Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Viral Packaging and Transduction
2.3. Isolation of MEFs
2.4. Derivation of Neonatal Fibroblasts
2.5. Magnetic Cell Sorting
2.6. Flow Cytometry
2.7. Immunocytochemistry (ICC)
2.8. Quantitative Real-Time qPCR
2.9. Western Blots
2.10. Chromatin Immunoprecipitation (ChIP) Followed with qPCR (ChIP-qPCR)
2.11. Statistical Analysis
3. Results
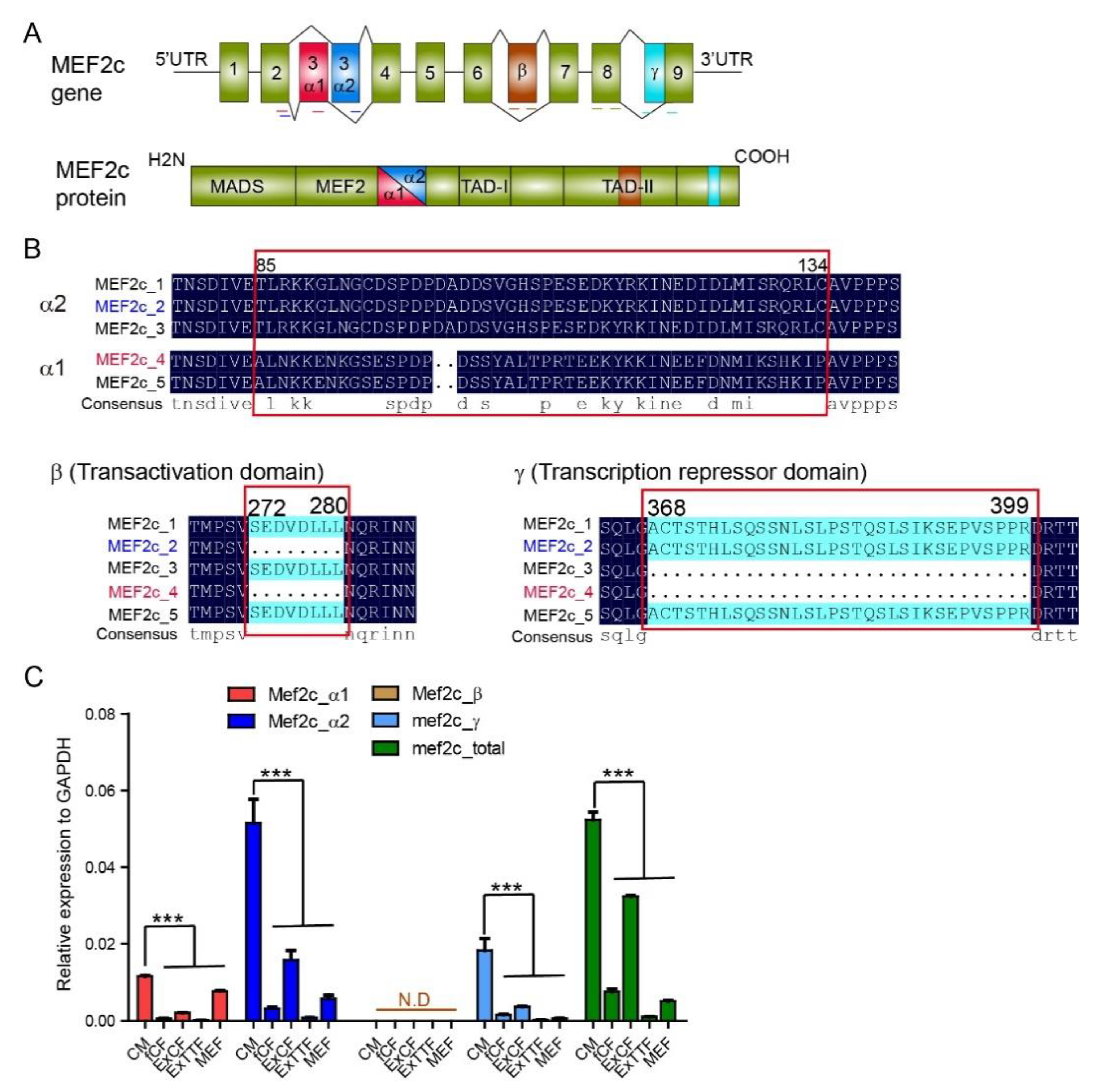
3.1. Expression of Mef2C Isoforms in Primary Cardiomyocytes and Fibroblasts
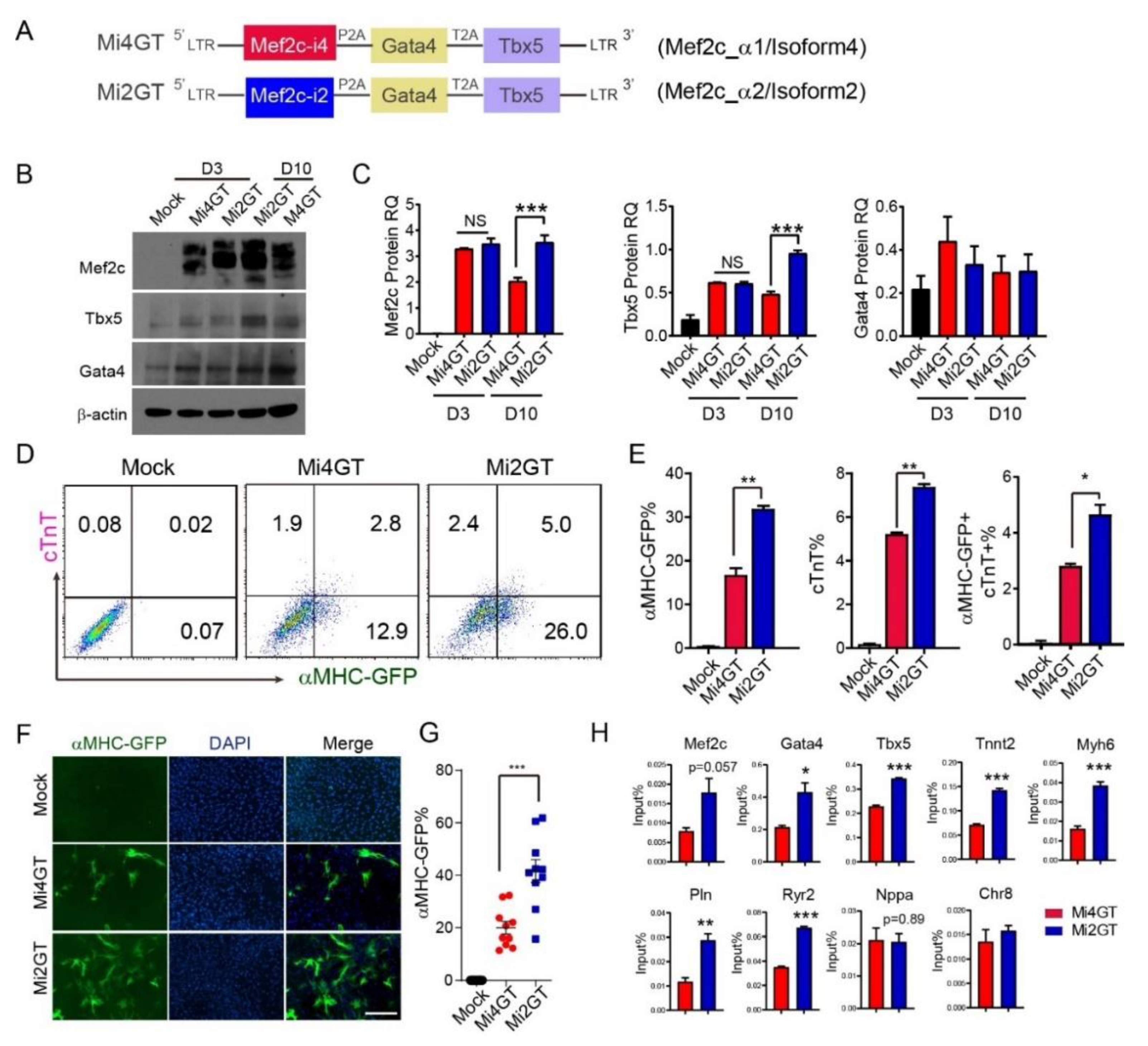
3.2. Mef2C Isoform 2 Induced Higher iCM Reprogramming Efficiency in MEFs When Using Polycistronic Construct
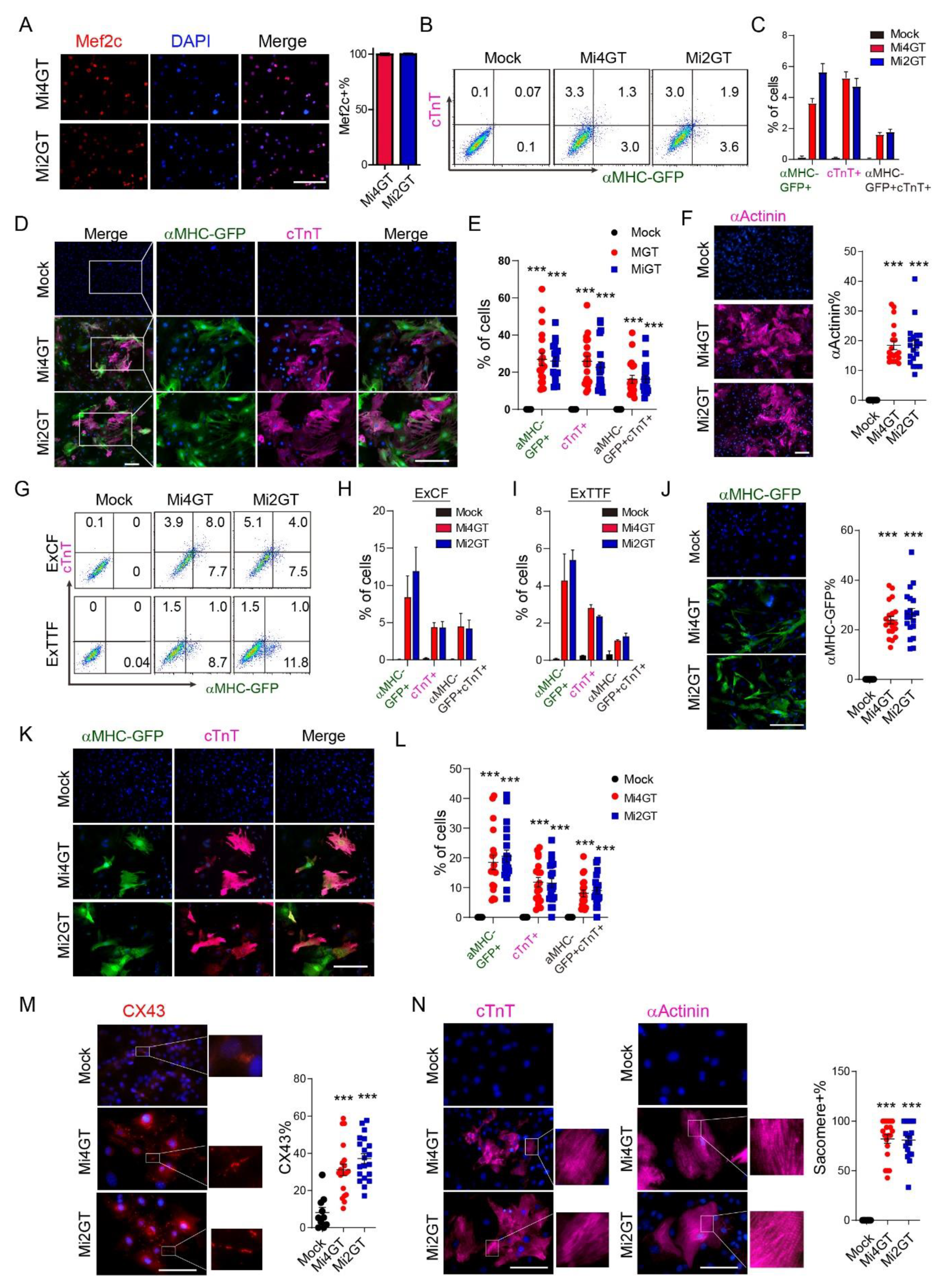
3.3. Mef2C Isoforms Yielded Similar Reprogramming Efficiency in CFs and TTFs Using Polycistronic Constructs
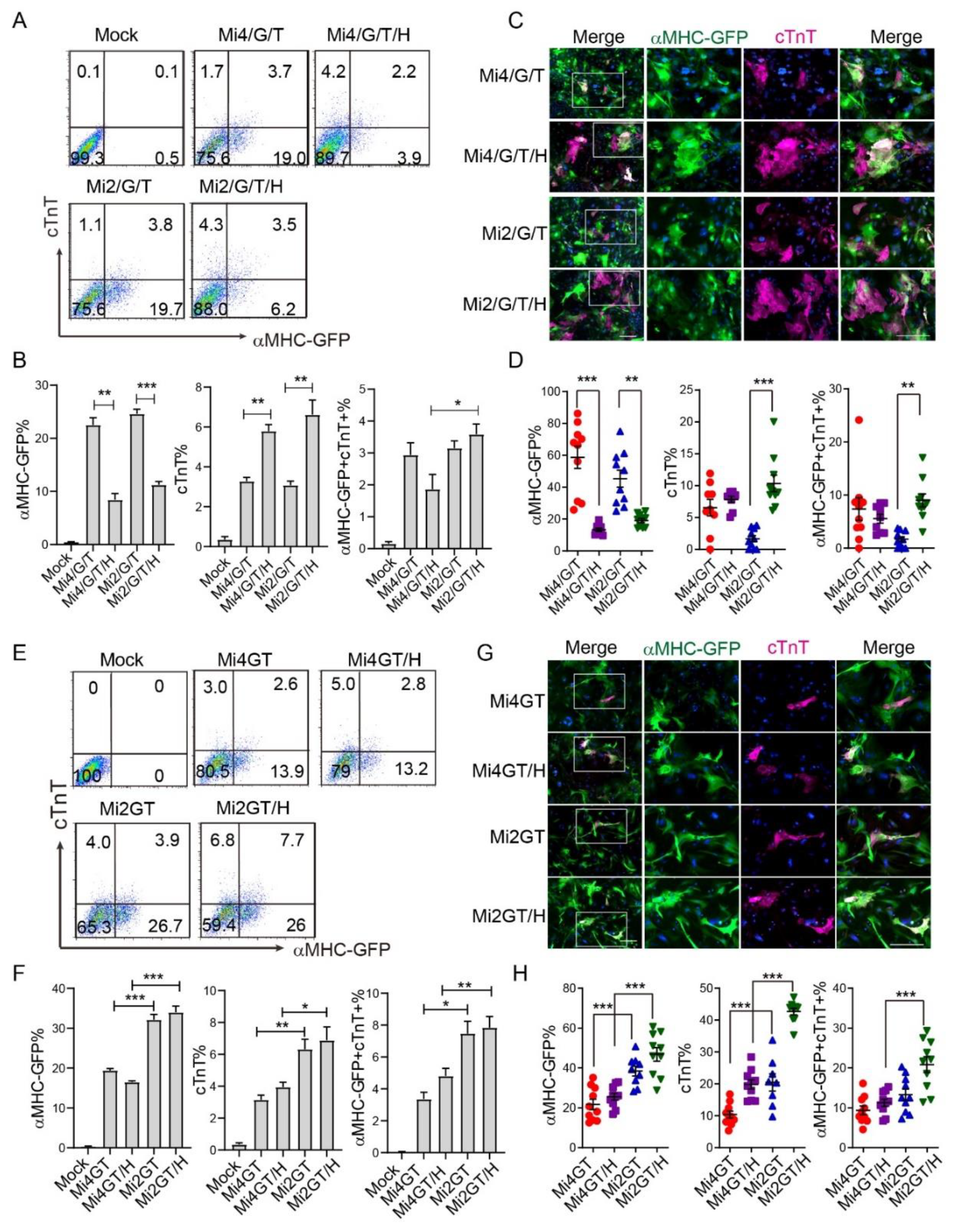
3.4. Addition of Hand2 Enhanced Both Mef2C Isoforms Mediated iCM Reprogramming when Using Separate Factors
3.5. Hand2 Enhanced Mef2C Isoform 2 but Not Isoform 4 Mediated iCM Reprogramming from MEF When Using Polycistronic Constructs
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Roth, G.A.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [Green Version]
- Muraoka, N.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Isomi, M.; Nakashima, H.; Akiyama, M.; Wada, R.; Inagawa, K.; et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014, 33, 1565–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; Dimaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.D.; Stone, N.R.; Liu, L.; Spencer, C.I.; Qian, L.; Hayashi, Y.; Delgado-Olguin, P.; Ding, S.