Cytokinin and Ethylene Cell Signaling Pathways from Prokaryotes to Eukaryotes
Abstract
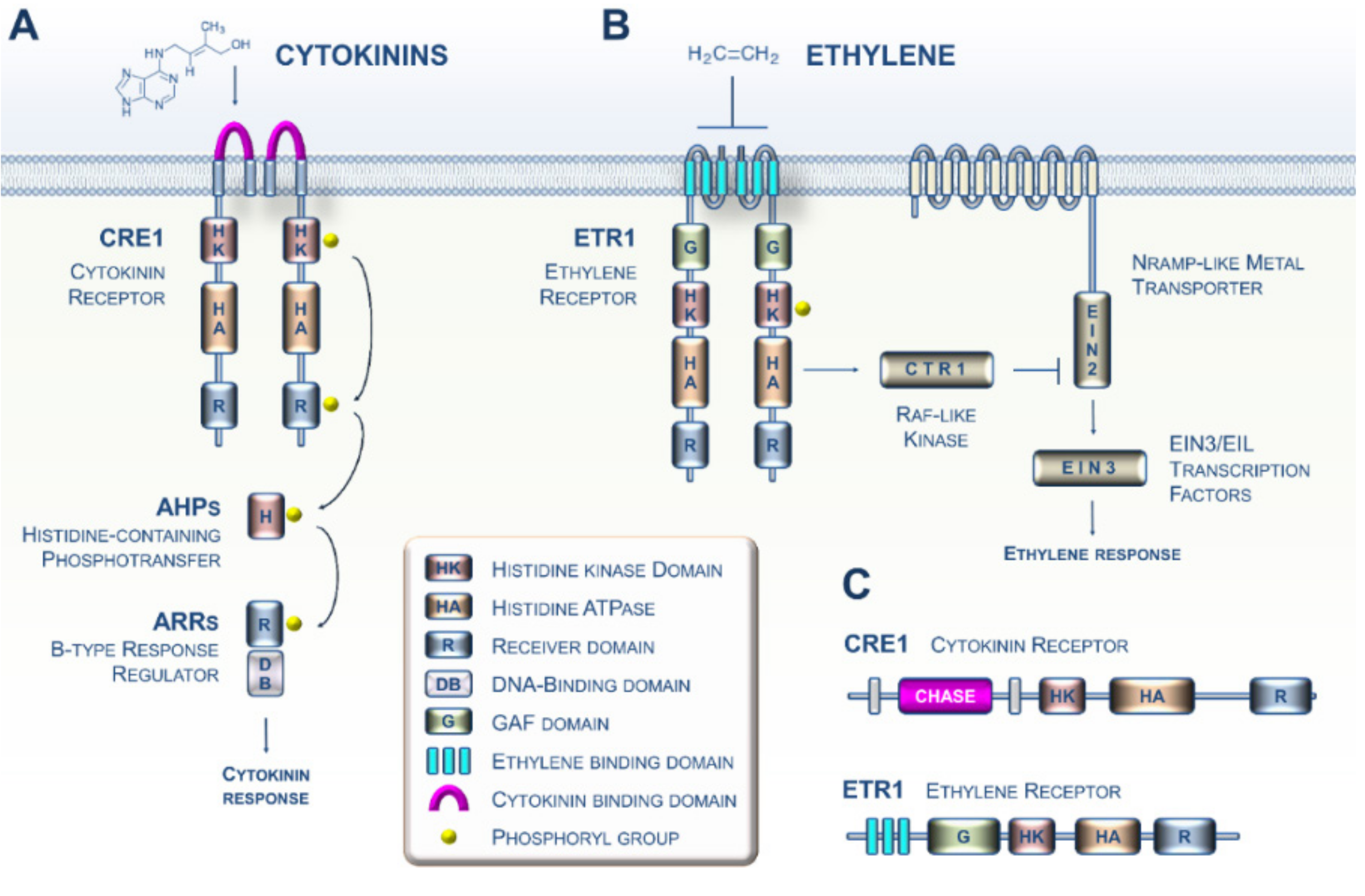
1. CK and ET Signaling in Plants: Beyond the Arabidopsis Paradigm
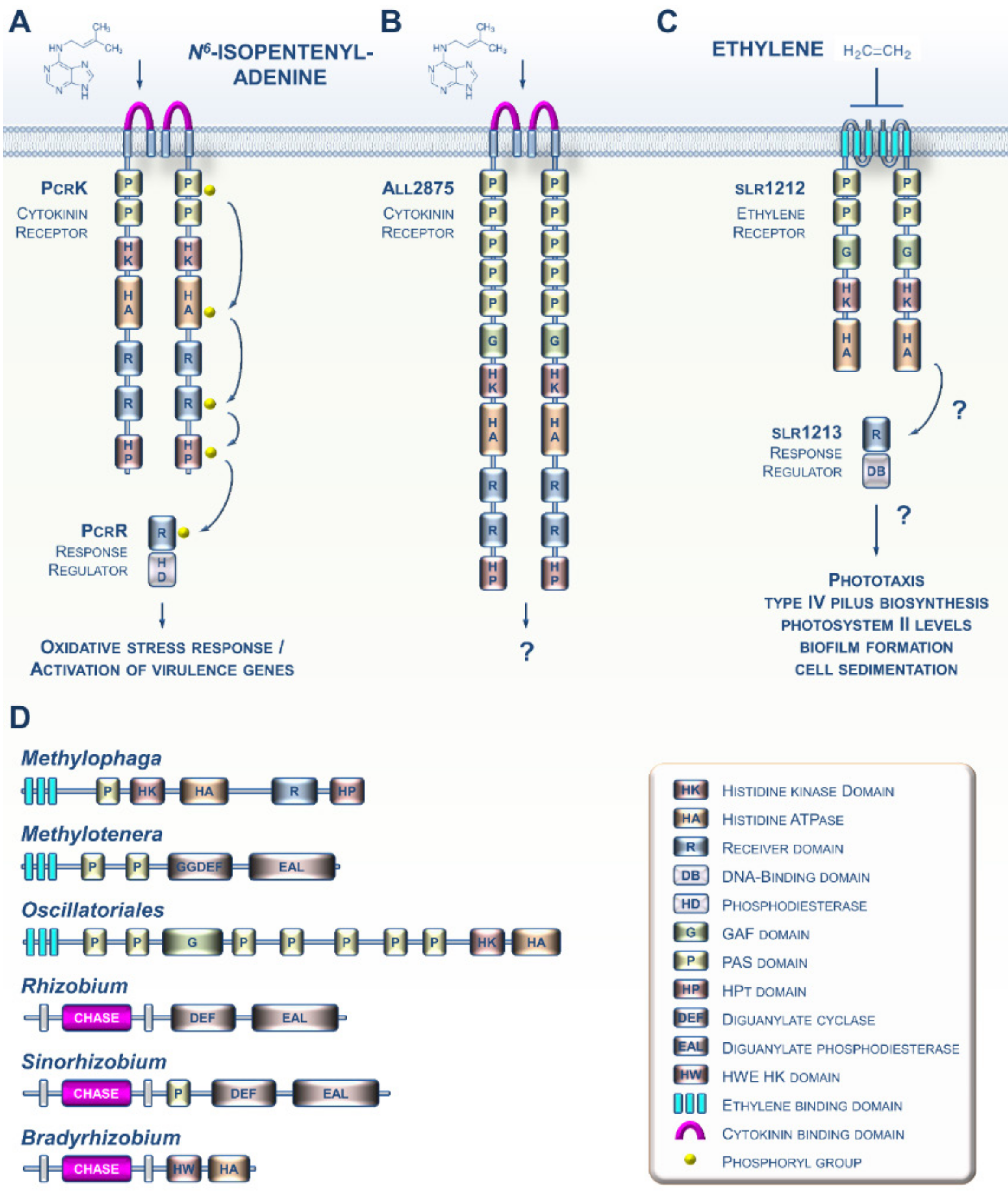
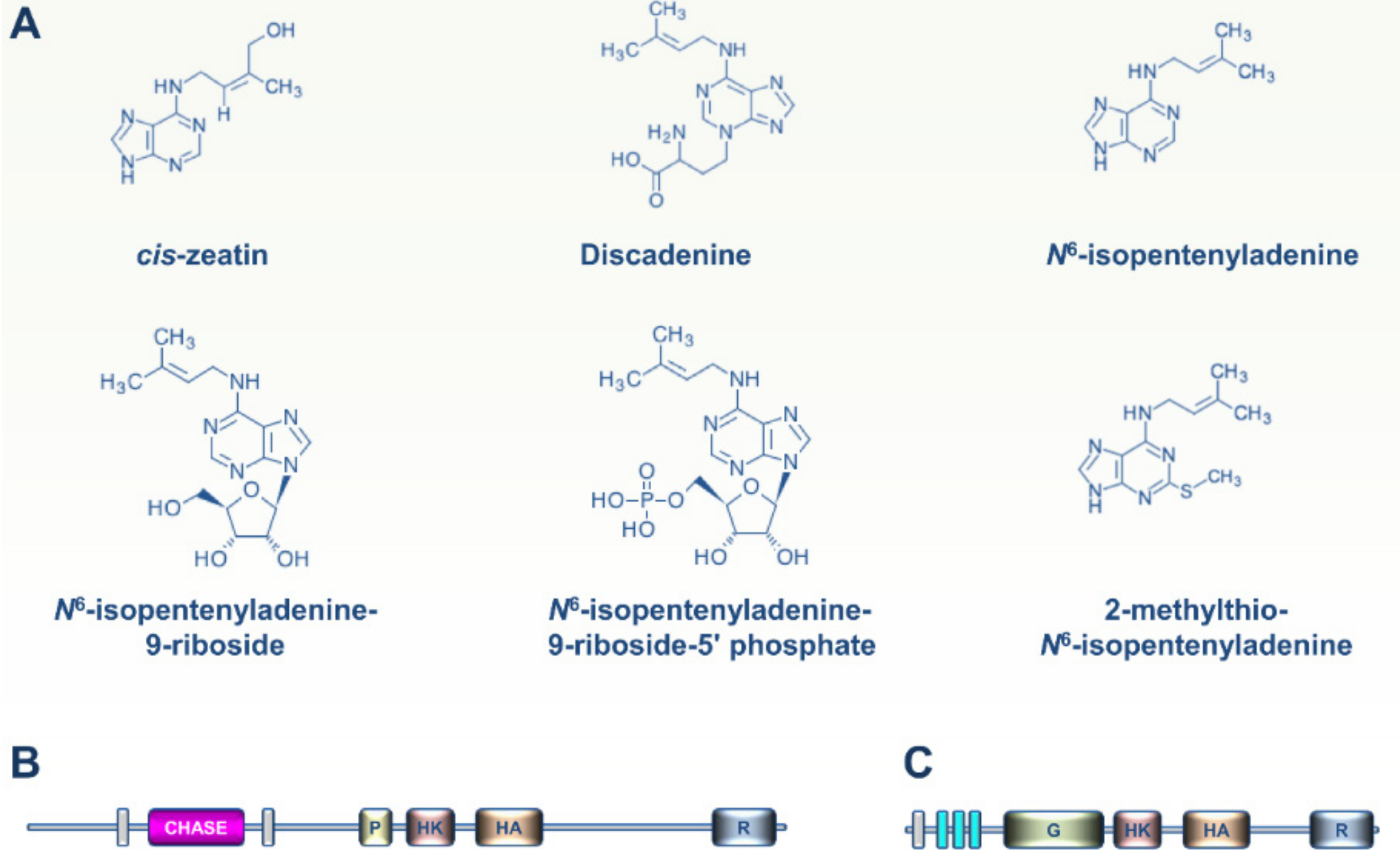
2. CK and ET Signaling in Bacteria
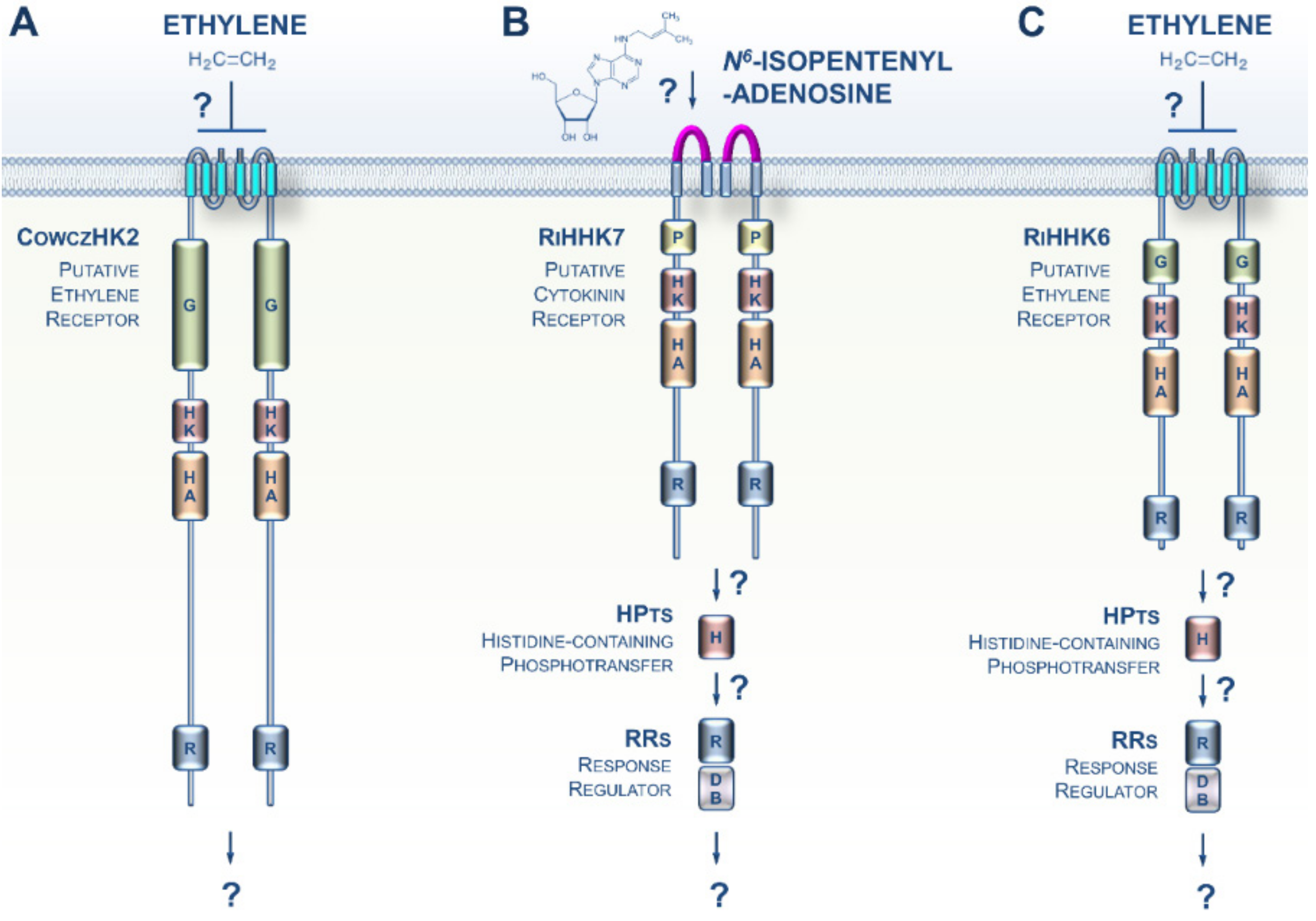
3. CK and ET Signaling in Opisthokonta
4. CKs and ET Signaling in Amoebozoa
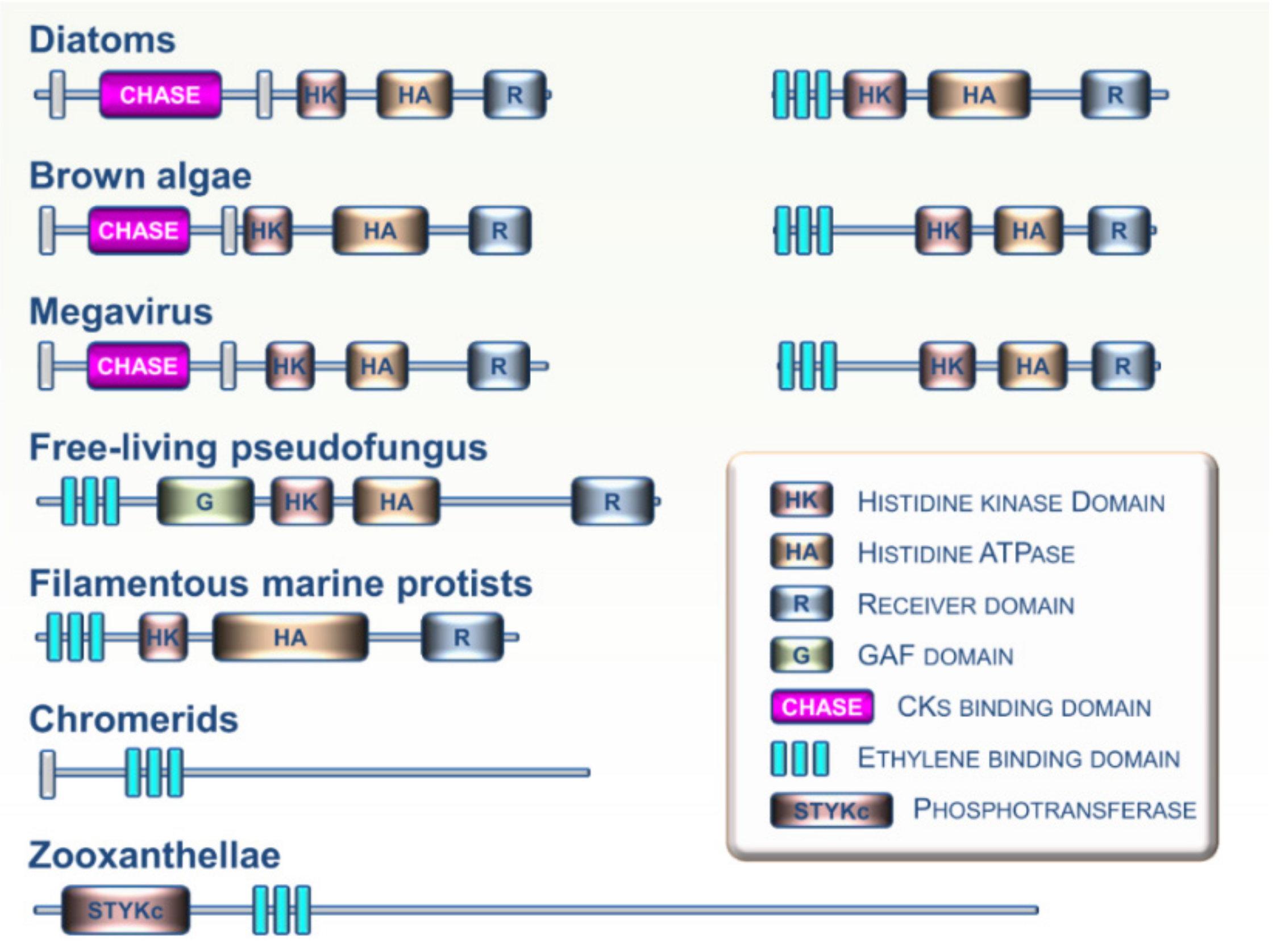
5. Viewpoint: An Evolutionary Perspective of CKs and ET Perception within the Tree of Life
Funding
Conflicts of Interest
References
