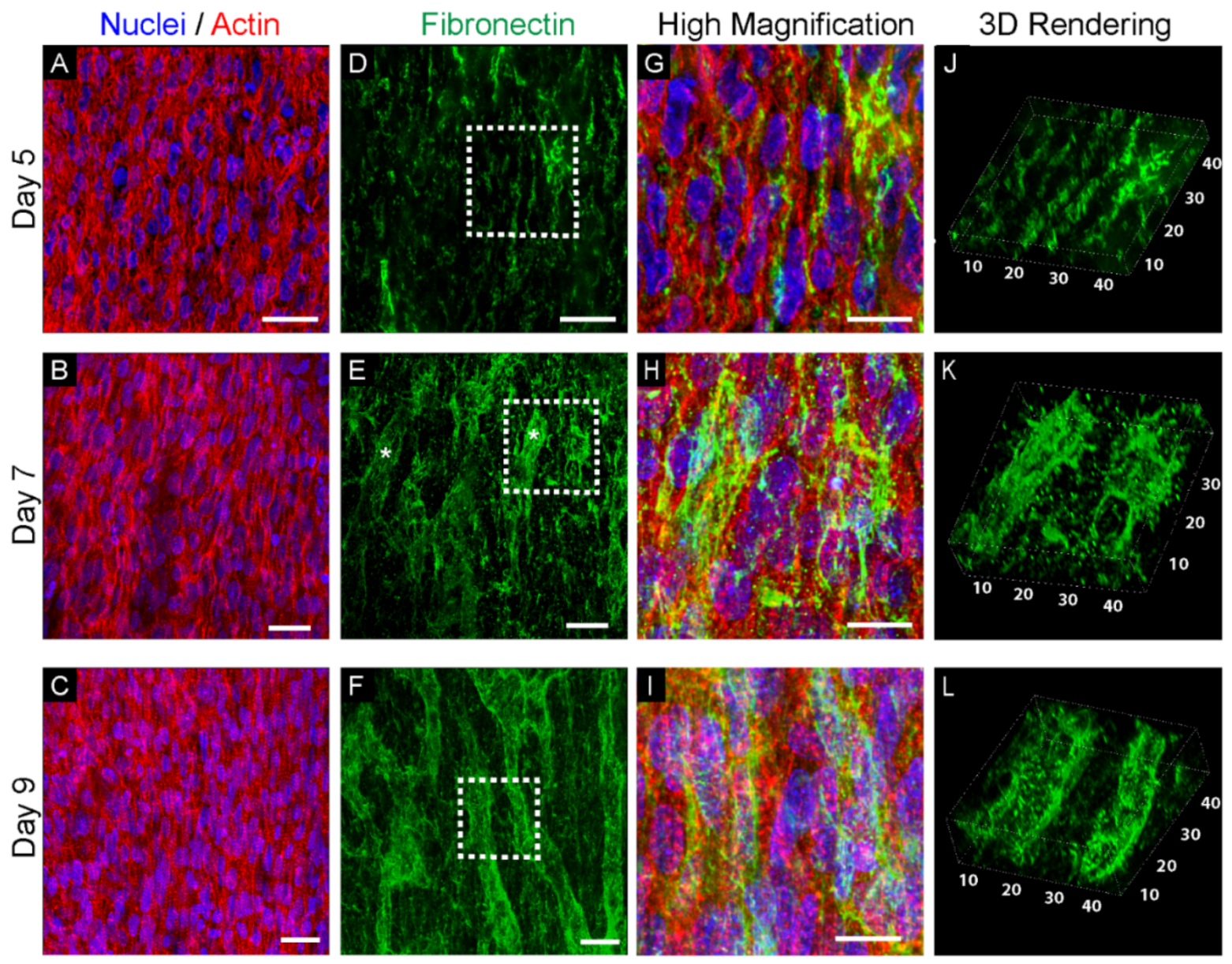
3.1. Fibronectin Is Associated with the Emergence of Cappilaries in the Left Ventricle Myocardium
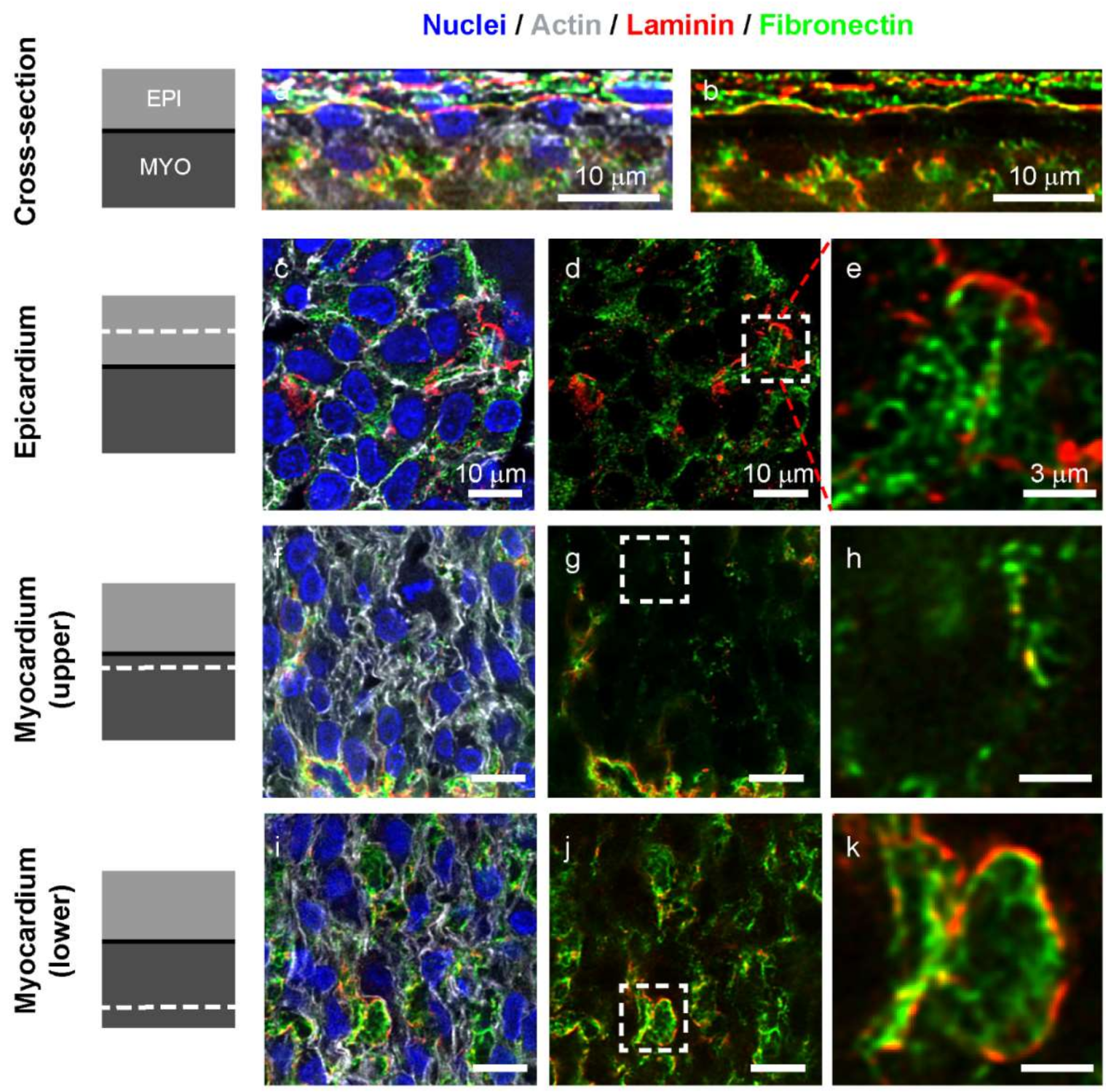
3.2. Laminin and Fibronectin, but not Collagen Type IV Are Found in the Left Ventricle Myocardium at Day 5
3.3. Fibronectin and Laminin are Colocalized in Intercellular Spaces in the Avascular Myocardium
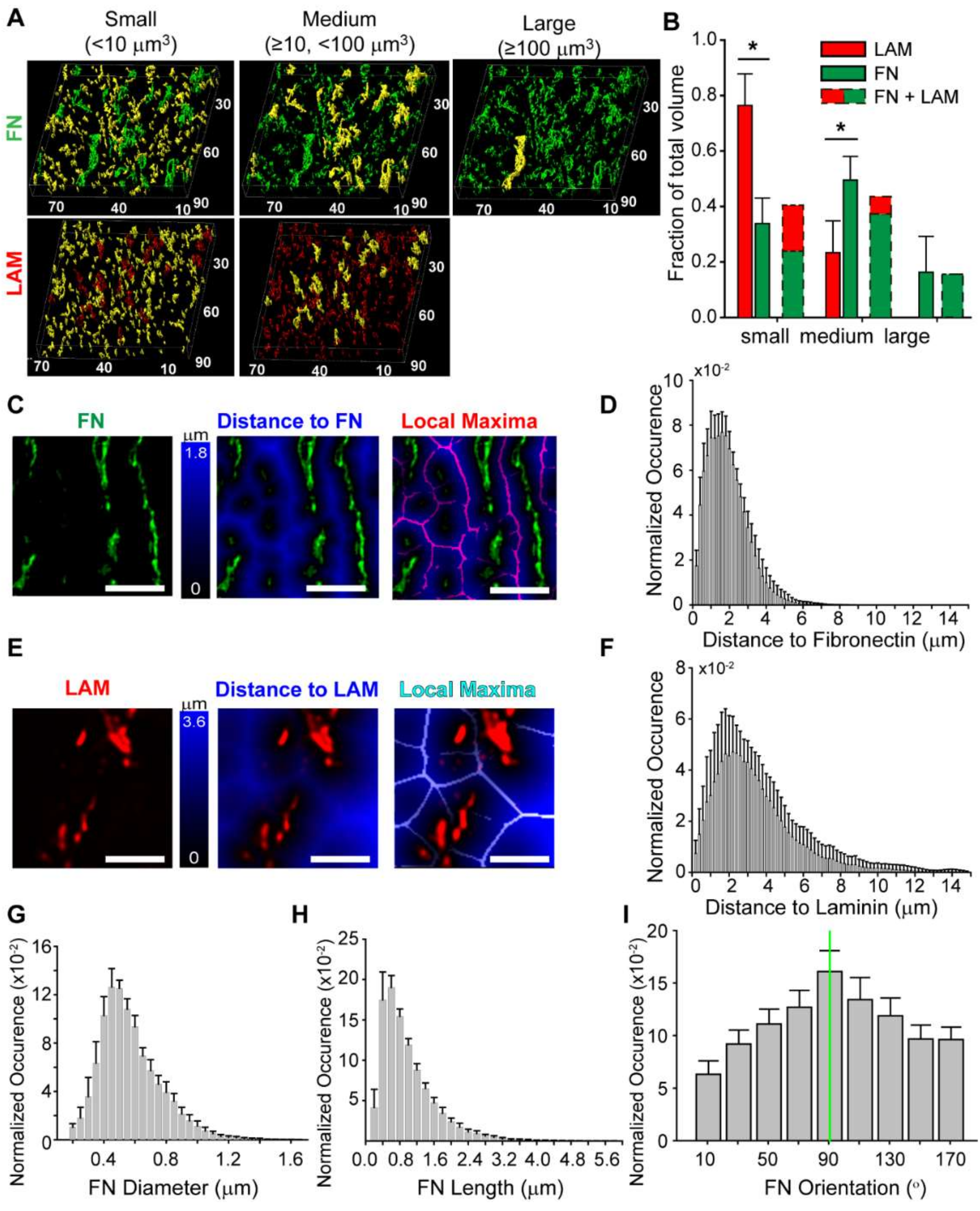
3.4. Quantitative Analysis of the Fibronectin and Laminin Matrix
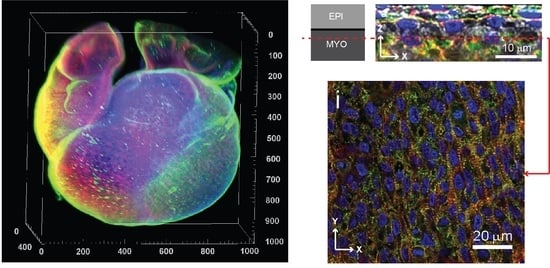
3.5. Fibronectin Localization at the Whole-Heart Scale
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Taber, L.A. Mechanical aspects of cardiac development. Prog. Biophys. Mol. Biol. 1998, 69, 237–255. [Google Scholar] [CrossRef]
- Harvey, R.P. Organogenesis: Patterning the vertebrate heart. Nat. Rev. Genet. 2002, 3, 544. [Google Scholar] [CrossRef]
- Sedmera, D.; Pexieder, T.; Vuillemin, M.; Thompson, R.P.; Anderson, R.H. Developmental patterning of the myocardium. Anat. Rec. 2000, 258, 319–337. [Google Scholar] [CrossRef]
- Olivey Harold, E.; Svensson Eric, C. Epicardial–Myocardial Signaling Directing Coronary Vasculogenesis. Circ. Res. 2010, 106, 818–832. [Google Scholar] [CrossRef] [PubMed]
- Red-Horse, K.; Ueno, H.; Weissman, I.L.; Krasnow, M.A. Coronary arteries form by developmental reprogramming of venous cells. Nature 2010, 464, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Olivey, H.E.; Compton, L.A.; Barnett, J.V. Coronary Vessel Development: The Epicardium Delivers. Trends Cardiovasc. Med. 2004, 14, 247–251. [Google Scholar] [PubMed]
- Pennisi, D.J.; Rentschler, S.; Gourdie, R.G.; Fishman, G.I.; Mikawa, T. Induction and patterning of the cardiac conduction system. Int. J. Dev. Biol. 2002, 46, 765–775. [Google Scholar]
