Experimental and Computational Approaches to Direct Cell Reprogramming: Recent Advancement and Future Challenges
Abstract
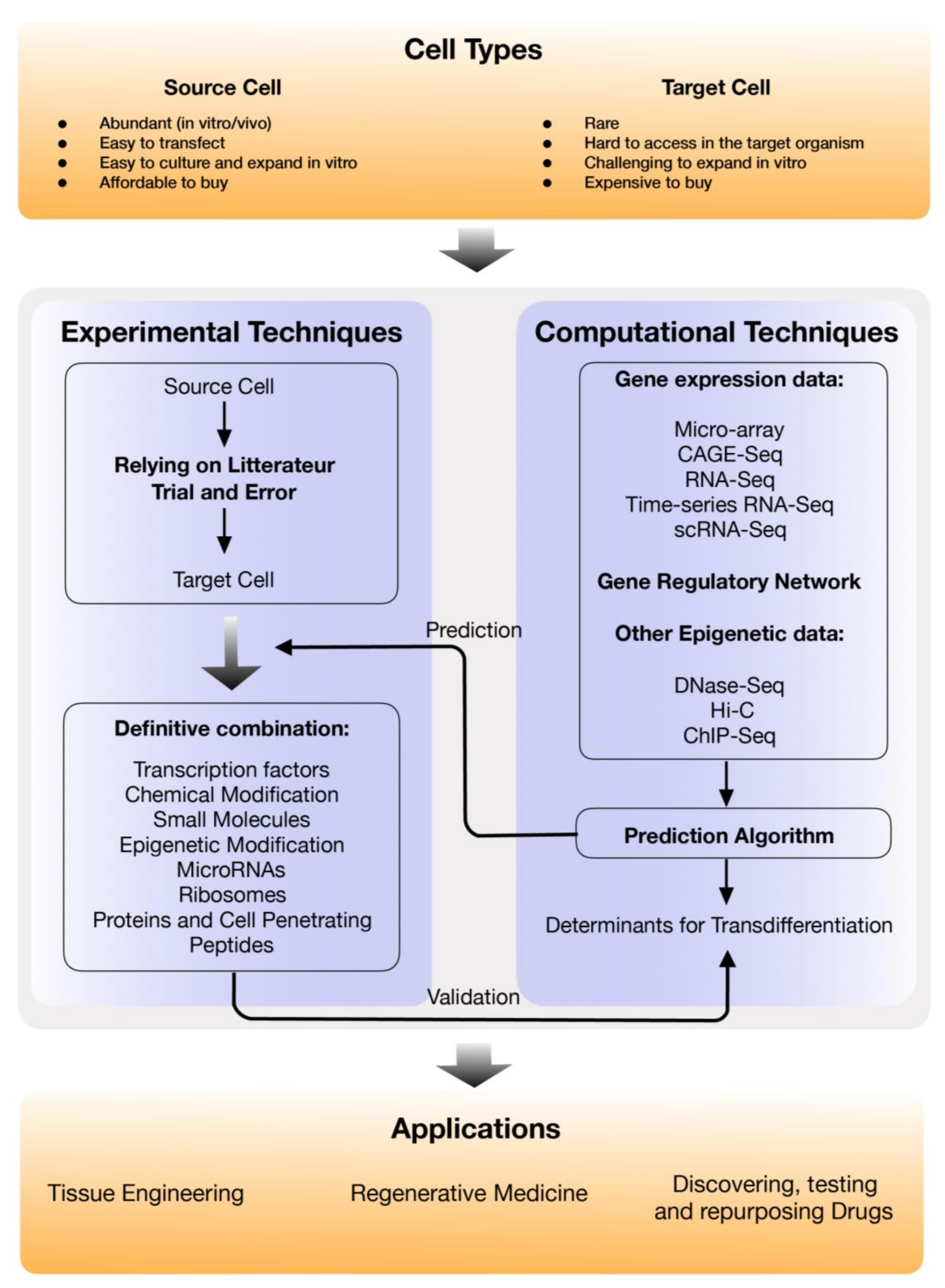
1. Introduction
2. Experimental Methods for Direct Cell Reprogramming and the Applications
2.1. Transcription Factors
2.2. Chemical Modification and Small Molecules
2.3. Epigenetic Modification
2.4. MicroRNA
2.5. Ribosomes
2.6. Proteins and Cell Penetrating Peptides
3. In-Silico Direct Reprogramming Factor Prediction
4. Major Applications of Direct Cell Reprogramming
5. Challenges and Prospective Solutions for Integrating Direct Cell Reprogramming in Clinics
Author Contributions
Funding
Conflicts of Interest
References
- Waddington, C.H. The Strategy of the Genes a Discussion of Some Aspects of Theoretical Biology; George Allen & Unwin: London, UK, 1957. [Google Scholar]
- Gurdon, J.B.; Elsdale, T.R.; Fischberg, M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 1958, 182, 64–65. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Tada, M.; Takahama, Y.; Abe, K.; Nakatsuji, N.; Tada, T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001, 11, 1553–1558. [Google Scholar] [CrossRef]
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H. Viable offspring derived from fetal and adult mammalian cells. Nature 1997, 385, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, S.; Subramaniam, S.; Shyy, J.Y.; Chien, S. Epigenetic Regulation: A New Frontier for Biomedical Engineers. Annu Rev. Biomed. Eng. 2017, 19, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.V. iPS cells: A more critical review. Stem Cells Dev. 2008, 17, 391–397. [Google Scholar] [CrossRef]
- Maherali, N.; Hochedlinger, K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell 2008, 3, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953. [Google Scholar] [CrossRef]
- Patel, M.; Yang, S. Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Rev. 2010, 6, 367–380. [Google Scholar] [CrossRef]
- Tweedell, K.S. New paths to pluripotent stem cells. Curr Stem Cell Res. 2008, 3, 151–162. [Google Scholar] [CrossRef]
- Singh, V.K.; Kalsan, M.; Kumar, N.; Saini, A.; Chandra, R. Induced pluripotent stem cells: Applications in regenerative medicine, disease modeling, and drug discovery. Front. Cell Dev. Biol. 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Atala, A. Regenerative medicine strategies. J. Pediatr Surg 2012, 47, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Brunt, K.R.; Weisel, R.D.; Li, R.K. Stem cells and regenerative medicine - future perspectives. Can. J. Physiol Pharm. 2012, 90, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Kavyasudha, C.; Macrin, D.; ArulJothi, K.N.; Joseph, J.P.; Harishankar, M.K.; Devi, A. Clinical Applications of Induced Pluripotent Stem Cells - Stato Attuale. Adv. Exp. Med. Biol. 2018, 1079, 127–149. [Google Scholar] [PubMed]
- Reardon, S.; Cyranoski, D. Japan stem-cell trial stirs envy. Nature 2014, 513, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Tabar, V.; Studer, L. Pluripotent stem cells in regenerative medicine: Challenges and recent progress. Nat. Rev. Genet. 2014, 15, 82–92. [Google Scholar] [CrossRef]
- Wu, S.M.; Hochedlinger, K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat. Cell Biol. 2011, 13, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Okano, H.; Nakamura, M.; Yoshida, K.; Okada, Y.; Tsuji, O.; Nori, S.; Ikeda, E.; Yamanaka, S.; Miura, K. Steps toward safe cell therapy using induced pluripotent stem cells. Circ. Res. 2013, 112, 523–533. [Google Scholar] [CrossRef]
