The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions
Abstract
1. Introduction
2. HHV-6 Epidemiology and Disease Association
3. HHV-6 Genome and U94 ORF
4. U94 and Virus Cycle
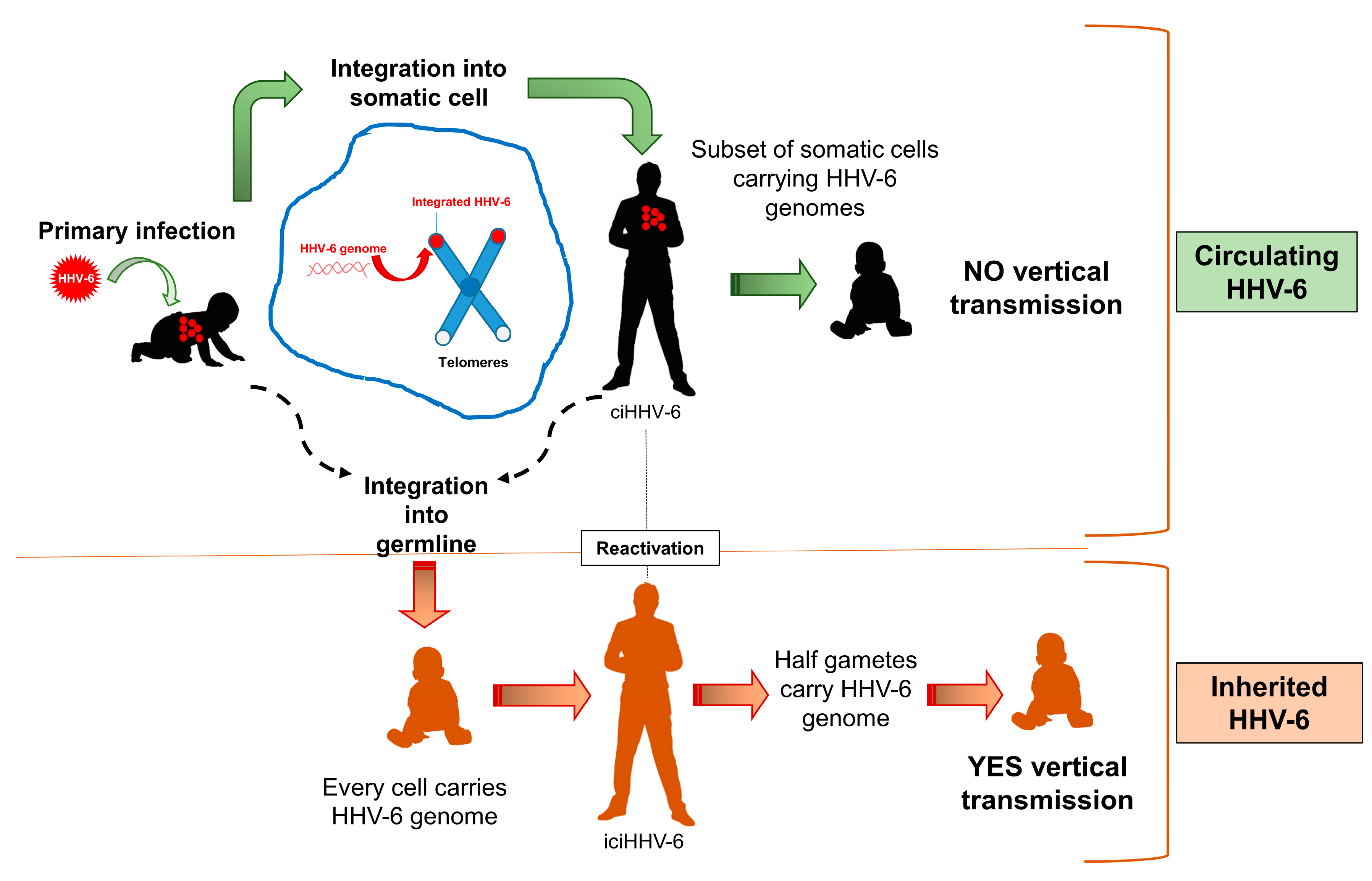
5. U94 and Virus Integration
6. U94 and Immunity
7. U94 Effects in Cells and Tissues
8. Conclusions
9. Patents
Author Contributions
Funding
Conflicts of Interest
References
- Salahuddin, S.Z.; Ablashi, D.V.; Markham, P.D.; Josephs, S.F.; Sturzenegger, S.; Kaplan, M.; Halligan, G.; Biberfeld, P.; Wong-Staal, F.; Kramarsky, B.; et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 1986, 234, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, K.; Shiraki, K.; Kondo, T.; Okuno, T.; Takahashi, M.; Asano, Y.; Kurata, T. Identification of Human Herpesvirus-6 as a Causal Agent for Exanthem Subitum. Lancet 1988, 331, 1065–1067. [Google Scholar] [CrossRef]
- Ablashi, D.; Agut, H.; Alvarez-Lafuente, R.; Clark, D.A.; Dewhurst, S.; DiLuca, D.; Flamand, L.; Frenkel, N.; Gallo, R.; Gompels, U.A.; et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch. Virol. 2014, 159, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Di Luca, D. Molecular biology and clinical associations of Roseoloviruses human herpesvirus 6 and human herpesvirus 7. New Microbiol. 2007, 30, 173–187. [Google Scholar] [PubMed]
- Yamanishi, K.; Mori, Y.; Pellett, P.E. Human Herpesvirus 6 and 7. In Virology, 6th ed.; Knipe, D.M., Howley, P., Eds.; Wolters Kluwer Health: Philadelphia, PA, USA, 2013; Volume 2, p. 2058. [Google Scholar]
- Zhang, E.; Bell, A.J.; Wilkie, G.S.; Suárez, N.M.; Batini, C.; Veal, C.D.; Armendáriz-Castillo, I.; Neumann, R.; Cotton, V.E.; Huang, Y.; et al. Inherited Chromosomally Integrated Human Herpesvirus 6 Genomes Are Ancient, Intact, and Potentially Able To Reactivate from Telomeres. J. Virol. 2017, 91, 91. [Google Scholar] [CrossRef] [PubMed]
- Greninger, A.L.; Knudsen, G.M.; Roychoudhury, P.; Hanson, D.J.; Sedlak, R.H.; Xie, H.; Guan, J.; Nguyen, T.; Peddu, V.; Boeckh, M.; et al. Comparative genomic, transcriptomic, and proteomic reannotation of human herpesvirus 6. BMC Genom. 2018, 19, 1–17. [Google Scholar] [CrossRef]
- Liu, X.; Kosugi, S.; Koide, R.; Kawamura, Y.; Ito, J.; Miura, H.; Matoba, N.; Matsuzaki, M.; Fujita, M.; Kamada, A.J.; et al. Endogenization and excision of human herpesvirus 6 in human genomes. PLoS Genet. 2020, 16, e1008915. [Google Scholar] [CrossRef]
- Ablashi, D.; Balachandran, N.; Josephs, S.; Hung, C.; Krueger, G.; Kramarsky, B.; Salahuddin, S.; Gallo, R. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 1991, 184, 545–552. [Google Scholar] [CrossRef]
- Aubin, J.-T.; Agut, H.; Collandre, H.; Yamanishi, K.; Chandran, B.; Montagnier, L.; Jean-Marie, H. Antigenic and genetic differentiation of the two putative types of human herpes virus 6. J. Virol. Methods 1993, 41, 223–234. [Google Scholar] [CrossRef]
- Dewhurst, S.; Chandran, B.; McIntyre, K.; Schnabel, K.; Hall, C. Phenotypic and genetic polymorphisms among human herpesvirus-6 isolates from North American infants. Virology 1992, 190, 490–493. [Google Scholar] [CrossRef]
- Achour, A.; Malet, I.; Le Gal, F.; Dehée, A.; Gautheret-Dejean, A.; Bonnafous, P.; Agut, H. Variability of gB and gH genes of human herpesvirus-6 among clinical specimens. J. Med. Virol. 2008, 80, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Santoro, F.; Kennedy, P.; Locatelli, G.; Malnati, M.S.; Berger, E.; Lusso, P. CD46 Is a Cellular Receptor for Human Herpesvirus 6. Cell 1999, 99, 817–827. [Google Scholar] [CrossRef]
- Tang, H.; Serada, S.; Kawabata, A.; Ota, M.; Hayashi, E.; Naka, T.; Yamanishi, K.; Mori, Y. CD134 is a cellular receptor specific for human herpesvirus-6B entry. Proc. Natl. Acad. Sci. USA 2013, 110, 9096–9099. [Google Scholar] [CrossRef] [PubMed]
- Kondo, K.; Kondo, T.; Okuno, T.; Takahashi, M.; Yamanishi, K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 1991, 72, 1401–1408. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Soffritti, I.; D’Accolti, M.; Bortolotti, D.; Di Luca, D.; Caselli, E. HHV-6A/6B Infection of NK Cells Modulates the Expression of miRNAs and Transcription Factors Potentially Associated to Impaired NK Activity. Front. Microbiol. 2017, 8, 2143. [Google Scholar] [CrossRef] [PubMed]
- Caruso, A.; Caselli, E.; Fiorentini, S.; Rotola, A.; Prandini, A.; Garrafa, E.; Saba, E.; Alessandri, G.; Cassai, E.; Di Luca, D. U94 of human herpesvirus 6 inhibits in vitro angiogenesis and lymphangiogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 20446–20451. [Google Scholar] [CrossRef]
