Stem Cell-Derived Exosomes as Therapeutic Approach for Neurodegenerative Disorders: From Biology to Biotechnology
Abstract
1. Extracellular Vesicles in Pathophysiology of Neurodegenerative Diseases
2. Extracellular Vesicles as Therapeutic Approach for Neurodegenerative Diseases
3. Stem Cells: The Main Source of Exosomes for Clinical Purpose
4. Cell-Free Therapy: A Novel Approach to Treat the Neurodegenerative Diseases
5. Brief History of Extracellular Vesicles Discovery
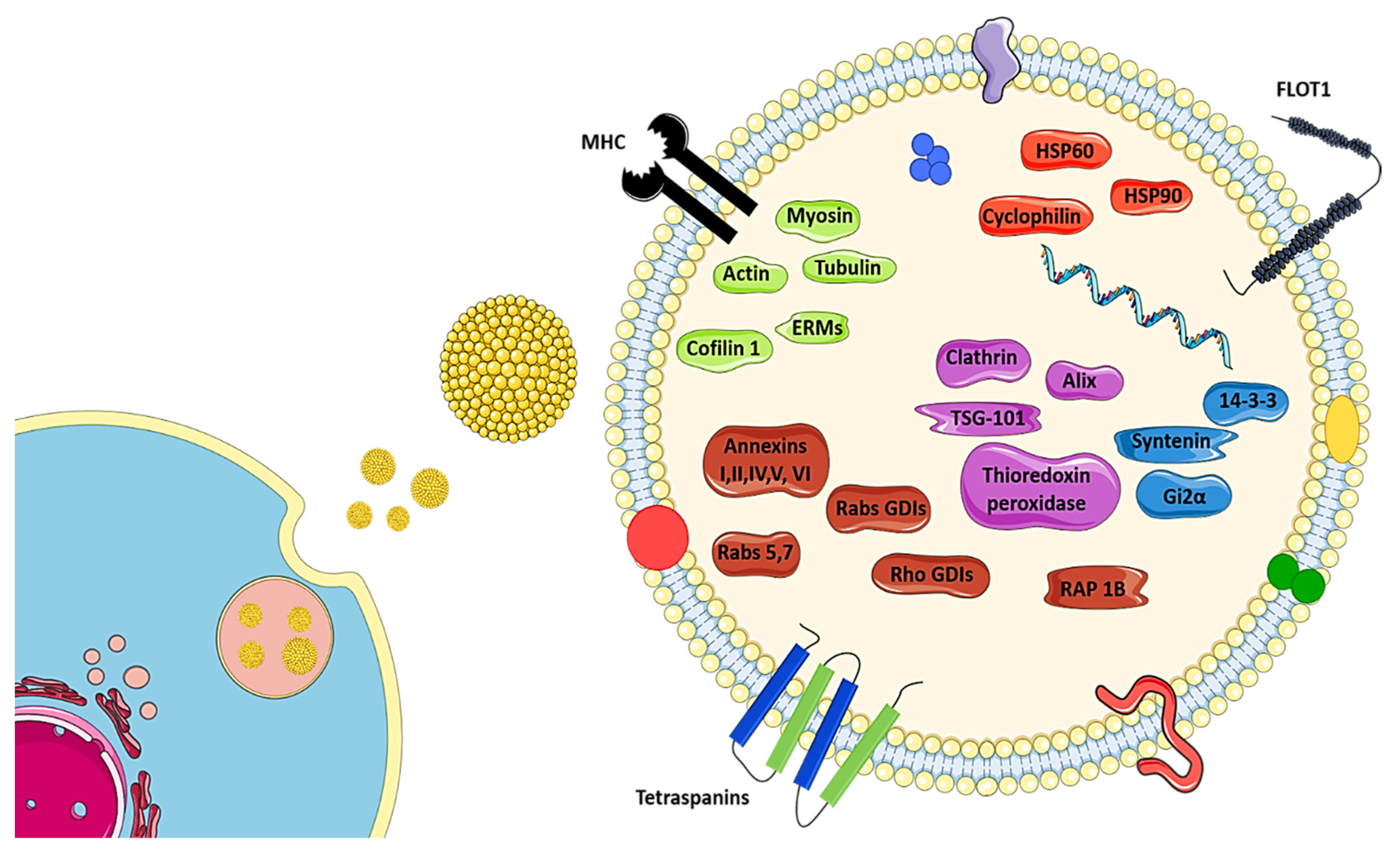
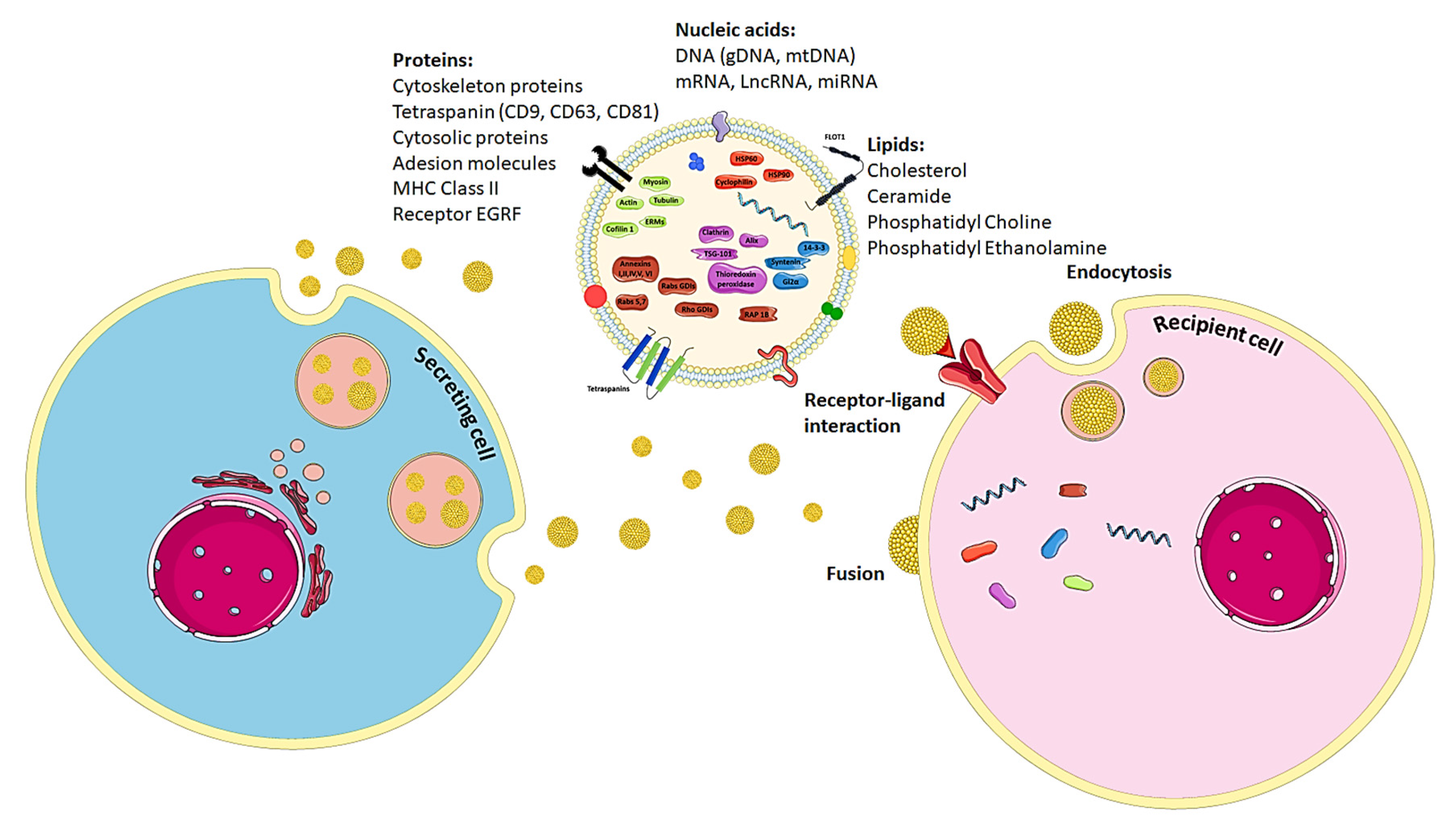
6. The Biology of Exosomes
7. Exosome-Mediated Horizontal Transfers of Nucleic Acids
8. Proteins Present in Exosomes
9. Strategies to Isolate Exosomes
10. Stem Cell-Derived Exosomes as a Therapeutic Approach for Neurodegenerative Diseases
11. Minimal Information to Study Exosomes
12. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Gitler, A.D.; Dhillon, P.; Shorter, J. Neurodegenerative disease: Models, mechanisms, and a new hope. Dis. Model. Mech. 2017, 10, 499–502. [Google Scholar] [CrossRef]
- Fratiglioni, L.; Qiu, C. Prevention of common neurodegenerative disorders in the elderly. Exp. Gerontol. 2009, 44, 46–50. [Google Scholar] [CrossRef]
- Batista, P.; Pereira, A. Quality of life in patients with neurodegenerative diseases. J. Neurol. Neurosci. 2016, 7, a028035. [Google Scholar] [CrossRef]
- Dugger, B.N.; Dickson, D.W. Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 2017, 9, 1–22. [Google Scholar] [CrossRef]
- Guzman-Martinez, L.; Maccioni, R.B.; Andrade, V.; Navarrete, L.P.; Pastor, M.G.; Ramos-Escobar, N. Neuroinflammation as a common feature of neurodegenerative disorders. Front. Pharmcol. 2019, 10, 1008. [Google Scholar] [CrossRef]
- Howitt, J.; Hill, A.F. Exosomes in the pathology of neurodegenerative diseases. J. Biol. Chem. 2016, 291, 26589–26597. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef]
- Ferrara, D.; Pasetto, L.; Bonetto, V.; Basso, M. Role of extracellular vesicles in Amyotrophic Lateral Sclerosis. Front. Neurosci. 2018, 12, 574. [Google Scholar] [CrossRef]
- Otake, K.; Kamiguchi, H.; Hirozane, Y. Identification of biomarkers for amyotrophic lateral sclerosis by comprehensive analysis of exosomal mRNAs in human cerebrospinal fluid. BMC Med. Genom. 2019, 12, 7. [Google Scholar] [CrossRef]
- Malm, T.; Loppi, S.; Kanninen, K.M. Exosomes in Alzheimer’s disease. Neurochem. Int. 2016, 97, 193–199. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell. Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlic, D.; Kuo, W.; Amphornrat, J.; Thilemann, S.; As, S.; Kirchoff, F.; Möbius, W.; Goebbels, S.; Nave, K.; et al. Neurotransmitter-Triggered Transfer of Exosomes Mediates Oligodendrocyte—Neuron Communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef]
- Lemprière, S. Exosomal α-synuclein as a biomarker for Parkinson disease. Nat. Rev. Neurol. 2020, 16, 242–243. [Google Scholar] [CrossRef]
- Hill, A.F. Extracellular Vesicles and Neurodegenerative Diseases. J. Neurosci. 2019, 39, 9269–9273. [Google Scholar] [CrossRef]
- Hartmann, A.; Muth, C.; Dabrowski, O.; Krasemann, S. Exosomes and the prion protein: More than one truth. Front. Neurosci. 2017, 11, 194. [Google Scholar] [CrossRef]
- Lee, J.C.; Kim, S.J.; Hong, S.; Kim, Y. Diagnosis of Alzheimer’s disease utilizing amyloid and tau as fluid biomarkers. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Sadigh-Eteghad, S.; Sabermarouf, B.; Majdi, A.; Talebi, M.; Farhoudi, M.; Mahmoudi, J. Amyloid-Beta: A Crucial Factor in Alzheimer’s Disease. Med. Princ. Pract. 2015, 24, 1–10. [Google Scholar] [CrossRef]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef]
- Basile, M.S.; Battaglia, G.; Bruno, V.; Mangano, K.; Fagone, P.; Petralia, M.C.; Nicoletti, F.; Cavalli, E. The dichotomic role of macrophage migration inhibitory factor in neurodegeneration. Int. J. Mol. Sci. 2020, 21, 3023. [Google Scholar] [CrossRef]
- Fujii, H.; Matsubara, K.; Sakai, K.; Ito, M.; Ohno, K.; Ueda, M.; Yamamoto, A. Dopaminergic differentiation of stem cells from human deciduous teeth and their therapeutic benefits for Parkinsonian rats. Brain Res. 2015, 1613, 59–72. [Google Scholar] [CrossRef]
- Braak, H.; Rüb, U.; Gai, W.P.; Tredici, K. Del Idiopathic Parkinson’s disease: Possible routes by which vulnerable neuronal types may be subject to neuroinvasion by an unknown pathogen. J. Neural Transm. 2003, 110, 517–536. [Google Scholar] [CrossRef]
- Porro, C.; Panaro, M.A.; Lofrumento, D.D.; Hasalla, E.; Trotta, T. The multiple roles of exosomes in Parkinson’s disease: An overview. Immunopharmacol. Immunotoxicol. 2019, 41, 469–476. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Chistiakov, A.A. α-Synuclein-carrying extracellular vesicles in Parkinson’s disease: Deadly transmitters. Acta Neurol. Belg. 2017, 117, 43–51. [Google Scholar] [CrossRef]
- Gao, H.-M.; Zhang, F.; Zhou, H.; Kam, W.; Wilson, B.; Hong, J.-S. Neuroinflammation and α-synuclein dysfunction potentiate each other, driving chronic progression of neurodegeneration in a mouse model of Parkinson’s disease. Environ. Health Perspect. 2011, 119, 807–814. [Google Scholar] [CrossRef]
- Yu, H.; Sun, T.; An, J.; Wen, L.; Liu, F.; Bu, Z.; Cui, Y.; Feng, J. Potential roles of exosomes in Parkinson’s disease: From pathogenesis, diagnosis, and treatment to prognosis. Front. Cell Dev. Biol. 2020, 8, 86. [Google Scholar] [CrossRef]