- Graf, T.; Enver, T. Forcing cells to change lineages. Nature 2009, 462, 587–594. [Google Scholar] [CrossRef]
- Xu, J.; Du, Y.; Deng, H. Direct lineage reprogramming: Strategies, mechanisms, and applications. Cell Stem Cell 2015, 16, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Grath, A.; Dai, G. Direct cell reprogramming for tissue engineering and regenerative medicine. J. Biol Eng. 2019, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Cahan, P.; Li, H.; Morris, S.A.; Da Rocha, E.L.; Daley, G.Q.; Collins, J.J. CellNet: Network biology applied to stem cell engineering. Cell 2014, 158, 903–915. [Google Scholar] [CrossRef] [PubMed]
- D’Alessio, A.C.; Fan, Z.P.; Wert, K.J.; Baranov, P.; Cohen, M.A.; Saini, J.S.; Cohick, E.; Charniga, C.; Dadon, D.; Hannett, N.M. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Rep. 2015, 5, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Okawa, S.; Nicklas, S.; Zickenrott, S.; Schwamborn, J.C.; Del Sol, A. A Generalized Gene-Regulatory Network Model of Stem Cell Differentiation for Predicting Lineage Specifiers. Stem Cell Rep. 2016, 7, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Rackham, O.J.; Firas, J.; Fang, H.; Oates, M.E.; Holmes, M.L.; Knaupp, A.S.; Consortium, F.; Suzuki, H.; Nefzger, C.M.; Daub, C.O.; et al. A predictive computational framework for direct reprogramming between human cell types. Nat. Genet. 2016, 48, 331–335. [Google Scholar] [CrossRef]
- Guerrero-Ramirez, G.I.; Valdez-Cordoba, C.M.; Islas-Cisneros, J.F.; Trevino, V. Computational approaches for predicting key transcription factors in targeted cell reprogramming. Mol. Med. Rep. 2018, 18, 1225–1237. [Google Scholar] [CrossRef]
- Schenborn, E.T.; Oler, J. Liposome-mediated transfection of mammalian cells. Methods Mol. Biol. 2000, 130, 155–164. [Google Scholar]
- Chesnoy, S.; Huang, L. Structure and function of lipid-DNA complexes for gene delivery. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 27–47. [Google Scholar] [CrossRef]
- Potter, H.; Heller, R. Transfection by electroporation. Curr. Protoc. Mol. Biol. 2018, 121, 9.3.1–9.3.13. [Google Scholar]
- Chow, Y.T.; Chen, S.; Wang, R.; Liu, C.; Kong, C.-w.; Li, R.A.; Cheng, S.H.; Sun, D. Single cell transfection through precise microinjection with quantitatively controlled injection volumes. Sci. Rep. 2016, 6, 24127. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Eberwine, J.H. Mammalian cell transfection: The present and the future. Anal. Bioanal. Chem. 2010, 397, 3173–3178. [Google Scholar] [CrossRef] [PubMed]
- Finer, M.; Glorioso, J. A Brief Account of Viral Vectors and Their Promise for Gene Therapy; Nature Publishing Group: London, UK, 2017. [Google Scholar]
- Nowakowski, A.; Andrzejewska, A.; Janowski, M.; Walczak, P.; Lukomska, B. Genetic engineering of stem cells for enhanced therapy. Acta Neurobiol Exp. (Wars) 2013, 73, 1–18. [Google Scholar] [PubMed]
- Cohen, S.; Au, S.; Panté, N. How viruses access the nucleus. Biochim. Et Biophys. Acta (Bba)-Mol. Cell Res. 2011, 1813, 1634–1645. [Google Scholar] [CrossRef]
- SM Wold, W.; Toth, K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr. Gene Ther. 2013, 13, 421–433. [Google Scholar] [CrossRef]