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013, 1, 235–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599–604. [Google Scholar] [CrossRef] [Green Version]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593–598. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Wang, L.; Yin, C.; Liu, J.; Qian, L. In vivo cardiac reprogramming using an optimal single polycistronic construct. Cardiovasc. Res. 2015, 108, 217–219. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.M.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 Influences the Efficiency and Quality of Induced Cardiac Myocyte Reprogramming. Circ. Res. 2015, 116, 237–244. [Google Scholar] [CrossRef]
- Christoforou, N.; Chellappan, M.; Adler, A.F.; Kirkton, R.D.; Wu, T.; Addis, R.C.; Bursac, N.; Leong, K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE 2013, 8, e63577. [Google Scholar] [CrossRef] [Green Version]
- Bichsel, C.; Neeld, D.; Hamazaki, T.; Chang, L.J.; Yang, L.J.; Terada, N.; Jin, S. Direct reprogramming of fibroblasts to myocytes via bacterial injection of MyoD protein. Cell. Reprogram. 2013, 15, 117–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Tu, W.; Fu, Y.; Wang, J.; Xie, X. Chemical-induced cardiac reprogramming in vivo. Cell Res. 2018, 28, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Ifkovits, J.L.; Addis, R.C.; Epstein, J.A.; Gearhart, J.D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE 2014, 9, e89678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, T.M.; Stone, N.R.; Berry, E.C.; Radzinsky, E.; Huang, Y.; Pratt, K.; Ang, Y.S.; Yu, P.; Wang, H.; Tang, S.; et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation 2017, 135, 978–995. [Google Scholar] [CrossRef]
- Yamakawa, H.; Muraoka, N.; Miyamoto, K.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Umei, T.; Akiyama, M.; Kuishi, Y.; et al. Fibroblast Growth Factors and Vascular Endothelial Growth Factor Promote Cardiac Reprogramming under Defined Conditions. Stem Cell Rep. 2015, 5, 1128–1142. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Londono, P.; Cao, Y.; Sharpe, E.J.; Proenza, C.; O’Rourke, R.; Jones, K.L.; Jeong, M.Y.; Walker, L.A.; Buttrick, P.M.; et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015, 6, 8243. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Morales, M.G.; Hashimoto, H.; Dickson, M.E.; Song, K.; Ye, W.; Kim, M.S.; Niederstrasser, H.; Wang, Z.; Chen, B.; et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017, 31, 1770–1783. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.D.; Wang, G.G.; Liu, J.; et al. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, K.; Akiyama, M.; Tamura, F.; Isomi, M.; Yamakawa, H.; Sadahiro, T.; Muraoka, N.; Kojima, H.; Haginiwa, S.; Kurotsu, S.; et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 2018, 22, 91–103.e105. [Google Scholar] [CrossRef] [Green Version]