- Gomes, C.; Keller, S.; Altevogt, P.; Costa, J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci. Lett. 2007, 428, 43–46. [Google Scholar] [CrossRef]
- Gagliardi, D.; Bresolin, N.; Comi, G.P.; Corti, S. Extracellular vesicles and amyotrophic lateral sclerosis: From misfolded protein vehicles to promising clinical biomarkers. Cell. Mol. Life Sci. 2020. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6478. [Google Scholar] [CrossRef]
- Basso, M.; Pozzi, S.; Tortarolo, M.; Fiordaliso, F.; Bisighini, C.; Pasetto, L.; Spaltro, G.; Lidonnici, D.; Gensano, F.; Battaglia, E.; et al. Mutant Copper-Zinc Superoxide Dismutase (SOD1) Induces Protein Secretion Pathway Alterations and Exosome Release in Astrocytes. J. Biol. Chem. 2013, 288, 15699–15711. [Google Scholar] [CrossRef]
- Grad, L.I.; Yerbury, J.J.; Turner, B.J.; Guest, W.C.; Pokrishevsky, E.; O’Neill, M.A.; Yanai, A.; Silverman, J.M.; Zeineddine, R.; Corcoran, L.; et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA 2014, 111, 3620–3625. [Google Scholar] [CrossRef]
- Iguchi, Y.; Eid, L.; Parent, M.; Soucy, G.; Bareil, C.; Riku, Y.; Kawai, K.; Takagi, S.; Yoshida, M.; Katsuno, M.; et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016, 139, 3187–3201. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef]
- Josephs, K.A.; Murray, M.E.; Whitwell, J.L.; Tosakulwong, N.; Weigand, S.D.; Petrucelli, L.; Liesinger, A.M.; Petersen, R.C.; Parisi, J.E.; Dickson, D.W. Updated TDP-43 in Alzheimer’s disease staging scheme. Acta Neuropathol. 2016, 131, 571–585. [Google Scholar] [CrossRef]
- Amador-Ortiz, C.; Lin, W.-L.; Ahmed, Z.; Personett, D.; Davies, P.; Duara, R.; Graff-Radford, N.R.; Hutton, M.L.; Dickson, D.W. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann. Neurol. 2007, 61, 435–445. [Google Scholar] [CrossRef]
- Shamir, C.; Venugopal, C.; Dhanushkodi, A. Dental pulp stem cells for treating neurodegenerative diseases. Neural Regen. Res. 2015, 10, 1910–1911. [Google Scholar] [CrossRef]
- Vilaça-Faria, H.; Salgado, A.J.; Teixeira, F.G. Mesenchymal stem cells -derived exosomes: A new possible therapeutic strategy for Parkinson’s disease? Cells 2019, 8, 118. [Google Scholar] [CrossRef]
- Sarko, D.K.; McKinney, C.E. Exosomes: Origins and therapeutic potential for neurodegenerative disease. Front. Neurosci. 2017, 11, 82. [Google Scholar] [CrossRef]
- Krämer-Albers, E.-M.; Hill, A.F. Extracellular vesicles: Interneural shuttles of complex messages. Curr. Opin. Neurobiol. 2016, 39, 101–107. [Google Scholar] [CrossRef]
- Kalani, A.; Tyagi, A.; Tyagi, N. Exosomes: Mediators of neurodegeneration, neuroprotection and therapeutics. Mol. Neurobiol. 2014, 49, 590–600. [Google Scholar] [CrossRef]
- András, I.E.; Toborek, M. Extracellular vesicles of the blood-brain barrier. Tissue Barriers 2016, 4, e1131804. [Google Scholar] [CrossRef]
- Jafari, D.; Shajari, S.; Jafari, R.; Mardi, N.; Gomari, H.; Ganji, F.; Forouzandeh Moghadam, M.; Samadikuchaksaraei, A. Designer exosomes: A new platform for biotechnology therapeutics. BioDrugs 2020, 34, 567–586. [Google Scholar] [CrossRef]
- Baek, G.; Choi, H.; Kim, Y.; Lee, H.C.; Choi, C. Mesenchymal stem cell-derived extracellular vesicles as therapeutics and as a drug delivery platform. Stem Cells Transl. Med. 2019, 8, 880–886. [Google Scholar] [CrossRef]
- Praveen Kumar, L.; Kandoi, S.; Misra, R.; Vijayalakshmi, S.; Rajagopal, K.; Verma, R.S. The mesenchymal stem cell secretome: A new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev. 2019, 46, 1–9. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, K.; Wu, S.; Cui, M.; Xu, T. Focus on mesenchymal stem cell-derived exosomes: Opportunities and challenges in cell-free therapy. Stem Cells Int. 2017, 2017, 6305295. [Google Scholar] [CrossRef]
- Evangelista, A.; Soares, M.B.P.; Villarreal, C.F. Cell-free therapy: A neuroregenerative approach to sensory neuropathy? Neural Regen. Res. 2019, 14, 1383–1384. [Google Scholar] [CrossRef]
- Bianco, P.; Robey, O.; Simmons, P. Mesenchymal stem cells: Revisiting history, concepts and assays. Cell Stem Cell 2008, 2, 313–319. [Google Scholar] [CrossRef]
- Fujita, Y.; Kadota, T.; Araya, J.; Ochiya, T.; Kuwano, K. Clinical application of mesenchymal stem cell-derived extracellular vesicle-based therapeutics for inflammatory lung diseases. J. Clin. Med. 2018, 7, 355. [Google Scholar] [CrossRef]
- Klymiuk, M.C.; Balz, N.; Elashry, M.I.; Heimann, M.; Wenisch, S.; Arnhold, S. Exosomes isolation and identification from equine mesenchymal stem cells 06 Biological Sciences 0601 Biochemistry and Cell Biology. BMC Vet. Res. 2019, 15, 42. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Chailakhyan, R.K.; Gerasimov, U.V. Bone marrow osteogenic stem cells: In vitro cultivation and transplantation in diffusion chambers. Cell Prolif. 1987, 20, 263–272. [Google Scholar] [CrossRef]
- Blurton-Jones, M.; Kitazawa, M.; Martinez-Coria, H.; Castello, N.A.; Müller, F.-J.; Loring, J.F.; Yamasaki, T.R.; Poon, W.W.; Green, K.N.; LaFerla, F.M. Neural stem cells improve cognition via BDNF in a transgenic model of Alzheimer disease. Proc. Natl. Acad. Sci. USA 2009, 106, 13594–13599. [Google Scholar] [CrossRef]
- Shi, Y.; Kirwan, P.; Smith, J.; MacLean, G.; Orkin, S.H.; Livesey, F.J. A Human Stem Cell Model of Early Alzheimer’s Disease Pathology in Down Syndrome. Sci. Transl. Med. 2012, 4, 124ra29. [Google Scholar] [CrossRef]
- Joyce, N.; Annett, G.; Wirthlin, L.; Olson, S.; Bauer, G.; Nolta, J.A. Mesenchymal stem cells for the treatment of neurodegenerative disease. Regen. Med. 2010, 5, 933–946. [Google Scholar] [CrossRef]