- Heinrich, C.; Blum, R.; Gascón, S.; Masserdotti, G.; Tripathi, P.; Sánchez, R.; Tiedt, S.; Schroeder, T.; Götz, M.; Berninger, B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010, 8, e1000373. [Google Scholar] [CrossRef] [PubMed]
- Karow, M.; Sánchez, R.; Schichor, C.; Masserdotti, G.; Ortega, F.; Heinrich, C.; Gascón, S.; Khan, M.A.; Lie, D.C.; Dellavalle, A. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 2012, 11, 471–476. [Google Scholar] [CrossRef]
- Corti, S.; Nizzardo, M.; Simone, C.; Falcone, M.; Donadoni, C.; Salani, S.; Rizzo, F.; Nardini, M.; Riboldi, G.; Magri, F. Direct reprogramming of human astrocytes into neural stem cells and neurons. Exp. Cell Res. 2012, 318, 1528–1541. [Google Scholar] [CrossRef]
- Forsberg, M.; Carlén, M.; Meletis, K.; Yeung, M.S.; Barnabé-Heider, F.; Persson, M.A.; Aarum, J.; Frisén, J. Efficient reprogramming of adult neural stem cells to monocytes by ectopic expression of a single gene. Proc. Natl. Acad. Sci. USA 2010, 107, 14657–14661. [Google Scholar] [CrossRef]
- Hendry, C.E.; Vanslambrouck, J.M.; Ineson, J.; Suhaimi, N.; Takasato, M.; Rae, F.; Little, M.H. Direct transcriptional reprogramming of adult cells to embryonic nephron progenitors. J. Am. Soc. Nephrol. 2013, 24, 1424–1434. [Google Scholar] [CrossRef]
- Lemper, M.; Leuckx, G.; Heremans, Y.; German, M.; Heimberg, H.; Bouwens, L.; Baeyens, L. Reprogramming of human pancreatic exocrine cells to β-like cells. Cell Death Differ. 2015, 22, 1117. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of myoblast to brown fat switch by a PRDM16–C/EBP-β transcriptional complex. Nature 2009, 460, 1154. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.-J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA 2013, 110, 5588–5593. [Google Scholar] [CrossRef] [PubMed]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA 2013, 110, 12667–12672. [Google Scholar] [CrossRef] [PubMed]
- Hiramatsu, K.; Sasagawa, S.; Outani, H.; Nakagawa, K.; Yoshikawa, H.; Tsumaki, N. Generation of hyaline cartilaginous tissue from mouse adult dermal fibroblast culture by defined factors. J. Clin. Investig. 2011, 121, 640–657. [Google Scholar] [CrossRef]
- Han, J.-K.; Chang, S.-H.; Cho, H.-J.; Choi, S.-B.; Ahn, H.-S.; Lee, J.; Jeong, H.; Youn, S.-W.; Lee, H.-J.; Kwon, Y.-W. Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation 2014, 130, 1168–1178. [Google Scholar] [CrossRef]
- Pereira, C.-F.; Chang, B.; Qiu, J.; Niu, X.; Papatsenko, D.; Hendry, C.E.; Clark, N.R.; Nomura-Kitabayashi, A.; Kovacic, J.C.; Ma’Ayan, A. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 2013, 13, 205–218. [Google Scholar] [CrossRef]
- Batta, K.; Florkowska, M.; Kouskoff, V.; Lacaud, G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014, 9, 1871–1884. [Google Scholar] [CrossRef]
- Szabo, E.; Rampalli, S.; Risueno, R.M.; Schnerch, A.; Mitchell, R.; Fiebig-Comyn, A.; Levadoux-Martin, M.; Bhatia, M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 2010, 468, 521. [Google Scholar] [CrossRef]
- Feng, R.; Desbordes, S.C.; Xie, H.; Tillo, E.S.; Pixley, F.; Stanley, E.R.; Graf, T. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6057–6062. [Google Scholar] [CrossRef]