- Abad, M.; Hashimoto, H.; Zhou, H.; Morales, M.G.; Chen, B.; Bassel-Duby, R.; Olson, E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Rep. 2017. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.; Yin, C.; Zhou, Y.; Liu, J.; Qian, L. Improved Generation of Induced Cardiomyocytes Using a Polycistronic Construct Expressing Optimal Ratio of Gata4, Mef2c and Tbx5. J. Vis. Exp. 2015, 105, e53426. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Chen, O.; Zheng, M.; Wang, L.; Zhou, Y.; Yin, C.; Liu, J.; Qian, L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016, 16, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.Z.; Valdez, M.R.; McAnally, J.; Richardson, J.; Olson, E.N. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development 2001, 128, 4623–4633. [Google Scholar]
- Suzuki, E.; Lowry, J.; Sonoda, G.; Testa, J.R.; Walsh, K. Structures and chromosome locations of the human MEF2A gene and a pseudogene MEF2AP. Cytogenet. Genome Res. 1996, 73, 244–249. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Olson, E.N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 1996, 93, 9366–9373. [Google Scholar] [CrossRef] [Green Version]
- Hakim, N.H.; Kounishi, T.; Alam, A.H.; Tsukahara, T.; Suzuki, H. Alternative splicing of Mef2c promoted by Fox-1 during neural differentiation in P19 cells. Genes Cells 2010, 15, 255–267. [Google Scholar] [CrossRef]