- Martinsen, B.J. Reference guide to the stages of chick heart embryology. Dev. Dyn. 2005, 233, 1217–1237. [Google Scholar] [CrossRef]
- Shiraishi, I.; Takamatsu, T.; Fujita, S. Three-dimensional observation with a confocal scanning laser microscope of fibronectin immunolabeling during cardiac looping in the chick embryo. Anat. Embryol. 1995, 191, 183–189. [Google Scholar] [CrossRef]
- Lockhart, M.; Wirrig, E.; Phelps, A.; Wessels, A. Extracellular matrix and heart development. Birth Defects. Res. Part. A: Clin. Mol. Teratol. 2011, 91, 535–550. [Google Scholar] [CrossRef]
- Hanson, K.P.; Jung, J.P.; Tran, Q.A.; Hsu, S.-P.P.; Iida, R.; Ajeti, V.; Campagnola, P.J.; Eliceiri, K.W.; Squirrell, J.M.; Lyons, G.E.; et al. Spatial and Temporal Analysis of Extracellular Matrix Proteins in the Developing Murine Heart: A Blueprint for Regeneration. Tissue Eng. Part. A 2012, 19, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Quinn, K.P.; Georgakoudi, I.; Black, L.D. Young developmental age cardiac extracellular matrix promotes the expansion of neonatal cardiomyocytes in vitro. Acta Biomater. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Davidson, L.A. Punctuated actin contractions during convergent extension and their permissive regulation by the non-canonical Wnt-signaling pathway. J. Cell Sci. 2011, 124, 635. [Google Scholar] [CrossRef]
- Besseling, T.H.; Jose, J.; Van Blaaderen, A. Methods to calibrate and scale axial distances in confocal microscopy as a function of refractive index. J. Microsc. 2015, 257, 142–150. [Google Scholar] [CrossRef]