- Caruso, A.; Favilli, F.; Rotola, A.; Comar, M.; Horejsh, D.; Alessandri, G.; Grassi, M.; Di Luca, D.; Fiorentini, S. Human herpesvirus-6 modulates RANTES production in primary human endothelial cell cultures. J. Med. Virol. 2003, 70, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Caruso, A.; Rotola, A.; Comar, M.; Favilli, F.; Galvan, M.; Tosetti, M.; Campello, C.; Caselli, E.; Alessandri, G.; Grassi, M.; et al. HHV-6 infects human aortic and heart microvascular endothelial cells, increasing their ability to secrete proinflammatory chemokines. J. Med. Virol. 2002, 67, 528–533. [Google Scholar] [CrossRef]
- Caselli, E.; Zatelli, M.C.; Rizzo, R.; Benedetti, S.; Martorelli, D.; Trasforini, G.; Cassai, E.; Uberti, E.C.D.; Di Luca, D.; Dolcetti, R. Virologic and immunologic evidence supporting an association between hhv-6 and hashimoto’s thyroiditis. PLoS Pathog. 2012, 8, e1002951. [Google Scholar] [CrossRef]
- Thomas, D.; Liakos, V.; Michou, V.; Kapranos, N.; Kaltsas, G.; Tsilivakos, V.; Tsatsoulis, A. Detection of Herpes Virus DNA in Post-operative Thyroid Tissue Specimens of Patients with Autoimmune Thyroid Disease. Exp. Clin. Endocrinol. Diabetes 2008, 116, 35–39. [Google Scholar] [CrossRef]
- Fox, J.; Briggs, M.; Ward, P.; Tedder, R. Human herpesvirus 6 in salivary glands. Lancet 1990, 336, 590–593. [Google Scholar] [CrossRef]
- Marci, R.; Gentili, V.; Bortolotti, D.; Monte, G.L.; Caselli, E.; Bolzani, S.; Rotola, A.; Di Luca, D.; Rizzo, R. Presence of HHV-6A in Endometrial Epithelial Cells from Women with Primary Unexplained Infertility. PLoS ONE 2016, 11, e0158304. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Soffritti, I.; D’Accolti, M.; Bortolotti, D.; Rizzo, R.; Sighinolfi, G.; Giuggioli, D.; Ferri, C. HHV-6A Infection and Systemic Sclerosis: Clues of a Possible Association. Microorganisms 2019, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Robert, C.; Aubin, J.-T.; Visse, B.; Fillet, A.-M.; Huraux, J.-M.; Agut, H. Difference in permissiveness of human fibroblast cells to variants A and B of human herpesvirus-6. Res. Virol. 1996, 147, 219–225. [Google Scholar] [CrossRef]
- Luppi, M.; Barozzi, P.; Maiorana, A.; Marasca, R.; Torelli, G. Human Herpesvirus 6 Infection in Normal Human Brain Tissue. J. Infect. Dis. 1994, 169, 943–944. [Google Scholar] [CrossRef]
- Levine, P.H.; Jahan, N.; Murari, P.; Manak, M.; Jaffe, E.S. Detection of Human Herpesvirus 6 in Tissues Involved by Sinus Histiocytosis with Massive Lymphadenopathy (Rosai-Dorfman Disease). J. Infect. Dis. 1992, 166, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Cone, R.W.; Huang, M.L.; Ashley, R.; Corey, L. Human herpesvirus 6 DNA in peripheral blood cells and saliva from immunocompetent individuals. J. Clin. Microbiol. 1993, 31, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Okuno, T.; Higashi, K.; Shiraki, K.; Yamanishi, K.; Takahashi, M.; Kokado, Y.; Ishibashi, M.; Takahara, S.; Sonoda, T.; Tanaka, K.; et al. Human Herpesvirus 6 Infection in Renal Transplantation. Transplantation 1990, 49, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Krueger, G.; Wassermann, K.; De Clerck, L.; Stevens, W.; Bourgeois, N.; Ablashi, D.; Josephs, S.; Balachandran, N. Latent herpesvirus-6 in salivary and bronchial glands. Lancet 1990, 336, 1255–1256. [Google Scholar] [CrossRef]
- Challoner, P.B.; Smith, K.T.; Parker, J.D.; MacLeod, D.L.; Coulter, S.N.; Rose, T.M.; Schultz, E.R.; Bennett, J.L.; Garber, R.L.; Chang, M. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc. Natl. Acad. Sci. USA 1995, 92, 7440–7444. [Google Scholar] [CrossRef]
- Crawford, J.R.; Santi, M.R.; Thorarinsdottir, H.K.; Cornelison, R.; Rushing, E.J.; Zhang, H.; Yao, K.; Jacobson, S.; Macdonald, T.J. Detection of human herpesvirus-6 variants in pediatric brain tumors: Association of viral antigen in low grade gliomas. J. Clin. Virol. 2009, 46, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.R.; Santi, M.R.; Cornelison, R.; Sallinen, S.-L.; Haapasalo, H.; Macdonald, T.J. Detection of human herpesvirus-6 in adult central nervous system tumors: Predominance of early and late viral antigens in glial tumors. J. Neuro Oncol. 2009, 95, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.J.; Jacobson, S.; Levine, P.H. An evaluation of HHV-6 as an etiologic agent in Hodgkin lymphoma and brain cancer using IARC criteria for oncogenicity. Infect. Agents Cancer 2019, 14, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kondo, T.; Torigoe, S.; Okada, S.; Mukai, T.; Yamanishi, K. Human herpesvirus 7: Another causal agent for roseola (exanthem subitum). J. Pediatr. 1994, 125, 1–5. [Google Scholar] [CrossRef]