- Sekiyama, Y.; Suzuki, H.; Tsukahara, T. Functional gene expression analysis of tissue-specific isoforms of Mef2c. Cell. Mol. Neurobiol. 2012, 32, 129–139. [Google Scholar] [CrossRef]
- Qian, L.; Berry, E.C.; Fu, J.D.; Ieda, M.; Srivastava, D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat. Protoc. 2013, 8, 1204–1215. [Google Scholar] [CrossRef]
- Saffitz, J.E.; Green, K.G.; Kraft, W.J.; Schechtman, K.B.; Yamada, K.A. Effects of diminished expression of connexin43 on gap junction number and size in ventricular myocardium. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H1662–H1670. [Google Scholar] [CrossRef]
- Liu, L.; Lei, I.; Karatas, H.; Li, Y.; Wang, L.; Gnatovskiy, L.; Dou, Y.; Wang, S.; Qian, L.; Wang, Z. Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discov. 2016, 2, 16036. [Google Scholar] [CrossRef]
- Li, Y.; Dal-Pra, S.; Mirotsou, M.; Jayawardena, T.M.; Hodgkinson, C.P.; Bursac, N.; Dzau, V.J. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci. Rep. 2016, 6, 38815. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D. Making or breaking the heart: From lineage determination to morphogenesis. Cell 2006, 126, 1037–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, A.; Tanos, R.; El-Hachem, N.; Kurban, M.; Bouvagnet, P.; Bitar, F.; Nemer, G. A HAND to TBX5 Explains the Link Between Thalidomide and Cardiac Diseases. Sci. Rep. 2017, 7, 1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, M.X.; Li, Y.; Xue, L.X.; Jia, H.T.; Jing, H. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J. Cell. Biochem. 2004, 93, 1255–1266. [Google Scholar] [CrossRef]