- Rakusan, K.; Flanagan, M.F.; Geva, T.; Southern, J.; Van Praagh, R. Morphometry of human coronary capillaries during normal growth and the effect of age in left ventricular pressure-overload hypertrophy. Circulation 1992, 86, 38–46. [Google Scholar] [CrossRef]
- Rongish, B.J.; Hinchman, G.; Doty, M.K.; Baldwin, H.S.; Tomanek, R.J. Relationship of the Extracellular Matrix to Coronary Neovascularization During Development. J. Mol. Cell. Cardiol. 1996, 28, 2203–2215. [Google Scholar] [CrossRef]
- Icardo, J.M.; Fernandez-Teran, A. Morphologic study of ventricular trabeculation in the embryonic chick heart. Acta Anat. 1987, 130, 264–274. [Google Scholar] [CrossRef]
- Williams, C.; Sullivan, K.; Black, L.D. Partially Digested Adult Cardiac Extracellular Matrix Promotes Cardiomyocyte Proliferation in Vitro. Adv. Healthc. Mater. 2015, 4, 1545–1554. [Google Scholar] [CrossRef]
- Astrof, S.; Hynes, R.O. Fibronectins in vascular morphogenesis. Angiogenesis 2009, 12, 165–175. [Google Scholar] [CrossRef]
- Astrof, S.; Crowley, D.; Hynes, R.O. Multiple cardiovascular defects caused by the absence of alternatively spliced segments of fibronectin. Dev. Biol. 2007, 311, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Riley, P.R.; Smart, N. Vascularizing the heart. Cardiovasc. Res. 2011, 91, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Reese, D.E.; Mikawa, T.; Bader, D.M. Development of the Coronary Vessel System. Circ. Res. 2002, 91, 761–768. [Google Scholar] [CrossRef]
- Wada, A.M.; Willet, S.G.; Bader, D. Coronary Vessel Development. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 2138–2145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Majesky, M.W. Development of Coronary Vessels. In Current Topics in Developmental Biology; Academic Press: Cambridge, MA, USA, 2004; Volume 62, pp. 225–259. [Google Scholar]