- Akashi, K.; Eizuru, Y.; Sumiyoshi, Y.; Minematsu, T.; Hara, S.; Harada, M.; Kikuchi, M.; Niho, Y.; Minamishima, Y. Brief report: Severe infectious mononucleosis-like syndrome and primary human herpesvirus 6 infection in an adult. N. Engl. J. Med. 1993, 329, 168–171. [Google Scholar] [CrossRef]
- Tesini, B.L.; Epstein, L.G.; Caserta, M.T. Clinical impact of primary infection with roseoloviruses. Curr. Opin. Virol. 2014, 9, 91–96. [Google Scholar] [CrossRef]
- Engdahl, E.; Gustafsson, R.; Huang, J.; Biström, M.; Bomfim, I.L.; Stridh, P.; Khademi, M.; Brenner, N.; Butt, J.; Michel, A.; et al. Increased Serological Response Against Human Herpesvirus 6A is Associated with Risk for Multiple Sclerosis. Front. Immunol. 2019, 10, 2715. [Google Scholar] [CrossRef]
- Scotet, E.; Peyrat, M.A.; Saulquin, X.; Retiere, C.; Couedel, C.; Davodeau, F.; Dulphy, N.; Toubert, A.; Bignon, J.D.; Lim, A.; et al. Frequent enrichment for cd8 t cells reactive against common herpes viruses in chronic inflammatory lesions: Towards a reassessment of the physiopathological significance of t cell clonal expansions found in autoimmune inflammatory processes. Eur. J. Immunol. 1999, 29, 973–985. [Google Scholar] [CrossRef]
- Agut, H.; Bonnafous, P.; Gautheret-Dejean, A. Laboratory and Clinical Aspects of Human Herpesvirus 6 Infections. Clin. Microbiol. Rev. 2015, 28, 313–335. [Google Scholar] [CrossRef]
- Pantry, S.N.; Medveczky, P.G. Latency, Integration, and Reactivation of Human Herpesvirus-6. Viruses 2017, 9, 194. [Google Scholar] [CrossRef]
- De Bolle, L.; Naesens, L.; De Clercq, E. Update on Human Herpesvirus 6 Biology, Clinical Features, and Therapy. Clin. Microbiol. Rev. 2005, 18, 217–245. [Google Scholar] [CrossRef] [PubMed]
- Cameron, B.; Flamand, L.; Juwana, H.; Middeldorp, J.M.; Naing, Z.; Rawlinson, W.D.; Ablashi, D.; Lloyd, A.R. Serological and virological investigation of the role of the herpesviruses EBV, CMV and HHV-6 in post-infective fatigue syndrome. J. Med. Virol. 2010, 82, 1684–1688. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Crawford, J.R.; Komaroff, A.L.; Ablashi, D.V.; Jacobson, S. Review part 2: Human herpesvirus-6 in central nervous system diseases. J. Med. Virol. 2010, 82, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.G.; Neely, M.N.; Gupta, S.; Lunn, M.R.; Loomis, K.S.; Pritchett, J.C.; Polsky, B.; Medveczky, P.G. Antiviral therapy of two patients with chromosomally-integrated human herpesvirus-6A presenting with cognitive dysfunction. J. Clin. Virol. 2012, 55, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Ablashi, D.V.; Lapps, W.; Kaplan, M.; Whitman, D.E.; Richert, J.R.; Pearson, G.R. Human herpesvirus-6 (hhv-6) infection in multiple sclerosis: A preliminary report. Mult. Scler. 1998, 4, 490–496. [Google Scholar] [CrossRef]
- Ablashi, D.; Eastman, H.; Owen, C.; Roman, M.; Friedman, J.; Zabriskie, J.; Peterson, D.; Pearson, G.; Whitman, J. Frequent HHV-6 reactivation in multiple sclerosis (MS) and chronic fatigue syndrome (CFS) patients. J. Clin. Virol. 2000, 16, 179–191. [Google Scholar] [CrossRef]
- Ben-Fredj, N.; Ben-Selma, W.; Rotola, A.; Nefzi, F.; Benedetti, S.; Frih-Ayed, M.; Di Luca, D.; Aouni, M.; Caselli, E. Prevalence of human herpesvirus U94/REP antibodies and DNA in Tunisian multiple sclerosis patients. J. NeuroVirol. 2013, 19, 42–47. [Google Scholar] [CrossRef]
- Caselli, E.; Boni, M.; Bracci, A.; Rotola, A.; Cermelli, C.; Castellazzi, M.; Di Luca, D.; Cassai, E. Detection of antibodies directed against human herpesvirus 6 U94/REP in sera of patients affected by multiple sclerosis. J. Clin. Microbiol. 2002, 40, 4131–4137. [Google Scholar] [CrossRef]
- Rizzo, R.; Gentili, V.; Casetta, I.; Caselli, E.; De Gennaro, R.; Granieri, E.; Cassai, E.; Di Luca, D.; Rotola, A. Altered natural killer cells’ response to herpes virus infection in multiple sclerosis involves KIR2DL2 expression. J. Neuroimmunol. 2012, 251, 55–64. [Google Scholar] [CrossRef]
- Broccolo, F.; Drago, F.; Cassina, G.; Fava, A.; Fusetti, L.; Matteoli, B.; Ceccherini-Nelli, L.; Sabbadini, M.G.; Lusso, P.; Parodi, A.; et al. Selective reactivation of human herpesvirus 6 in patients with autoimmune connective tissue diseases. J. Med. Virol. 2013, 85, 1925–1934. [Google Scholar] [CrossRef]
- Broccolo, F.; Drago, F.; Paolino, S.; Cassina, G.; Gatto, F.; Fusetti, L.; Matteoli, B.; Zaccaria, E.; Parodi, A.; Lusso, P.; et al. Reactivation of human herpesvirus 6 (HHV-6) infection in patients with connective tissue diseases. J. Clin. Virol. 2009, 46, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Broccolo, F.; Fusetti, L.; Ceccherini-Nelli, L. Possible Role of Human Herpesvirus 6 as a Trigger of Autoimmune Disease. Sci. World J. 2013, 2013, 1–7. [Google Scholar] [CrossRef]