- Ayo, C.M.; Dalalio, M.M.d.O.; Visentainer, J.E.L.; Reis, P.G.; Sippert, E.Â.; Jarduli, L.R.; Alves, H.V.; Sell, A.M. Genetic susceptibility to Chagas disease: An overview about the infection and about the association between disease and the immune response genes. Biomed Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Ang, C.E.; Davila, J.; Pak, C.; Mall, M.; Lee, Q.Y.; Ahlenius, H.; Jung, S.W.; Südhof, T.C.; Wernig, M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 2014, 3, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Zheng, Q.; Wu, J.; Xu, Z.; Wang, L.; Li, W.; Zhang, H.; Zhao, X.-Y.; Liu, L.; Wang, Z. Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 2012, 22, 208. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011, 475, 386. [Google Scholar] [CrossRef] [PubMed]
- Marro, S.; Pang, Z.P.; Yang, N.; Tsai, M.-C.; Qu, K.; Chang, H.Y.; Südhof, T.C.; Wernig, M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 2011, 9, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011, 475, 390. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.-d.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.-D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. nature 2008, 455, 627. [Google Scholar] [CrossRef]
- Banga, A.; Akinci, E.; Greder, L.V.; Dutton, J.R.; Slack, J.M. In vivo reprogramming of Sox9+ cells in the liver to insulin-secreting ducts. Proc. Natl. Acad. Sci. USA 2012, 109, 15336–15341. [Google Scholar] [CrossRef] [PubMed]
- Torper, O.; Pfisterer, U.; Wolf, D.A.; Pereira, M.; Lau, S.; Jakobsson, J.; Björklund, A.; Grealish, S.; Parmar, M. Generation of induced neurons via direct conversion in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 7038–7043. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Zang, T.; Zou, Y.; Fang, S.; Smith, D.K.; Bachoo, R.; Zhang, C.-L. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 2013, 15, 1164. [Google Scholar] [CrossRef] [PubMed]
- Rouaux, C.; Arlotta, P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nat. Cell Biol. 2013, 15, 214. [Google Scholar] [CrossRef] [PubMed]
- Riddell, J.; Gazit, R.; Garrison, B.S.; Guo, G.; Saadatpour, A.; Mandal, P.K.; Ebina, W.; Volchkov, P.; Yuan, G.-C.; Orkin, S.H. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 2014, 157, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795. [Google Scholar] [CrossRef] [PubMed]
- Baranek, M.; Belter, A.; Naskręt-Barciszewska, M.; Stobiecki, M.; Markiewicz, W.; Barciszewski, J. Effect of small molecules on cell reprogramming. Mol. Biosyst. 2017, 13, 277–313. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, L.-h.; Xie, X. iPSCs and small molecules: A reciprocal effort towards better approaches for drug discovery. Acta Pharmacol. Sin. 2013, 34, 765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, W.; Laurent, T.; Ding, S. Small molecules, big roles–the chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci 2012, 125, 5609–5620. [Google Scholar] [CrossRef]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Yin, X.; Yang, W.; Du, Y.; Hou, P.; Ge, J.; Liu, C.; Zhang, W.; Zhang, X. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011, 21, 196. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Liu, K.; Tang, S.; Ding, S. Chemical approaches to cell reprogramming. Curr. Opin. Genet. Dev. 2014, 28, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Qiu, B.; Guan, W.; Wang, Q.; Wang, M.; Li, W.; Gao, L.; Shen, L.; Huang, Y.; Xie, G.; et al. Direct Conversion of Normal and Alzheimer's Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell 2015, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yin, J.C.; Yeh, H.; Ma, N.X.; Lee, G.; Chen, X.A.; Wang, Y.; Lin, L.; Chen, L.; Jin, P.; et al. Small Molecules Efficiently Reprogram Human Astroglial Cells into Functional Neurons. Cell Stem Cell 2015, 17, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Huang, C.; Xu, X.; Gu, H.; Ye, Y.; Jiang, C.; Qiu, Z.; Xie, X. Direct reprogramming of mouse fibroblasts into cardiomyocytes with chemical cocktails. Cell Res. 2015, 25, 1013. [Google Scholar] [CrossRef] [PubMed]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.-D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef]
- Xie, X.; Fu, Y.; Liu, J. Chemical reprogramming and transdifferentiation. Curr. Opin. Genet. Dev. 2017, 46, 104–113. [Google Scholar] [CrossRef]
- Sakurada, K. Environmental epigenetic modifications and reprogramming-recalcitrant genes. Stem Cell Res. 2010, 4, 157–164. [Google Scholar] [CrossRef][Green Version]
- Paksa, A.; Rajagopal, J. The epigenetic basis of cellular plasticity. Curr. Opin. Cell Biol. 2017, 49, 116–122. [Google Scholar] [CrossRef]
- Barneda-Zahonero, B.; Román-González, L.; Collazo, O.; Rafati, H.; Islam, A.B.; Bussmann, L.H.; Di Tullio, A.; De Andres, L.; Graf, T.; López-Bigas, N. HDAC7 is a repressor of myeloid genes whose downregulation is required for transdifferentiation of pre-B cells into macrophages. PLoS Genet. 2013, 9, e1003503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, L.; Vaseghi, H.R.; Liu, Z.; Lu, R.; Alimohamadi, S.; Yin, C.; Fu, J.-D.; Wang, G.G.; Liu, J. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell 2016, 18, 382–395. [Google Scholar] [CrossRef] [PubMed]
- Bramswig, N.C.; Everett, L.J.; Schug, J.; Dorrell, C.; Liu, C.; Luo, Y.; Streeter, P.R.; Naji, A.; Grompe, M.; Kaestner, K.H. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J. Clin. Investig. 2013, 123, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Björklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef]
- Son, E.Y.; Ichida, J.K.; Wainger, B.J.; Toma, J.S.; Rafuse, V.F.; Woolf, C.J.; Eggan, K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 2011, 9, 205–218. [Google Scholar] [CrossRef]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Ohta, K. Reprogramming of human somatic cells by bacteria. Dev. Growth Differ. 2015, 57, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.N.; Cate, J.H.D. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012, 4, a011536. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003, 19, 415–417. [Google Scholar] [CrossRef]
- Ohta, K.; Kawano, R.; Ito, N. Lactic acid bacteria convert human fibroblasts to multipotent cells. PLoS ONE 2012, 7, e51866. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Katoh, K.; Kushige, H.; Saito, Y.; Umemoto, T.; Matsuzaki, Y.; Kiyonari, H.; Kobayashi, D.; Soga, M.; Era, T. Ribosome incorporation into somatic cells promotes lineage transdifferentiation towards multipotency. Sci. Rep. 2018, 8, 1634. [Google Scholar] [CrossRef] [PubMed]
- Collas, P.; Håkelien, A.-M. Teaching cells new tricks. Trends Biotechnol. 2003, 21, 354–361. [Google Scholar] [CrossRef]
- Håkelien, A.-M.; Landsverk, H.B.; Robl, J.M.; Skålhegg, B.S.; Collas, P. Reprogramming fibroblasts to express T-cell functions using cell extracts. Nat. Biotechnol. 2002, 20, 460. [Google Scholar] [CrossRef]