- Manisastry, S.M.; Zaal, K.J.M.; Horowits, R. Myofibril assembly visualized by imaging N-RAP, alpha-actinin, and actin in living cardiomyocytes. Exp. Cell Res. 2009, 315, 2126–2139. [Google Scholar] [CrossRef]
- De Tombe, P.P. Cardiac myofilaments: Mechanics and regulation. J. Biomech. 2003, 36, 721–730. [Google Scholar] [CrossRef]
- Sabatier, L.; Chen, D.; Fagotto-Kaufmann, C.; Hubmacher, D.; McKee, M.D.; Annis, D.S.; Mosher, D.F.; Reinhardt, D.P. Fibrillin Assembly Requires Fibronectin. Mol. Biol. Cell 2008, 20, 846–858. [Google Scholar] [CrossRef]
- Cox, G.; Sheppard, C.J.R. Practical limits of resolution in confocal and non-linear microscopy. Microsc. Res. Tech. 2004, 63, 18–22. [Google Scholar] [CrossRef]
- Erickson, H.P.; Carrell, N.; McDonagh, J. Fibronectin molecule visualized in electron microscopy: A long, thin, flexible strand. J. Cell Biol. 1981, 91, 673–678. [Google Scholar] [CrossRef]
- Erickson, H.P. Stretching fibronectin. J. Muscle Res. Cell Motil 2002, 23, 575–580. [Google Scholar] [CrossRef]
- Szymanski, J.M.; Sevcik, E.N.; Zhang, K.; Feinberg, A.W. Stretch-dependent changes in molecular conformation in fibronectin nanofibers. Biomater. Sci. 2017, 5, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, T.; Kiehart, D.P.; Erickson, H.P. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc. Natl. Acad. Sci. USA 1999, 96, 2153–2158. [Google Scholar] [CrossRef] [PubMed]
- Diao, J.; Maniotis, A.J.; Folberg, R.; Tajkhorshid, E. Interplay of mechanical and binding properties of Fibronectin type I. Chem. Acc. 2010, 125, 397–405. [Google Scholar] [CrossRef]