- Caselli, E.; D’Accolti, M.; Soffritti, I.; Zatelli, M.C.; Rossi, R.; Degli Uberti, E.; Di Luca, D. HHV-6A in vitro infection of thyrocytes and T cells alters the expression of miRNA associated to autoimmune thyroiditis. Virol. J. 2017, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Caselli, E.; Bortolotti, D.; Marci, R.; Rotola, A.; Gentili, V.; Soffritti, I.; D’Accolti, M.; Monte, G.L.; Sicolo, M.; Barao, I.; et al. HHV-6A Infection of Endometrial Epithelial Cells Induces Increased Endometrial NK Cell-Mediated Cytotoxicity. Front. Microbiol. 2017, 8, 2525. [Google Scholar] [CrossRef] [PubMed]
- Komaroff, A.L. Is human herpesvirus-6 a trigger for chronic fatigue syndrome? J. Clin. Virol. 2006, 37, S39–S46. [Google Scholar] [CrossRef]
- Miyagawa, F.; Nakamura, Y.; Miyashita, K.; Iioka, H.; Himuro, Y.; Ogawa, K.; Nishimura, C.; Nishikawa, M.; Mitsui, Y.; Ito, Y.; et al. Preferential expression of CD134, an HHV-6 cellular receptor, on CD4T cells in drug-induced hypersensitivity syndrome (DIHS)/drug reaction with eosinophilia and systemic symptoms (DRESS). J. Dermatol. Sci. 2016, 83, 151–154. [Google Scholar] [CrossRef]
- Suzuki, Y.; Inagi, R.; Aono, T.; Yamanishi, K.; Shiohara, T. Human Herpesvirus 6 Infection as a Risk Factor for the Development of Severe Drug-Induced Hypersensitivity Syndrome. Arch. Dermatol. 1998, 134, 1108–1112. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.-V.; Funk, C.C.; Richards, M.A.; Shannon, P.; Haroutunian, V.; Sano, M.; Liang, W.S.; Beckmann, N.D.; Price, N.D.; et al. Multiscale Analysis of Independent Alzheimer’s Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron 2018, 99, 64–82.e7. [Google Scholar] [CrossRef]
- Rizzo, R.; Bortolotti, D.; Gentili, V.; Rotola, A.; Bolzani, S.; Caselli, E.; Tola, M.R.; Di Luca, D. Kir2ds2/kir2dl2/hla-c1 haplotype is associated with alzheimer’s disease: Implication for the role of herpesvirus infections. J. Alzheimers Dis. 2019, 67, 1379–1389. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, T.; Liu, M.; Zhang, L.; Wang, F.; Zhao, S.; Liu, H.; Xia, H.; Wang, Y.; Li, L. Positive detection of SARS-Cov-2 combined hsv1 and hhv6b virus nucleic acid in tear and conjunctival secretions of a non-conjunctivitis Covid-19 patient with obstruction of common lacrimal duct. Acta Ophthalmol. 2020. [Google Scholar] [CrossRef]
- Piper, C.; Zhou, V.; Komorowski, R.; Szabo, A.; Vincent, B.; Serody, J.S.; Alegre, M.-L.; Edelson, B.T.; Taneja, R.; Drobyski, W.R. Pathogenic Bhlhe40+ GM-CSF+ CD4+ T cells promote indirect alloantigen presentation in the GI tract during GVHD. Blood 2020, 135, 568–581. [Google Scholar] [CrossRef] [PubMed]
- Dursun, R.; Temiz, S.A. The clinics of HHV-6 infection in COVID-19 pandemic: Pityriasis rosea and Kawasaki disease. Dermatol. Ther. 2020, 33, e13730. [Google Scholar] [CrossRef] [PubMed]
- Finkel, Y.; Schmiedel, D.; Tai-Schmiedel, J.; Nachshon, A.; Winkler, R.; Dobesova, M.; Schwartz, M.; Mandelboim, O.; Stern-Ginossar, N. Comprehensive annotations of human herpesvirus 6A and 6B genomes reveal novel and conserved genomic features. eLife 2020, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Gompels, U.; Nicholas, J.; Lawrence, G.; Jones, M.; Thomson, B.; Martin, M.; Efstathiou, S.; Craxton, M.; Macaulay, H. The DNA Sequence of Human Herpesvirus-6: Structure, Coding Content, and Genome Evolution. Virology 1995, 209, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Sanyal, A.; Pirzer, F.; Gravel, A.; Mori, Y.; Flamand, L.; Kaufer, B.B. The Telomeric Repeats of Human Herpesvirus 6A (HHV-6A) Are Required for Efficient Virus Integration. PLoS Pathog. 2016, 12, e1005666. [Google Scholar] [CrossRef] [PubMed]
- Gilbert-Girard, S.; Gravel, A.; Collin, V.; Wight, D.J.; Kaufer, B.B.; Lazzerini-Denchi, E.; Flamand, L. Role for the shelterin protein TRF2 in human herpesvirus 6A/B chromosomal integration. PLoS Pathog. 2020, 16, e1008496. [Google Scholar] [CrossRef]
- Aimola, G.; Beythien, G.; Aswad, A.; Kaufer, B.B. Current understanding of human herpesvirus 6 (HHV-6) chromosomal integration. Antivir. Res. 2020, 176, 104720. [Google Scholar] [CrossRef]
- Stanton, R.J.; Wilkinson, G.W.G.; Fox, J.D. Analysis of human herpesvirus-6 IE1 sequence variation in clinical samples. J. Med. Virol. 2003, 71, 578–584. [Google Scholar] [CrossRef]