- Haider, K.H.; Aramini, B. Mircrining the injured heart with stem cell-derived exosomes: An emerging strategy of cell-free therapy. Stem Cell Res. Ther. 2020, 11, 23. [Google Scholar] [CrossRef]
- Sakai, K.; Yamamoto, A.; Matsubara, K.; Nakamura, S.; Naruse, M.; Yamagata, M.; Sakamoto, K.; Tauchi, R.; Wakao, N.; Imagama, S.; et al. Human dental pulp-derived stem cells promote locomotor recovery after complete transection of the rat spinal cord by multiple neuro-regenerative mechanisms. J. Clin. Investig. 2012, 122. [Google Scholar] [CrossRef]
- Brooke, G.; Tong, H.; Levesque, J.-P.; Atkinson, K. Molecular trafficking mechanisms of multipotent mesenchymal stem cells derived from human bone marrow and placenta. Stem Cells Dev. 2008, 17, 929–940. [Google Scholar] [CrossRef]
- Karaöz, E.; Demircan, P.C.; Sağlam, Ö.; Aksoy, A.; Kaymaz, F.; Duruksu, G. Human dental pulp stem cells demonstrate better neural and epithelial stem cell properties than bone marrow-derived mesenchymal stem cells. Histochem. Cell Biol. 2011, 136, 455. [Google Scholar] [CrossRef]
- Gomzikova, M.O.; Rizvanov, A.A. Current trends in regenerative medicine: From cell to cell-free therapy. Bionanoscience 2017, 7, 240–245. [Google Scholar] [CrossRef]
- Duffield, J.S.; Park, K.M.; Hsiao, L.-L.; Kelley, V.R.; Scadden, D.T.; Ichimura, T.; Bonventre, J.V. Restoration of tubular epithelial cells during repair of the postischemic kidney occurs independently of bone marrow-derived stem cells. J. Clin. Investig. 2005, 115, 1743–1755. [Google Scholar] [CrossRef]
- Biancone, L.; Bruno, S.; Deregibus, M.C.; Tetta, C.; Camussi, G. Therapeutic potential of mesenchymal stem cell-derived microvesicles. Nephrol. Dial. Transpl. 2012, 27, 3037–3042. [Google Scholar] [CrossRef]
- Gao, J.; Dennis, J.E.; Muzic, R.F.; Lundberg, M.; Caplan, A.I. The dynamic in vivo distribution of bone marrow-derived mesenchymal stem cells after infusion. Cells Tissues Organs 2001, 169, 12–20. [Google Scholar] [CrossRef]
- Lee, R.H.; Pulin, A.A.; Seo, M.J.; Kota, D.J.; Ylostalo, J.; Larson, B.L.; Semprun-Prieto, L.; Delafontaine, P.; Prockop, D.J. Intravenous hMSCs improve myocardial Infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell 2009, 5, 54–63. [Google Scholar] [CrossRef]
- Eggenhofer, E.; Benseler, V.; Kroemer, A.; Popp, F.C.; Geissler, E.K.; Schlitt, H.J.; Baan, C.C.; Dahlke, M.H.; Hoogduijn, M.J. Mesenchymal stem cells are short-lived and do not migrate beyond the lungs after intravenous infusion. Front. Immunol. 2012, 3, 297. [Google Scholar] [CrossRef]
- Song, Y.S.; Lee, H.J.; Doo, S.H.; Lee, S.J.; Lim, I.; Chang, K.-T.; Kim, S.U. Mesenchymal stem cells overexpressing hepatocyte growth factor (HGF) inhibit collagen deposit and improve bladder function in rat model of bladder outlet obstruction. Cell Transpl. 2012, 21, 1641–1650. [Google Scholar] [CrossRef]
- Takahashi, M.; Li, T.-S.; Suzuki, R.; Kobayashi, T.; Ito, H.; Ikeda, Y.; Matsuzaki, M.; Hamano, K. Cytokines produced by bone marrow cells can contribute to functional improvement of the infarcted heart by protecting cardiomyocytes from ischemic injury. Am. J. Physiol. Circ. Physiol. 2006, 291, H886–H893. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Stoppato, M.; Sere, Y.Y.; Coles, A.; Didiot, M.C.; Wollacott, R.; Sapp, E.; Dubuke, M.L.; Li, X.; et al. Exosomes produced from 3D cultures of MSCs by tangential flow filtration show higher yield and improved activity. Mol. Ther. 2018, 26, 2838–2847. [Google Scholar] [CrossRef]
- Vizoso, F.J.; Eiro, N.; Cid, S.; Schneider, J.; Perez-Fernandez, R. Mesenchymal stem cell secretome: Toward cell-free therapeutic strategies in regenerative medicine. Int. J. Mol. Sci. 2017, 18, 1852. [Google Scholar] [CrossRef]
- Baraniak, P.R.; McDevitt, T.C. Stem cell paracrine actions and tissue regeneration. Regen. Med. 2010, 5, 121–143. [Google Scholar] [CrossRef]
- Hu, P.; Yang, Q.; Wang, Q.; Shi, C.; Wang, D.; Armato, U.; Prà, I.D.; Chiarini, A. Mesenchymal stromal cells-exosomes: A promising cell-free therapeutic tool for wound healing and cutaneous regeneration. Burn. Trauma 2019, 7, 1–10. [Google Scholar] [CrossRef]
- Han, C.; Sun, X.; Liu, L.; Jiang, H.; Shen, Y.; Xu, X.; Li, J.; Zhang, G.; Huang, J.; Lin, Z.; et al. Exosomes and their therapeutic potentials of stem cells. Stem Cells Int. 2016, 2016, 7653489. [Google Scholar] [CrossRef]
- Bu, H.; He, D.; He, X.; Wang, K. Exosomes: Isolation, Analysis, and Applications in Cancer Detection and Therapy. ChemBioChem 2019, 20, 451–461. [Google Scholar] [CrossRef]
- Boltze, J. The dark side of the force—Constraints and complications of cell therapies for stroke. Front. Neurol. 2015, 6, 155. [Google Scholar] [CrossRef]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar]
- Wolf, P. The Nature and Ssgnificance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Chen, Y.; Li, G.; Liu, M.-L. Microvesicles as emerging biomarkers and therapeutic targets in cardiometabolic diseases. Genom. Proteom. Bioinform. 2018, 16, 50–62. [Google Scholar] [CrossRef]
- Caruso, S.; Poon, I.K.H. Apoptotic cell-derived extracellular vesicles: More than just debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef]
- Shaimardanova, A.; Solovyeva, V.; Chulpanova, D.; James, V.; Kitaeva, K.; Rizvanov, A. Extracellular vesicles in the diagnosis and treatment of central nervous system diseases. Neural Regen. Res. 2020, 15, 586. [Google Scholar] [CrossRef]