- Landsverk, H.B.; Håkelien, A.M.; Küntziger, T.; Robl, J.M.; Skålhegg, B.S.; Collas, P. Reprogrammed gene expression in a somatic cell-free extract. Embo Rep. 2002, 3, 384–389. [Google Scholar] [CrossRef]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Elliott, G.; O'Hare, P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 1997, 88, 223–233. [Google Scholar] [CrossRef]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Islas, J.F.; Liu, Y.; Weng, K.-C.; Robertson, M.J.; Zhang, S.; Prejusa, A.; Harger, J.; Tikhomirova, D.; Chopra, M.; Iyer, D. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc. Natl. Acad. Sci. USA 2012, 109, 13016–13021. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Guo, Y.; Wang, C.; Liu, Q.; Yang, Y.; Li, S.; Guo, X.; Lian, R.; Yu, R.; Liu, H. Non-genetic direct reprogramming and biomimetic platforms in a preliminary study for adipose-derived stem cells into corneal endothelia-like cells. PLoS ONE 2014, 9, e109856. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Chen, R.; Teesalu, T.; Ruoslahti, E.; Clegg, D.O. Reprogramming human retinal pigmented epithelial cells to neurons using recombinant proteins. Stem Cells Transl. Med. 2014, 3, 1526–1534. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.S.; Geras-Raaka, E.; Gershengorn, M.C. Reprogramming adult human dermal fibroblasts to islet-like cells by epigenetic modification coupled to transcription factor modulation. Stem Cells Dev. 2013, 22, 2551–2560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lin, Y.-H.; Sun, Y.J.; Zhu, S.; Zheng, J.; Liu, K.; Cao, N.; Li, K.; Huang, Y.; Ding, S. Pharmacological reprogramming of fibroblasts into neural stem cells by signaling-directed transcriptional activation. Cell Stem Cell 2016, 18, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Kamaraj, U.S.; Gough, J.; Polo, J.M.; Petretto, E.; Rackham, O.J. Computational methods for direct cell conversion. Cell Cycle 2016, 15, 3343–3354. [Google Scholar] [CrossRef]
- Morris, S.A.; Cahan, P.; Li, H.; Zhao, A.M.; San Roman, A.K.; Shivdasani, R.A.; Collins, J.J.; Daley, G.Q. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 2014, 158, 889–902. [Google Scholar] [CrossRef]
- Radley, A.H.; Schwab, R.M.; Tan, Y.; Kim, J.; Lo, E.K.W.; Cahan, P. Assessment of engineered cells using CellNet and RNA-seq. Nat. Protoc. 2017, 12, 1089. [Google Scholar] [CrossRef]
- Ronquist, S.; Patterson, G.; Muir, L.A.; Lindsly, S.; Chen, H.; Brown, M.; Wicha, M.S.; Bloch, A.; Brockett, R.; Rajapakse, I. Algorithm for cellular reprogramming. Proc. Natl. Acad. Sci. USA 2017, 114, 11832–11837. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Lizio, M.; Harshbarger, J.; Shimoji, H.; Severin, J.; Kasukawa, T.; Sahin, S.; Abugessaisa, I.; Fukuda, S.; Hori, F.; Ishikawa-Kato, S. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Addis, R.C.; Ifkovits, J.L.; Pinto, F.; Kellam, L.D.; Esteso, P.; Rentschler, S.; Christoforou, N.; Epstein, J.A.; Gearhart, J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013, 60, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Forrest, A.R.; Van Nimwegen, E.; Daub, C.O.; Balwierz, P.J.; Irvine, K.M.; Lassmann, T.; Ravasi, T.; Hasegawa, Y.; De Hoon, M.J. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009, 41, 553. [Google Scholar] [PubMed]
- Rosvall, M.; Bergstrom, C.T. An information-theoretic framework for resolving community structure in complex networks. Proc. Natl. Acad. Sci. USA 2007, 104, 7327–7331. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R. On testing the significance of sets of genes. Ann. Appl. Stat. 2007, 1, 107–129. [Google Scholar] [CrossRef]