- Dominguez, G.; Dambaugh, T.R.; Stamey, F.R.; Dewhurst, S.; Inoue, N.; Pellett, P.E. Human Herpesvirus 6B Genome Sequence: Coding Content and Comparison with Human Herpesvirus 6A. J. Virol. 1999, 73, 8040–8052. [Google Scholar] [CrossRef]
- Gravel, A.; Gosselin, J.; Flamand, L. Human Herpesvirus 6 Immediate-Early 1 Protein Is a Sumoylated Nuclear Phosphoprotein Colocalizing with Promyelocytic Leukemia Protein-associated Nuclear Bodies. J. Biol. Chem. 2002, 277, 19679–19687. [Google Scholar] [CrossRef]
- Rapp, J.C.; Krug, L.T.; Inoue, N.; Dambaugh, T.R.; Pellett, P.E. U94, the Human Herpesvirus 6 Homolog of the Parvovirus Nonstructural Gene, Is Highly Conserved among Isolates and Is Expressed at Low mRNA Levels as a Spliced Transcript. Virology 2000, 268, 504–516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vink, C.; Beuken, E.; Bruggeman, C.A.; Preston, C.M.; Harman, A.N.; Nicholl, M.J. Complete DNA Sequence of the Rat Cytomegalovirus Genome. J. Virol. 2000, 74, 7656–7665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Todd, S.; Tachedjian, M.; Barr, J.A.; Luo, M.; Yu, M.; Marsh, G.A.; Crameri, G.; Wang, L.-F. A Novel Bat Herpesvirus Encodes Homologues of Major Histocompatibility Complex Classes I and II, C-Type Lectin, and a Unique Family of Immune-Related Genes. J. Virol. 2012, 86, 8014–8030. [Google Scholar] [CrossRef] [PubMed]
- Voigt, S.; Sandford, G.R.; Hayward, G.S.; Burns, W.H. The English strain of rat cytomegalovirus (CMV) contains a novel captured CD200 (vOX2) gene and a spliced CC chemokine upstream from the major immediate-early region: Further evidence for a separate evolutionary lineage from that of rat CMV Maastricht. J. Gen. Virol. 2005, 86, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Van Cleef, K.W.R.; Scaf, W.M.A.; Maes, K.; Kaptein, S.J.F.; Beuken, E.; Beisser, P.S.; Stassen, F.R.; Grauls, G.E.L.M.; Bruggeman, C.A.; Vink, C. The rat cytomegalovirus homologue of parvoviral rep genes, r127, encodes a nuclear protein with single- and double-stranded DNA-binding activity that is dispensable for virus replication. J. Gen. Virol. 2004, 85, 2001–2013. [Google Scholar] [CrossRef]
- Thomson, B.J.; Efstathiou, S.; Honess, R.W. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nat. Cell Biol. 1991, 351, 78–80. [Google Scholar] [CrossRef]
- Arbuckle, J.H.; Medveczky, P.G. The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect. 2011, 13, 731–741. [Google Scholar] [CrossRef]
- Im, D.S.; Muzyczka, N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J. Virol. 1992, 66, 1119–1128. [Google Scholar] [CrossRef]
- Im, D.-S.; Muzyczka, N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 1990, 61, 447–457. [Google Scholar] [CrossRef]
- Linden, R.M.; Ward, P.; Giraud, C.; Winocour, E.; Berns, K.I. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 1996, 93, 11288–11294. [Google Scholar] [CrossRef]
- Linden, R.M.; Winocour, E.; Berns, K.I. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. USA 1996, 93, 7966–7972. [Google Scholar] [CrossRef]
- Thomson, B.J.; Weindler, F.W.; Gray, D.; Schwaab, V.; Heilbronn, R. Human Herpesvirus 6 (HHV-6) Is a Helper Virus for Adeno-Associated Virus Type 2 (AAV-2) and the AAV-2 rep Gene Homologue in HHV-6 Can Mediate AAV-2 DNA Replication and Regulate Gene Expression. Virology 1994, 204, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Nash, K.; Chen, W.; Salganik, M.; Muzyczka, N. Identification of Cellular Proteins That Interact with the Adeno-Associated Virus Rep Protein. J. Virol. 2008, 83, 454–469. [Google Scholar] [CrossRef]
- Mori, Y.; Dhepakson, P.; Shimamoto, T.; Ueda, K.; Gomi, Y.; Tani, H.; Matsuura, Y.; Yamanishi, K. Expression of human herpesvirus 6b rep within infected cells and binding of its gene product to the tata-binding protein in vitro and in vivo. J. Virol. 2000, 74, 6096–6104. [Google Scholar] [CrossRef]
- Araujo, J.C.; Doniger, J.; Kashanchi, F.; Hermonat, P.L.; Thompson, J.; Rosenthal, L.J. Human herpesvirus 6A ts suppresses both transformation by H-ras and transcription by the H-ras and human immunodeficiency virus type 1 promoters. J. Virol. 1995, 69, 4933–4940. [Google Scholar] [CrossRef]
- Caselli, E.; Bracci, A.; Galvan, M.; Boni, M.; Rotola, A.; Bergamini, C.; Cermelli, C.; Monte, P.D.; Gompels, U.A.; Cassai, E.; et al. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology 2006, 346, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Dhepakson, P.; Mori, Y.; Jiang, Y.B.; Huang, H.L.; Akkapaiboon, P.; Okuno, T.; Yamanishi, K. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J. Gen. Virol. 2002, 83, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Trempe, F.; Gravel, A.; Dubuc, I.; Wallaschek, N.; Collin, V.; Gilbert-Girard, S.; Morissette, G.; Kaufer, B.B.; Flamand, L. Characterization of human herpesvirus 6A/B U94 as ATPase, helicase, exonuclease and DNA-binding proteins. Nucleic Acids Res. 2015, 43, 6084–6098. [Google Scholar] [CrossRef]