- Cao, L.; Zeller, M.K.; Fiore, V.F.; Strane, P.; Bermudez, H.; Barker, T.H. Phage-based molecular probes that discriminate force-induced structural states of fibronectin in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, 7251–7256. [Google Scholar] [CrossRef]
- Liu, A.P.; Chaudhuri, O.; Parekh, S.H. New advances in probing cell–extracellular matrix interactions. Integr. Biol. 2017, 9, 383–405. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.W.; Alford, P.W.; Jin, H.; Ripplinger, C.M.; Werdich, A.A.; Sheehy, S.P.; Grosberg, A.; Parker, K.K. Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture. Biomaterials 2012, 33, 5732–5741. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.W.; Feigel, A.; Shevkoplyas, S.S.; Sheehy, S.; Whitesides, G.M.; Parker, K.K. Muscular Thin Films for Building Actuators and Powering Devices. Science 2007, 317, 1366–1370. [Google Scholar] [CrossRef]
- Kim, D.-H.; Lipke, E.A.; Kim, P.; Cheong, R.; Thompson, S.; Delannoy, M.; Suh, K.-Y.; Tung, L.; Levchenko, A. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc. Natl. Acad. Sci. USA 2010, 107, 565–570. [Google Scholar] [CrossRef]
- Feinberg, A.W.; Ripplinger, C.M.; van der Meer, P.; Sheehy, S.P.; Domian, I.; Chien, K.R.; Parker, K.K. Functional Differences in Engineered Myocardium from Embryonic Stem Cell-Derived versus Neonatal Cardiomyocytes. Stem Cell Rep. 2013, 1, 387–396. [Google Scholar] [CrossRef]
- Domian, I.J.; Chiravuri, M.; van der Meer, P.; Feinberg, A.W.; Shi, X.; Shao, Y.; Wu, S.M.; Parker, K.K.; Chien, K.R. Generation of Functional Ventricular Heart Muscle from Mouse Ventricular Progenitor Cells. Science 2009, 326, 426–429. [Google Scholar] [CrossRef]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Hudson, A.R.; Shiwarski, D.J.; Tashman, J.W.; Hinton, T.J.; Yerneni, S.; Bliley, J.M.; Campbell, P.G.; Feinberg, A.W. 3D bioprinting of collagen to rebuild components of the human heart. Science 2019, 365, 482–487. [Google Scholar] [CrossRef]