- Yuan, Q.; Li, X.; Zhang, S.; Wang, H.; Wang, Y. Extracellular vesicles in neurodegenerative diseases: Insights and new perspectives. Genes Dis. 2019. [Google Scholar] [CrossRef]
- Malik, Z.A.; Kott, K.S.; Poe, A.J.; Kuo, T.; Chen, L.; Ferrara, K.W.; Knowlton, A.A. Cardiac myocyte exosomes: Stability, HSP60, and proteomics. Am. J. Physiol. Circ. Physiol. 2013, 304, H954–H965. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol. Cancer Res. Treat. 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Loebinger, M.R.; Janes, S.M. Stem cells as vectors for antitumour therapy. Thorax 2010, 65, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef]
- Vázquez-Ríos, A.J.; Molina-Crespo, Á.; Bouzo, B.L.; López-López, R.; Moreno-Bueno, G.; de la Fuente, M. Exosome-mimetic nanoplatforms for targeted cancer drug delivery. J. Nanobiotechnol. 2019, 17, 85. [Google Scholar] [CrossRef]
- Harding, C.; Stahl, P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem. Biophys. Res. Commun. 1983, 113, 650–658. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammonds, J.R.; Turbide, C. Vesicle formation during reticulocyte maturation. J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar]
- Johnstone, R.M. Revisiting the road to the discovery of exosomes. Blood Cells Mol. Dis. 2005, 34, 214–219. [Google Scholar] [CrossRef]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef]
- Antounians, L.; Tzanetakis, A.; Pellerito, O.; Catania, V.D.; Sulistyo, A.; Montalva, L.; McVey, M.J.; Zani, A. The regenerative potential of amniotic fluid stem cell extracellular vesicles: Lessons learned by comparing different isolation techniques. Sci. Rep. 2019, 9, 1837. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Exosomes, E.; Vivo, I.; Verweij, F.J.; Revenu, C.; Arras, G.; Del Bene, F.; Van Niel, G.; Verweij, F.J.; Revenu, C.; Arras, G.; et al. Live Tracking of Inter-organ Communication by Resource Live Tracking of Inter-organ Communication by Endogenous Exosomes In Vivo. Dev. Cell 2019, 573–589. [Google Scholar] [CrossRef]
- Kourembanas, S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015, 77, 13–27. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular e ndosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Juan, T.; Fürthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Fader, C.M.; Colombo, M.I. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2009, 16, 70–78. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles. Traffic 2011, 1659–1668. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Heusermann, W.; Hean, J.; Trojer, D.; Steib, E.; von Bueren, S.; Graff-Meyer, A.; Genoud, C.; Martin, K.; Pizzato, N.; Voshol, J.; et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell Biol. 2016, 213, 173–184. [Google Scholar] [CrossRef]
- Lee, H.K.; Finniss, S.; Cazacu, S.; Xiang, C.; Brodie, C. Mesenchymal stem cells deliver exogenous miRNAs to neural cells and induce their differentiation and glutamate transporter expression. Stem Cells Dev. 2014, 23, 2851–2861. [Google Scholar] [CrossRef]
- Wei, H.; Xu, Y.; Chen, Q.; Chen, H.; Zhu, X.; Li, Y. Mesenchymal stem cell-derived exosomal miR-223 regulates neuronal cell apoptosis. Cell Death Dis. 2020, 11, 290. [Google Scholar] [CrossRef]
- Cui, G.; Wu, J.; Mou, F.; Xie, W.; Wang, F.; Wang, Q.; Fang, J.; Xu, Y.; Dong, Y.; Liu, J.; et al. Exosomes derived from hypoxia-preconditioned mesenchymal stromal cells ameliorate cognitive decline by rescuing synaptic dysfunction and regulating inflammatory responses in APP/PS1 mice. FASEB J. 2018, 32, 654–668. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; Connell, J.T.O.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, L.; Li, F.; Yang, J.; Wan, X.; Ouyang, M. microRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J. Cell. Physiol. 2019, 234, 21380–21394. [Google Scholar] [CrossRef]
- Che, Y.; Shi, X.; Shi, Y.; Jiang, X.; Ai, Q.; Shi, Y.; Gong, F.; Jiang, W. Exosomes derived from miR-143-overexpressing MSCs inhibit cell migration and invasion in human prostate cancer by downregulating TFF3. Mol. Ther. Nucleic Acids 2019, 18, 232–244. [Google Scholar] [CrossRef]
- Ma, T.; Chen, Y.; Chen, Y.; Meng, Q.; Sun, J.; Shao, L.; Yu, Y.; Huang, H.; Hu, Y.; Yang, Z.; et al. MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 2018, 2018, 3290372. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, Y.; Ding, X.; Li, Q.; Qiu, F.; Wang, M.; Shen, Z.; Zheng, H.; Fu, G. Bone marrow mesenchymal stem cell-secreted exosomes carrying microRNA-125b protect against myocardial ischemia reperfusion injury via targeting SIRT7. Mol. Cell. Biochem. 2020, 465, 103–114. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes. J. Biol. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumor microvesicles contain retrotransposons elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef]
- Guescini, M.; Guidolin, D.; Vallorani, L.; Casadei, L.; Gioacchini, A.M.; Tibollo, P.; Battistelli, M.; Falcieri, E.; Battistin, L.; Agnati, L.F.; et al. C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp. Cell Res. 2010, 316, 1977–1984. [Google Scholar] [CrossRef]
- Sisquella, X.; Ofir-Birin, Y.; Pimentel, M.A.; Cheng, L.; Abou Karam, P.; Sampaio, N.G.; Penington, J.S.; Connolly, D.; Giladi, T.; Scicluna, B.J.; et al. Malaria parasite DNA-harbouring vesicles activate cytosolic immune sensors. Nat. Commun. 2017, 8, 1985. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 275–280. [Google Scholar] [CrossRef]