- Mirandola, P.; Menegazzi, P.; Merighi, S.; Ravaioli, T.; Cassai, E.; Di Luca, D. Temporal Mapping of Transcripts in Herpesvirus 6 Variants. J. Virol. 1998, 72, 3837–3844. [Google Scholar] [CrossRef]
- Rotola, A.; Ravaioli, T.; Gonelli, A.; Dewhurst, S.; Cassai, E.; Di Luca, D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. USA 1998, 95, 13911–13916. [Google Scholar] [CrossRef]
- Turner, S.; DiLuca, D.; Gompels, U. Characterisation of a human herpesvirus 6 variant A ’amplicon’ and replication modulation by U94-Rep ’latency gene’. J. Virol. Methods 2002, 105, 331–341. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Akimoto, S.; Nishimura, N.; Ozaki, T.; Ihira, M.; Ohashi, M.; Morooka, M.; Suga, S.; Asano, Y.; Takemoto, M.; et al. Evaluation of active human herpesvirus 6 infection by reverse transcription-PCR. J. Med. Virol. 2003, 70, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Rotola, A.; Cassai, E.; Tola, M.R.; Granieri, E.; Di Luca, D. Human herpesvirus 6 is latent in peripheral blood of patients with relapsing-remitting multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 1999, 67, 529–531. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kondo, K.; Shimada, K.; Sashihara, J.; Tanaka-Taya, K.; Yamanishi, K. Identification of Human Herpesvirus 6 Latency-Associated Transcripts. J. Virol. 2002, 76, 4145–4151. [Google Scholar] [CrossRef]
- Luppi, M.; Marasca, R.; Barozzi, P.; Ferrari, S.; Ceccherini-Nelli, L.; Batoni, G.; Merelli, E.; Torelli, G. Three cases of human herpesvirus-6 latent infection: Integration of viral genome in peripheral blood mononuclear cell DNA. J. Med. Virol. 1993, 40, 44–52. [Google Scholar] [CrossRef]
- Luppi, M.; Barozzi, P.; Marasca, R.; Torelli, G. Integration of human herpesvirus-6 (HHV-6) genome in chromosome 17 in two lymphoma patients. Leukemia 1994, 8, 41–45. [Google Scholar]
- Ahlqvist, J.; Fotheringham, J.; Akhyani, N.; Yao, K.; Fogdell-Hahn, A.; Jacobson, S. Differential tropism of human herpesvirus 6 (HHV-6) variants and induction of latency by HHV-6A in oligodendrocytes. J. NeuroVirol. 2005, 11, 384–394. [Google Scholar] [CrossRef]
- Luppi, M.; Barozzi, P.; Morris, C.; Maiorana, A.; Garber, R.; Bonacorsi, G.; Donelli, A.; Marasca, R.; Tabilio, A.; Torelli, G. Human Herpesvirus 6 Latently Infects Early Bone Marrow Progenitors In Vivo. J. Virol. 1999, 73, 754–759. [Google Scholar] [CrossRef]
- Saviola, A.J.; Zimmermann, C.; Mariani, M.P.; Signorelli, S.A.; Gerrard, D.L.; Boyd, J.R.; Wight, D.J.; Morissette, G.; Gravel, A.; Dubuc, I.; et al. Chromatin Profiles of Chromosomally Integrated Human Herpesvirus-6A. Front. Microbiol. 2019, 10, 1408. [Google Scholar] [CrossRef]
- Kaufer, B.B.; Flamand, L. Chromosomally integrated HHV-6: Impact on virus, cell and organismal biology. Curr. Opin. Virol. 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Aswad, A.; Katzourakis, A. The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses. PLoS Genet. 2014, 10, e1004332. [Google Scholar] [CrossRef] [PubMed]
- Gulve, N.; Frank, C.; Klepsch, M.; Prusty, B.K. Chromosomal integration of HHV-6A during non-productive viral infection. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gravel, A.; Dubuc, I.; Morissette, G.; Sedlak, R.H.; Jerome, K.R.; Flamand, L. Inherited chromosomally integrated human herpesvirus 6 as a predisposing risk factor for the development of angina pectoris. Proc. Natl. Acad. Sci. USA 2015, 112, 8058–8063. [Google Scholar] [CrossRef] [PubMed]
- Gaccioli, F.; Lager, S.; De Goffau, M.C.; Sovio, U.; Dopierala, J.; Gong, S.; Cook, E.; Sharkey, A.; Moffett, A.; Lee, W.K.; et al. Fetal inheritance of chromosomally integrated human herpesvirus 6 predisposes the mother to pre-eclampsia. Nat. Microbiol. 2020, 5, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Strenger, V.; Caselli, E.; Lautenschlager, I.; Schwinger, W.; Aberle, S.W.; Loginov, R.; Gentili, V.; Nacheva, E.; Di Luca, D.; Urban, C. Detection of HHV-6-specific mRNA and antigens in PBMCs of individuals with chromosomally integrated HHV-6 (ciHHV-6). Clin. Microbiol. Infect. 2014, 20, 1027–1032. [Google Scholar] [CrossRef]
- Politikos, I.; McMasters, M.; Bryke, C.; Avigan, D.; Boussiotis, V.A. Possible reactivation of chromosomally integrated human herpesvirus 6 after treatment with histone deacetylase inhibitor. Blood Adv. 2018, 2, 1367–1370. [Google Scholar] [CrossRef]