- Margariti, A.; Winkler, B.; Karamariti, E.; Zampetaki, A.; Tsai, T.-n.; Baban, D.; Ragoussis, J.; Huang, Y.; Han, J.-D.J.; Zeng, L. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc. Natl. Acad. Sci. USA 2012, 109, 13793–13798. [Google Scholar] [CrossRef]
- Hong, X.; Margariti, A.; Le Bras, A.; Jacquet, L.; Kong, W.; Hu, Y.; Xu, Q. Transdifferentiated human vascular smooth muscle cells are a new potential cell source for endothelial regeneration. Sci. Rep. 2017, 7, 5590. [Google Scholar] [CrossRef]
- Ni, X.; Gao, Y.; Wu, Z.; Ma, L.; Chen, C.; Wang, L.; Lin, Y.; Hui, L.; Pan, G. Functional human induced hepatocytes (hiHeps) with bile acid synthesis and transport capacities: A novel in vitro cholestatic model. Sci. Rep. 2016, 6, 38694. [Google Scholar] [CrossRef]
- DeMaagd, G.; Philip, A. Parkinson’s disease and its management: Part 1: Disease entity, risk factors, pathophysiology, clinical presentation, and diagnosis. Pharm. Ther. 2015, 40, 504. [Google Scholar]
- Ruggieri, M.; Riboldi, G.; Brajkovic, S.; Bucchia, M.; Bresolin, N.; Comi, G.P.; Corti, S. Induced neural stem cells: Methods of reprogramming and potential therapeutic applications. Prog. Neurobiol. 2014, 114, 15–24. [Google Scholar] [CrossRef]
- Tang, Y.; Yu, P.; Cheng, L. Current progress in the derivation and therapeutic application of neural stem cells. Cell Death Dis. 2017, 8, e3108. [Google Scholar] [CrossRef] [PubMed]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral vectors: A look back and ahead on gene transfer technology. New Microbiol. 2013, 36, 1–22. [Google Scholar]
- Hacein-Bey-Abina, S.; Von Kalle, C.; Schmidt, M.; McCormack, M.; Wulffraat, N.; Leboulch, P.a.; Lim, A.; Osborne, C.; Pawliuk, R.; Morillon, E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003, 302, 415–419. [Google Scholar] [CrossRef]
- Zhang, J.; Chu, L.-F.; Hou, Z.; Schwartz, M.P.; Hacker, T.; Vickerman, V.; Swanson, S.; Leng, N.; Nguyen, B.K.; Elwell, A. Functional characterization of human pluripotent stem cell-derived arterial endothelial cells. Proc. Natl. Acad. Sci. USA 2017, 114, E6072–E6078. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Miroshnikova, Y.A.; Nava, M.M.; Wickström, S.A. Emerging roles of mechanical forces in chromatin regulation. J. Cell Sci 2017, 130, 2243–2250. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gam, R.; Sung, M.; Prasad Pandurangan, A. Experimental and Computational Approaches to Direct Cell Reprogramming: Recent Advancement and Future Challenges. Cells 2019, 8, 1189. https://doi.org/10.3390/cells8101189
Gam R, Sung M, Prasad Pandurangan A. Experimental and Computational Approaches to Direct Cell Reprogramming: Recent Advancement and Future Challenges. Cells. 2019; 8(10):1189. https://doi.org/10.3390/cells8101189
Chicago/Turabian StyleGam, Rihab, Minkyung Sung, and Arun Prasad Pandurangan. 2019. "Experimental and Computational Approaches to Direct Cell Reprogramming: Recent Advancement and Future Challenges" Cells 8, no. 10: 1189. https://doi.org/10.3390/cells8101189
APA StyleGam, R., Sung, M., & Prasad Pandurangan, A. (2019). Experimental and Computational Approaches to Direct Cell Reprogramming: Recent Advancement and Future Challenges. Cells, 8(10), 1189. https://doi.org/10.3390/cells8101189