- Qu, X.; Li, Q.; Yang, J.; Zhao, H.; Wang, F.; Zhang, F.; Zhang, S.; Zhang, H.; Wang, R.; Wang, Q.; et al. Double-stranded DNA in exosomes of malignant pleural effusions as a novel DNA source for EGFR mutation detection in lung adenocarcinoma. Front. Oncol. 2019, 9, 931. [Google Scholar] [CrossRef]
- Pires, A.O.; Mendes-Pinheiro, B.; Teixeira, F.G.; Anjo, S.I.; Ribeiro-Samy, S.; Gomes, E.D.; Serra, S.C.; Silva, N.A.; Manadas, B.; Sousa, N.; et al. Unveiling the differences of secretome of human bone marrow mesenchymal stem cells, adipose tissue-derived stem cells, and human umbilical cord perivascular cells: A proteomic analysis. Stem Cells Dev. 2016, 25, 1073–1083. [Google Scholar] [CrossRef]
- Assoni, A.; Coatti, G.; Valadares, M.C.; Beccari, M.; Gomes, J.; Pelatti, M.; Mitne-Neto, M.; Carvalho, V.M.; Zatz, M. Different donors mesenchymal stromal cells secretomes reveal heterogeneous profile of relevance for therapeutic use. Stem Cells Dev. 2016, 26, 206–214. [Google Scholar] [CrossRef]
- Phinney, D.G.; Pittenger, M.F. Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells 2017, 35, 851–858. [Google Scholar] [CrossRef]
- Garcia, N.A.; Moncayo-Arlandi, J.; Sepulveda, P.; Diez-Juan, A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc. Res. 2016, 109, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.E.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef]
- Levy, S.; Todd, S.C.; Maecker, H.T. Cd81 (Tapa-1): A Molecule Involved in Signal Transduction and Cell Adhesion in the Immune System. Annu. Rev. Immunol. 1998, 16, 89–109. [Google Scholar] [CrossRef]
- Kang, D.; Oh, S.; Ahn, S.-M.; Lee, B.-H.; Moon, M.H. Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography−tandem mass spectrometry. J. Proteome Res. 2008, 7, 3475–3480. [Google Scholar] [CrossRef]
- Caponnetto, F.; Manini, I.; Skrap, M.; Palmai-Pallag, T.; Di Loreto, C.; Beltrami, A.P.; Cesselli, D.; Ferrari, E. Size-dependent cellular uptake of exosomes. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1011–1020. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Belting, M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J. Extracell. Vesicles 2013, 2, 20304. [Google Scholar] [CrossRef]
- Beninson, L.A.; Fleshner, M. Exosomes: An emerging factor in stress-induced immunomodulation. Semin. Immunol. 2014, 26, 394–401. [Google Scholar] [CrossRef]
- Bartaula-Brevik, S. Secretome of mesenchymal stem cells grown in hypoxia accelerates wound healing and vessel formation in vitro. Int. J. Stem Cell Res. Ther. 2017, 4. [Google Scholar] [CrossRef]
- Park, K.S.; Bandeira, E.; Shelke, G.V.; Lässer, C.; Lötvall, J. Enhancement of therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Stem Cell Res. Ther. 2019, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lee, Y.; Johansson, H.J.; Mäger, I.; Vader, P.; Nordin, J.Z.; Wiklander, O.P.B.; Lehtiö, J.; Wood, M.J.A.; Andaloussi, S. EL Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J. Extracell. Vesicles 2015, 4, 26883. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Rojas, M.; Badewien-Rentzsch, B.; Plendl, J.; Kohn, B.; Einspanier, R. Exploration of serum- and cell culture-derived exosomes from dogs. BMC Vet. Res. 2018, 14, 179. [Google Scholar] [CrossRef] [PubMed]
- Duong, P.; Chung, A.; Bouchareychas, L.; Raffai, R.L. Cushioned-density gradient ultracentrifugation (C-DGUC) improves the isolation efficiency of extracellular vesicles. PLoS ONE 2019, 14, e0215324. [Google Scholar] [CrossRef]
- Narbute, K.; Piļipenko, V.; Pupure, J.; Dzirkale, Z.; Jonavičė, U.; Tunaitis, V.; Kriaučiūnaitė, K.; Jarmalavičiūtė, A.; Jansone, B.; Kluša, V.; et al. Intranasal administration of extracellular vesicles derived from human teeth stem cells improves motor symptoms and normalizes tyrosine hydroxylase expression in the substantia nigra and striatum of the 6-hydroxydopamine-treated rats. Stem Cells Transl. Med. 2019, 8, 490–499. [Google Scholar] [CrossRef]
- Li, D.; Zhang, D.; Tang, B.; Zhou, Y.; Guo, W.; Kang, Q.; Wang, Z.; Shen, L.; Wei, G.; He, D. Exosomes from human umbilical cord mesenchymal stem cells reduce damage from oxidative stress and the epithelial-mesenchymal transition in renal epithelial cells exposed to oxalate and calcium oxalate monohydrate. Stem Cells Int. 2019, 2019, 6935806. [Google Scholar] [CrossRef]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef]
- Li, J.; He, X.; Deng, Y.; Yang, C. An update on isolation methods for proteomic studies of extracellular vesicles in biofluids. Molecules 2019, 24, 3516. [Google Scholar] [CrossRef]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef]
- Patel, G.K.; Khan, M.A.; Zubair, H.; Srivastava, S.K.; Khushman, M.; Singh, S.; Singh, A.P. Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 2019, 9, 440. [Google Scholar] [CrossRef]