- Endo, A.; Watanabe, K.; Ohye, T.; Suzuki, K.; Matsubara, T.; Shimizu, N.; Kurahashi, H.; Yoshikawa, T.; Katano, H.; Inoue, N.; et al. Molecular and Virological Evidence of Viral Activation From Chromosomally Integrated Human Herpesvirus 6A in a Patient With X-Linked Severe Combined Immunodeficiency. Clin. Infect. Dis. 2014, 59, 545–548. [Google Scholar] [CrossRef]
- Gomples, U.A.; Macaulay, H.A. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J. Gen. Virol. 1995, 76, 451–458. [Google Scholar] [CrossRef]
- Thomson, B.J.; Dewhurst, S.; Gray, D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J. Virol. 1994, 68, 3007–3014. [Google Scholar] [CrossRef]
- Arbuckle, J.H.; Medveczky, M.M.; Luka, J.; Hadley, S.H.; Luegmayr, A.; Ablashi, D.; Lund, T.C.; Tolar, J.; De Meirleir, K.; Montoya, J.G.; et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 5563–5568. [Google Scholar] [CrossRef]
- Huang, Y.; Hidalgo-Bravo, A.; Zhang, E.; Cotton, V.E.; Mendez-Bermudez, A.; Wig, G.; Medina-Calzada, Z.; Neumann, R.; Jeffreys, A.J.; Winney, B.; et al. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res. 2013, 42, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.B.; Caserta, M.T.; Schnabel, K.C.; Boettrich, C.; McDermott, M.P.; Lofthus, G.K.; Carnahan, J.A.; Dewhurst, S. Congenital infections with human herpesvirus 6 (HHV6) and human herpesvirus 7 (HHV7). J. Pediatr. 2004, 145, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Wallaschek, N.; Gravel, A.; Flamand, L.; Kaufer, B.B. The putative U94 integrase is dispensable for human herpesvirus 6 (HHV-6) chromosomal integration. J. Gen. Virol. 2016, 97, 1899–1903. [Google Scholar] [CrossRef] [PubMed]
- Wight, D.J.; Wallaschek, N.; Sanyal, A.; Weller, S.K.; Flamand, L.; Kaufer, B.B. Viral Proteins U41 and U70 of Human Herpesvirus 6A Are Dispensable for Telomere Integration. Viruses 2018, 10, 656. [Google Scholar] [CrossRef]
- Collin, V.; Gravel, A.; Kaufer, B.B.; Flamand, L. The Promyelocytic Leukemia Protein facilitates human herpesvirus 6B chromosomal integration, immediate-early 1 protein multiSUMOylation and its localization at telomeres. PLoS Pathog. 2020, 16, e1008683. [Google Scholar] [CrossRef]
- Kumata, R.; Ito, J.; Sato, K. Inherited chromosomally integrated HHV-6 possibly modulates human gene expression. Virus Genes 2020, 56, 386–389. [Google Scholar] [CrossRef]
- Rizzo, R.; Zatelli, M.C.; Rotola, A.; Cassai, E.; Degli Uberti, E.; Di Luca, D.; Caselli, E. Increase in Peripheral CD3−CD56brightCD16−Natural Killer Cells in Hashimoto’s Thyroiditis Associated with HHV-6 Infection. Retin. Degener. Dis. 2015, 897, 113–120. [Google Scholar] [CrossRef]
- Hunt, J.S.; Langat, D.K.; McIntire, R.H.; Morales, P.J. The role of HLA-G in human pregnancy. Reprod. Biol. Endocrinol. 2006, 4, S10. [Google Scholar] [CrossRef]
- Caselli, E.; Campioni, D.; Cavazzini, F.; Gentili, V.; Bortolotti, D.; Cuneo, A.; Di Luca, D.; Rizzo, R. Acute human herpesvirus-6A infection of human mesothelial cells modulates HLA molecules. Arch. Virol. 2015, 160, 2141–2149. [Google Scholar] [CrossRef]
- Rizzo, R.; D’Accolti, M.; Bortolotti, D.; Caccuri, F.; Caruso, A.; Di Luca, D.; Caselli, E. Human Herpesvirus 6A and 6B inhibit in vitro angiogenesis by induction of Human Leukocyte Antigen G. Sci. Rep. 2018, 8, 17683. [Google Scholar] [CrossRef]
- Campbell, A.; Hogestyn, J.M.; Folts, C.J.; Lopez, B.; Pröschel, C.; Mock, D.J.; Mayer-Pröschel, M. Expression of the Human Herpesvirus 6A Latency-Associated Transcript U94A Disrupts Human Oligodendrocyte Progenitor Migration. Sci. Rep. 2017, 7, 3978. [Google Scholar] [CrossRef] [PubMed]
- Rotola, A.; Ricotta, D.; Muneretto, C.; Di Luca, D.; Cassai, E.; Giulio, A.; Turano, A.; Caruso, A. Human Herpesvirus 6 Infects and Replicates in Aortic Endothelium. J. Clin. Microbiol. 2000, 38, 3135–3136. [Google Scholar] [CrossRef] [PubMed]
- Fons, P.; Chabot, S.; Cartwright, J.E.; Lenfant, F.; L’Faqihi, F.; Giustiniani, J.; Herault, J.-P.; Gueguen, G.; Bono, F.; Savi, P.; et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood 2006, 108, 2608–2615. [Google Scholar] [CrossRef] [PubMed]
- Araujo, J.C.; Doniger, J.; Stöppler, H.; Sadaie, M.R.; Rosenthal, L.J. Cell lines containing and expressing the human herpesvirus 6A ts gene are protected from both H-ras and BPV-1 transformation. Oncogene 1997, 14, 937–943. [Google Scholar] [CrossRef][Green Version]