- Dai, Y.S.; Cserjesi, P.; Markham, B.E.; Molkentin, J.D. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem. 2002, 277, 24390–24398. [Google Scholar] [CrossRef] [Green Version]
- Umei, T.C.; Yamakawa, H.; Muraoka, N.; Sadahiro, T.; Isomi, M.; Haginiwa, S.; Kojima, H.; Kurotsu, S.; Tamura, F.; Osakabe, R.; et al. Single-Construct Polycistronic Doxycycline-Inducible Vectors Improve Direct Cardiac Reprogramming and Can Be Used to Identify the Critical Timing of Transgene Expression. Int. J. Mol. Sci. 2017, 18, 1805. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Huang, P.; Near, D.; Ravi, K.; Xu, Y.; Liu, J.; Qian, L. Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming. Cells 2020, 9, 268. https://doi.org/10.3390/cells9020268
Wang L, Huang P, Near D, Ravi K, Xu Y, Liu J, Qian L. Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming. Cells. 2020; 9(2):268. https://doi.org/10.3390/cells9020268
Chicago/Turabian StyleWang, Li, Peisen Huang, David Near, Karan Ravi, Yangxi Xu, Jiandong Liu, and Li Qian. 2020. "Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming" Cells 9, no. 2: 268. https://doi.org/10.3390/cells9020268
APA StyleWang, L., Huang, P., Near, D., Ravi, K., Xu, Y., Liu, J., & Qian, L. (2020). Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming. Cells, 9(2), 268. https://doi.org/10.3390/cells9020268