- Pardridge, W.M. Drug Transport across the blood–brain barrier. J. Cereb. Blood Flow Metab. 2012, 32, 1959–1972. [Google Scholar] [CrossRef] [PubMed]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’ s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Gorabi, A.M.; Kiaie, N.; Barreto, G.E.; Read, M.I.; Tafti, H.A.; Sahebkar, A. The therapeutic potential of mesenchymal stem cell–derived exosomes in treatment of neurodegenerative diseases. Mol. Neurobiol. 2019, 56, 8157–8167. [Google Scholar] [CrossRef] [PubMed]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.-E.; El-Baba, M.D.; Saxena, P.; Ausländer, S.; Tan, K.R.; Fussenegger, M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Lin, Q.; Huang, L.; Fu, Y.; Wang, L.; He, S.; Fu, Y.; Yang, S.; Zhang, Z.; Zhang, L.; et al. Dopamine-loaded blood exosomes targeted to brain for better treatment of Parkinson’s disease. J. Control. Release 2018, 287, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-X.; Liang, F.-C.; Gu, P.; Xu, B.-L.; Xu, H.-J.; Wang, W.-T.; Hou, J.-Y.; Xie, D.-X.; Chai, X.-Q.; An, S.-J. Exosomes derived from mesenchymal stem cells repair a Parkinson’s disease model by inducing autophagy. Cell Death Dis. 2020, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Yin, Z.; Chen, F.; Lei, P. Mesenchymal stem cell-derived exosome: A promising alternative in the therapy of Alzheimer’s disease. Alzheimers. Res. Ther. 2020, 12, 109. [Google Scholar] [CrossRef]
- Reza-Zaldivar, E.; Hernández-Sapiéns, M.; Gutiérrez-Mercado, Y.; Sandoval-Ávila, S.; Gomez-Pinedo, U.; Márquez-Aguirre, A.; Vázquez-Méndez, E.; Padilla-Camberos, E.; Canales-Aguirre, A. Mesenchymal stem cell-derived exosomes promote neurogenesis and cognitive function recovery in a mouse model of Alzheimer’s disease. Neural Regen. Res. 2019, 14, 1626. [Google Scholar] [CrossRef]
- Ding, M.; Shen, Y.; Wang, P.; Xie, Z.; Xu, S.; Zhu, Z.; Wang, Y.; Lyu, Y.; Wang, D.; Xu, L.; et al. Exosomes isolated from human umbilical cord mesenchymal stem cellsaAlleviate neuroinflammation and reduce amyloid-beta deposition by modulating microglial activation in Alzheimer’s disease. Neurochem. Res. 2018, 43, 2165–2177. [Google Scholar] [CrossRef]
- Elia, C.A.; Tamborini, M.; Rasile, M.; Desiato, G.; Marchetti, S.; Swuec, P.; Mazzitelli, S.; Clemente, F.; Anselmo, A.; Matteoli, M.; et al. Intracerebral injection of extracellular vesicles from mesenchymal stem cells exerts reduced Aβ plaque durden in early stages of a preclinical model of Alzheimer’s disease. Cells 2019, 8, 1059. [Google Scholar] [CrossRef]
- Nakano, M.; Kubota, K.; Kobayashi, E.; Chikenji, T.S.; Saito, Y.; Konari, N.; Fujimiya, M. Bone marrow-derived mesenchymal stem cells improve cognitive impairment in an Alzheimer’s disease model by increasing the expression of microRNA-146a in hippocampus. Sci. Rep. 2020, 10, 10772. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-S.; Jia, J.; Wang, Z. Mesenchymal stem cell-derived extracellular vesicles suppresses iNOS expression and ameliorates neural impairment in Alzheimer’s disease mice. J. Alzheimer’s Dis. 2018, 61, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Guo, H.; Li, H.; Zhai, Y.; Gong, Z.; Wu, J.; Liu, J.; Dong, Y.; Hou, S.; Liu, J. RVG-modified exosomes derived from mesenchymal stem cells rescue memory deficits by regulating inflammatory responses in a mouse model of Alzheimer’s disease. Immun. Ageing 2019, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Zhang, S.; Barthe, S.; Kumar, P.; Pivetti, C.; Kreutzberg, N.; Reed, C.; Wang, Y.; Paxton, Z.; Farmer, D.; et al. Placental mesenchymal stem cell-derived extracellular vesicles promote myelin regeneration in an animal model of multiple sclerosis. Cells 2019, 8, 1497. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, F.; He, X.; Yang, X.; Shan, F.; Feng, J. Exosomes derived from mesenchymal stem cells attenuate inflammation and demyelination of the central nervous system in EAE rats by regulating the polarization of microglia. Int. Immunopharmacol. 2019, 67, 268–280. [Google Scholar] [CrossRef]
- Laso-García, F.; Ramos-Cejudo, J.; Carrillo-Salinas, F.J.; Otero-Ortega, L.; Feliú, A.; Gómez-de Frutos, M.; Mecha, M.; Díez-Tejedor, E.; Guaza, C.; Gutiérrez-Fernández, M. Therapeutic potential of extracellular vesicles derived from human mesenchymal stem cells in a model of progressive multiple sclerosis. PLoS ONE 2018, 13, e0202590. [Google Scholar] [CrossRef]
- Bonafede, R.; Scambi, I.; Peroni, D.; Potrich, V.; Boschi, F.; Benati, D.; Bonetti, B.; Mariotti, R. Exosome derived from murine adipose-derived stromal cells: Neuroprotective effect on in vitro model of amyotrophic lateral sclerosis. Exp. Cell Res. 2016, 340, 150–158. [Google Scholar] [CrossRef]
- Lee, M.; Ban, J.-J.; Kim, K.Y.; Jeon, G.S.; Im, W.; Sung, J.-J.; Kim, M. Adipose-derived stem cell exosomes alleviate pathology of amyotrophic lateral sclerosis in vitro. Biochem. Biophys. Res. Commun. 2016, 479, 434–439. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Emelyanov, A.; Shtam, T.; Kamyshinsky, R.; Garaeva, L.; Verlov, N.; Miliukhina, I.; Kudrevatykh, A.; Gavrilov, G.; Zabrodskaya, Y.; Pchelina, S.; et al. Cryo-electron microscopy of extracellular vesicles from cerebrospinal fluid. PLoS ONE 2020, 15, e0227949. [Google Scholar] [CrossRef]
- Soo, C.Y.; Song, Y.; Zheng, Y.; Campbell, E.C.; Riches, A.C.; Gunn-Moore, F.; Powis, S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 2012, 136, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Øvstebø, R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 2017, 6, 1344087. [Google Scholar] [CrossRef] [PubMed]
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010, 27, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Frokjaer, S.; Otzen, D.E. Protein drug stability: A formulation challenge. Nat. Rev. Drug Discov. 2005, 4, 298–306. [Google Scholar] [CrossRef]
- Philo, J. A critical review of methods for size characterization of non-particulate protein aggregates. Curr. Pharm. Biotechnol. 2009, 10, 359–372. [Google Scholar] [CrossRef]