- Lee, A.; Feinberg, A.W. 3D bioprinting of cardiac muscle tissue. In 3D Bioprinting in Regenerative Engineering: Principles and Applications, 1st ed.; Khademhosseini, A., Camci-Unal, G., Eds.; CRC Press: Boca Raton, FL, USA, 2018; p. 22. [Google Scholar] [CrossRef]
- Garreta, E.; Oria, R.; Tarantino, C.; Pla-Roca, M.; Prado, P.; Fernández-Avilés, F.; Campistol, J.M.; Samitier, J.; Montserrat, N. Tissue engineering by decellularization and 3D bioprinting. Mater. Today 2017, 20, 166–178. [Google Scholar] [CrossRef]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.-K.; Black, L.D.; Kren, S.M.; Netoff, T.I.; Taylor, D.A. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jallerat, Q.; Feinberg, A.W. Extracellular Matrix Structure and Composition in the Early Four-Chambered Embryonic Heart. Cells 2020, 9, 285. https://doi.org/10.3390/cells9020285
Jallerat Q, Feinberg AW. Extracellular Matrix Structure and Composition in the Early Four-Chambered Embryonic Heart. Cells. 2020; 9(2):285. https://doi.org/10.3390/cells9020285
Chicago/Turabian StyleJallerat, Quentin, and Adam W. Feinberg. 2020. "Extracellular Matrix Structure and Composition in the Early Four-Chambered Embryonic Heart" Cells 9, no. 2: 285. https://doi.org/10.3390/cells9020285
APA StyleJallerat, Q., & Feinberg, A. W. (2020). Extracellular Matrix Structure and Composition in the Early Four-Chambered Embryonic Heart. Cells, 9(2), 285. https://doi.org/10.3390/cells9020285