- Ifon, E.T.; Pang, A.L.; Johnson, W.E.; Cashman, K.; Zimmerman, S.; Muralidhar, S.; Chan, W.-Y.; Casey, J.L.; Rosenthal, L.J. U94 alters FN1 and ANGPTL4 gene expression and inhibits tumorigenesis of prostate cancer cell line PC3. Cancer Cell Int. 2005, 5, 19. [Google Scholar] [CrossRef][Green Version]
- Caccuri, F.; Ronca, R.; Laimbacher, A.S.; Berenzi, A.; Steimberg, N.; Campilongo, F.; Mazzuca, P.; Giacomini, A.; Mazzoleni, G.; Benetti, A.; et al. U94 of human herpesvirus 6 down-modulates Src, promotes a partial mesenchymal-to-epithelial transition and inhibits tumor cell growth, invasion and metastasis. Oncotarget 2017, 8, 44533–44549. [Google Scholar] [CrossRef][Green Version]
- Gu, B.; Li, L.; Li, M.; Wang, J.; Zhang, G.; Yao, K.; Wang, S. U94/rep of human herpesvirus 6 inhibits proliferation, invasion, and angiogenesis of glioma. Cancer Manag. Res. 2018, 10, 5991–6001. [Google Scholar] [CrossRef]
- Caccuri, F.; Sommariva, M.; Marsico, S.; Giordano, F.; Zani, A.; Giacomini, A.; Fraefel, C.; Balsari, A.; Caruso, A. Inhibition of DNA Repair Mechanisms and Induction of Apoptosis in Triple Negative Breast Cancer Cells Expressing the Human Herpesvirus 6 U94. Cancers 2019, 11, 1006. [Google Scholar] [CrossRef]


| Field of Action | Context | Results | Condition | Reference |
|---|---|---|---|---|
| Humoral Immunity | Anti-U94 IgG | Increased | Acutely HHV-6 infected subjects | [49] |
| Anti-U94 IgG | Increased prevalence/titer compared to control population | Multiple sclerosis patients (serum/plasma) | [49] | |
| Anti-U94 IgG | Increased prevalence/titer compared to control population | Hashimoto’s thyroiditis patients (serum/plasma) | [20] | |
| Anti-U94 IgG | Increased prevalence/titer compared to control population | Systemic sclerosis patients (serum/plasma) | [24] | |
| Cell-mediated immunity | Anti-U94 CD4+/CD8+ T-cells | Increased number | Hashimoto’s thyroiditis patients (purified PBMCs) | [20] |
| Innate immunity | NK cells | Decreased NK activation | Systemic sclerosis patients (purified PBMCs) | [24] |
| Mesothelium | Mesothelial cells | HLA-G induction | In vitro impairment of NK response against infected cells | [120] |
| Demyelination (multiple sclerosis) | Oligodendrocytes progenitors | Inhibition of migration of hOPCs | In vitro studies; animal model (NSG mice) | [122] |
| Angiogenesis | Vascular and lymphatic endothelial cells | Inhibition of angiogenesis | In vitro studies | [17] |
| Vascular endothelial cells (HUVEC) | HLA-G induction (via ATF3 activation) | In vitro studies | [121] | |
| Rat aortic rings | Inhibition of migration | Ex vivo studies | [17] | |
| Tumorigenesis | NIH3T3 cells | Suppression of transformation by ras oncogene | In vitro studies | [126] |
| PC3 cells | Inhibition of tumorigenesis | Animal model (athymic nude mice) | [127] | |
| MDA-MB-231 cells | Down-modulation of src oncogene; inhibition of cell motility and invasion; mesenchymal-to-epithelial transition | In vitro studies | [125] | |
| MDA-MB-231 cells | Down-regulation of TWIST, N-cadherin, Snail1 and MMP2 | In vitro studies | [125] | |
| MDA-MB-231 cells | Decrease of tumor growth | Animal model (NOD/SCID mice) | ||
| Glioma | Down-regulation of proangiogenic factors and MMPs | Ex vivo studies (human gliomas) | [128] | |
| Triple-negative breast cancer (TNBC) cells | Inhibition of DMA repair mechanisms, induction of apoptosis; synergistic action with anticancer drugs | In vitro studies | [129] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caselli, E.; D’Accolti, M.; Caccuri, F.; Soffritti, I.; Gentili, V.; Bortolotti, D.; Rotola, A.; Cassai, E.; Fiorentini, S.; Zani, A.; et al. The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions. Cells 2020, 9, 2608. https://doi.org/10.3390/cells9122608
Caselli E, D’Accolti M, Caccuri F, Soffritti I, Gentili V, Bortolotti D, Rotola A, Cassai E, Fiorentini S, Zani A, et al. The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions. Cells. 2020; 9(12):2608. https://doi.org/10.3390/cells9122608
Chicago/Turabian StyleCaselli, Elisabetta, Maria D’Accolti, Francesca Caccuri, Irene Soffritti, Valentina Gentili, Daria Bortolotti, Antonella Rotola, Enzo Cassai, Simona Fiorentini, Alberto Zani, and et al. 2020. "The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions" Cells 9, no. 12: 2608. https://doi.org/10.3390/cells9122608
APA StyleCaselli, E., D’Accolti, M., Caccuri, F., Soffritti, I., Gentili, V., Bortolotti, D., Rotola, A., Cassai, E., Fiorentini, S., Zani, A., Caruso, A., Rizzo, R., & Di Luca, D. (2020). The U94 Gene of Human Herpesvirus 6: A Narrative Review of Its Role and Potential Functions. Cells, 9(12), 2608. https://doi.org/10.3390/cells9122608