- Sebaihi, N.; De Boeck, B.; Yuana, Y.; Nieuwland, R.; Pétry, J. Dimensional characterization of extracellular vesicles using atomic force microscopy. Meas. Sci. Technol. 2017, 28, 34006. [Google Scholar] [CrossRef]
- Chuo, S.T.-Y.; Chien, J.C.-Y.; Lai, C.P.-K. Imaging extracellular vesicles: Current and emerging methods. J. Biomed. Sci. 2018, 25, 91. [Google Scholar] [CrossRef]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.W.M.M.; Coumans, F.A.W. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1555419. [Google Scholar] [CrossRef]
- Jung, M.K.; Mun, J.Y. Sample preparation and imaging of exosomes by transmission electron microscopy. J. Vis. Exp. 2018, 131, e56482. [Google Scholar] [CrossRef]
- Deng, J.-H.; Li, Z.-J.; Wang, Z.-X.; Feng, J.; Huang, X.-J.; Zeng, Z.-M. Electron Microscopy-Based Comparison and Investigation of the Morphology of Exosomes Derived from Hepatocellular Carcinoma Cells Isolated at Different Centrifugal Speeds. Microsc. Microanal. 2020, 26, 310–318. [Google Scholar] [CrossRef]
- Zhou, M.; Weber, S.R.; Zhao, Y.; Chen, H.; Sundstrom, J.M. Methods for exosome isolation and characterization. In Exosomes; Elsevier: Amsterdam, The Netherlands, 2020; pp. 23–38. [Google Scholar]
- Peak, T.C.; Praharaj, P.P.; Panigrahi, G.K.; Doyle, M.; Su, Y.; Schlaepfer, I.R.; Singh, R.; Vander Griend, D.J.; Alickson, J.; Hemal, A.; et al. Exosomes secreted by placental stem cells selectively inhibit growth of aggressive prostate cancer cells. Biochem. Biophys. Res. Commun. 2018, 499, 1004–1010. [Google Scholar] [CrossRef] [PubMed]
- Chernyshev, V.S.; Rachamadugu, R.; Tseng, Y.H.; Belnap, D.M.; Jia, Y.; Branch, K.J.; Butterfield, A.E.; Pease, L.F.; Bernard, P.S.; Skliar, M. Size and shape characterization of hydrated and desiccated exosomes. Anal. Bioanal. Chem. 2015, 407, 3285–3301. [Google Scholar] [CrossRef] [PubMed]
- Rocha, B.; Calamia, V.; Blanco, F.J.; Ruiz-Romero, C. Identification of Factors Produced and Secreted by Mesenchymal Stromal Cells with the SILAC Method. Methods Mol. Biol. 2016, 1416, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Riis, S.; Stensballe, A.; Emmersen, J.; Pennisi, C.P.; Birkelund, S.; Zachar, V. Mass spectrometry analysis of adipose-derived stem cells reveals a significant effect of hypoxia on pathways regulating extracellular matrix. Stem Cell Res. Ther. 2016, 7, 52. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of extracellular vesicles: State-of-the-art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef]
- Prendergast, E.N.; De Souza Fonseca, M.A.; Dezem, F.S.; Lester, J.; Karlan, B.Y.; Noushmehr, H.; Lin, X.; Lawrenson, K. Optimizing exosomal RNA isolation for RNA-Seq analyses of archival sera specimens. PLoS ONE 2018, 13, e0196913. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Durães, F.; Pinto, M.; Sousa, E. Old drugs as new treatments for neurodegenerative diseases. Pharmaceuticals 2018, 11, 44. [Google Scholar] [CrossRef]
- Jafarinia, M.; Alsahebfosoul, F.; Salehi, H.; Eskandari, N.; Ganjalikhani-Hakemi, M. Mesenchymal stem cell-derived extracellular vesicles: A novel cell-free therapy. Immunol. Investig. 2020, 49, 758–780. [Google Scholar] [CrossRef]
| Noncommercial Methods | |||
|---|---|---|---|
| Methods | Procedures | Time | Reference |
| UC | Clearance: CM is collected and centrifuged at 2700× g for 5 min and afterwards filtered using a 0.2 μm filter. Isolation: Supernatant is centrifuged at 100,000× g overnight at 4 °C | >16 h | Klymiuk et al. [49] |
| UC | Clearance: CM is collected and filtered using 0.2 μm filter. Filtrate is subjected to two centrifugation steps: (1) 2000× g for 10 min and (2) 10,000× g for 70 min. Isolation: Supernatant is centrifuged at 100,000× g for 70 min and afterwards, washed in PBS and centrifuged at 100,000× g for 70 min | >3 h | Klymiuk et al. [49] |
| UC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 300× g for 10 min and (2) 10,000× g for 30 min. Next, the supernatant is filtered using a 0.2 μm filter. Isolation: Filtrate is centrifuged at 100,000× g for 90 min, discarding the supernatant. Exosome pellet is washed in PBS and then centrifuged 100,000× g for 90 min | ≈3 h | Haraszti et al. [67] |
| UC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 300× g and (2) 1200× g, both for 10 min. Supernatant is filtered using 0.2 μm filter. Isolation: Filtrate is centrifuged at 100,000× g for 14 h at 4 °C. | >14 h | Antounians et al. [89] |
| UC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 400× g for 10 min and (2) 2000× g for 20 min. Supernatant is filtrated using a 0.2 μm filter. Isolation: Filtrate is centrifuged at 100,000 × g for 3 h | ≈4 h | Duong et al. [134] |
| UC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 300× g and (2) 1200× g, both for 10 min. Isolation: Filtrate is centrifuged at 100,000× g for 14 h | ≈15 h | Antounias et al. [89] |
| UC | Clearance: CM is collected and centrifuged at 2700× g for 5 min and afterwards filtered using a 0.2 μm filter. Filtrate is subjected to two centrifugation steps: (1) 2000× g for 10 min and (2) 10,000× g for 30 min. Isolation: Supernatant is centrifuged at 100,000× g for 70 min, discarding the supernatant. Exosome pellet is washed in PBS and then centrifuged 100,000× g for 70 min. | ≈4 h | Klymiuk et al. [49] |
| UC | Clearance: CM is collected and subjected to three centrifugation steps: (1) 300× g for 10 min, (2) 2000× g for 30 min, and (3) 20,000 for 30 min. Isolation: Supernatant is centrifuged at 100,000× g for 70 min, discarding the supernatant. Exosome pellet is washed in PBS and then centrifuged 100,000× g for 70 min. | ≈4 h | Narbute et al. [135] |
| CUC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 400× g for 10 min and (2) 2000× g for 20 min. Supernatant iscollected Isolation: Filtrate is loaded on 60% iodixanol cushion. Sample is centrifuges at 100,000× g for 3 h. | ≈4 h | Duong et al. [134] |
| CUC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 300× g and (2) 6000× g, both for 30 min. Supernatant is filtrated using a using a 0.2 μm filter. Isolation: Filtrate is loaded on 30% sucrose/D2O cushion. Samples is centrifuged at 120,000× g for 90 min, discarding the supernatant. Exosome pellet is washed in PBS and next, centrifuged at 120,000× g for 90 min. | ≈6 h | Li et al. [136] |
| DGUC | Clearance: CM is collected and subjected to two centrifugation steps: (1) 400× g for 10 min and (2) 2000× g for 20 min. Supernatant is filtrated using a 0.2 μm filter. Isolation: Exosomes, previously isolated using UC method, is loaded on 5, 10 or 20% iodixanol gradient solution with 0.25 mm sucrose, 1 mM Tris-HCl pH 7.4 and centrifuged at 100,000× g for 18 h | ≈20 h | Duong et al. [134] |
| PEG-B | Clearance: not applied Isolation: CM is collected and transferred to centrifugation tubes containing 1:4 (v/v) of 50% PEG 6000 with 375 mm NaCl—CM. Sample is incubated for 18 h at 4 °C and next, centrifuged at 1500× g for 30 min. | ≈19 h | Duong et al. [134] |
| PEG-B | Clearance: CM is collected and centrifuged at 3000× g for 20 min. Supernatant is filtrated using a 0.2 μm filter. Isolation: Filtrate is incubated with 50% PEG 35.000 and 2% protaminesulfate for 1 h at 4 °C and, next, centrifuged at 12,000× g for 10 min. | ≈2 h | Klymiuk et al. [49] |
| UF | Clearance: CM is collected and centrifuged at 3000× g for 20 min. Supernatant is filtrated using a 0.2 μm filter. Isolation: Filtrate obtained is filtrated using 3K Ultra Filter and then, centrifuged at 2700× g for 10 min. Pellet is washed with PBS and, next, centrifuged at 2700× g for 10 min. | ≈0.5 h | Klymiuk et al. [49] |
| TFF | Isolation: CM is subjected to ultrafiltration in a tangential flow filtration (TFF) system, using a 500 kDa cutoff TFF cartridge with a flow rate of 120 mL/min and a transmembrane pressure < 3.5 psi. Then, the filtrate is concentrated 9-fold and exchange with 6× volume of PBS. Finally, the exosomes are filtrated using a 0.2 μm filter. | ≈2 h | Haraszti et al. [67] |
| Commercial Methods | |||
| Methods | Procedures | Time | Reference |
| Total Exosome Isolation (TEI) | Procedure: CM is collected and centrifuged at 2000× g for 30 min at 4 °C. Supernatant is incubated overnight at 4 °C with the Total Exosome Isolation reagent. Then, the sample is centrifuged at 10,000× g for 60 min at 4 °C. | >20 h | Thermo Fischer Scientific |
| ExoQuick Preciptation Solution | Procedure: CM is collected and centrifuged at 3000× g for 15 min at 4 °C. Supernatant is filtered using a 0.2 μm filter. Filtrated is incubated overnight at 4 °C with the ExoQuick reagent. Then, the sample is centrifuged at 1500× g for 5 min at 4 °C. | >20 h | System Biosciences |
| ExoMAX Opti Enhancer Reagent | Procedure: CM is collected and centrifuged at 3000× g for 30 min (clearance). Supernatant is incubated overnight with ExoMAX Opti Enhancer Reagent. Then, the sample is centrifuged at 1500× g for 30 min. Pellet is washed and centrifuged at 75,000× g for 70 min. | >20 h | BioCat |
| qEV Exosome Isolation | Procedure: CM is collected and centrifuged at 16,000× g for 30 min at 4 °C (clearance). Supernatant is incubated overnight at 4 °C with the Exo-Spin reagent. Then, the sample is centrifuged at 16,000× g for 60 min at 4 °C. | >20 h | Cell Guidance Systems |
| MagCaptureTM Exosome Isolation Kit | Procedure: CM is collected and loaded on a stationary phase consisting of porous resin particles, in which particles between 35–350 nm enter into the pores. | ≈2 h | IZON |
| Exo-SpinTM Exosome Purification | Procedure: CM is collected and centrifuged at 300× g for 10 min. Next, the supernatant is filtered using a 0.2 μm filter (clearance). Filtrate is incubated overnight with Tim4 protein solidified magnetic beads able to bind to phosphatidylserine on the surface of extracellular vesicles. Finally, exosomes are captured using a magnetic rack. | >20 h | Wako |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araldi, R.P.; D’Amelio, F.; Vigerelli, H.; de Melo, T.C.; Kerkis, I. Stem Cell-Derived Exosomes as Therapeutic Approach for Neurodegenerative Disorders: From Biology to Biotechnology. Cells 2020, 9, 2663. https://doi.org/10.3390/cells9122663
Araldi RP, D’Amelio F, Vigerelli H, de Melo TC, Kerkis I. Stem Cell-Derived Exosomes as Therapeutic Approach for Neurodegenerative Disorders: From Biology to Biotechnology. Cells. 2020; 9(12):2663. https://doi.org/10.3390/cells9122663
Chicago/Turabian StyleAraldi, Rodrigo Pinheiro, Fernanda D’Amelio, Hugo Vigerelli, Thatiana Correa de Melo, and Irina Kerkis. 2020. "Stem Cell-Derived Exosomes as Therapeutic Approach for Neurodegenerative Disorders: From Biology to Biotechnology" Cells 9, no. 12: 2663. https://doi.org/10.3390/cells9122663
APA StyleAraldi, R. P., D’Amelio, F., Vigerelli, H., de Melo, T. C., & Kerkis, I. (2020). Stem Cell-Derived Exosomes as Therapeutic Approach for Neurodegenerative Disorders: From Biology to Biotechnology. Cells, 9(12), 2663. https://doi.org/10.3390/cells9122663