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef] [PubMed]
- Abeles, F.; Morgan, P.; Saltveit, M.J. Ethylene in Plant Biology, 2nd ed.; Academic Press: San Diego, CA, USA, 1992. [Google Scholar]
- Rashotte, A.M. The evolution of cytokinin signaling and its role in development before Angiosperms. Semin. Cell Dev. Biol. 2020, 29. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Shiu, S.-H.; Armitage, J.P. Two-Component Systems and Their Co-Option for Eukaryotic Signal Transduction. Curr. Biol. 2011, 21, R320–R330. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Stock, A.M. Two-component systems. Curr. Biol. 2019, 29, R724–R725. [Google Scholar] [CrossRef]
- Inoue, T.; Higuchi, M.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Kato, T.; Tabata, S.; Shinozaki, K.; Kakimoto, T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nat. Cell Biol. 2001, 409, 1060–1063. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Kwok, S.F.; Bleecker, A.B.; Meyerowitz, E.M. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 1993, 262, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Aravind, L. The CHASE domain: A predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem. Sci. 2001, 26, 579–582. [Google Scholar] [CrossRef]
- Mougel, C.; Zhulin, I.B. CHASE: An extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 2001, 26, 582–584. [Google Scholar] [CrossRef]
- Wang, W.; Esch, J.J.; Shiu, S.-H.; Agula, H.; Binder, B.M.; Chang, C.; Patterson, S.E.; Bleecker, A.B. Identification of Important Regions for Ethylene Binding and Signaling in the Transmembrane Domain of the ETR1 Ethylene Receptor of Arabidopsis. Plant Cell 2006, 18, 3429–3442. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.; Van De Poel, B.; Cooper, E.D.; Thierer, J.H.; Gibbons, T.R.; Delwiche, C.F.; Chang, C. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 2015, 1, 14004. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Xian, W.; Fu, Y.; Marin, B.; Keller, J.; Wu, T.; Sun, W.; Li, X.; Xu, Y.; Zhang, Y.; et al. Genomes of Subaerial Zygnematophyceae Provide Insights into Land Plant Evolution. Cell 2019, 179, 1057–1067.e14. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, L.; Li, H.; Sahu, S.K.; Wang, H.; Xu, Y.; Xian, W.; Song, B.; Liang, H.; Cheng, S.; et al. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 2020, 6, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, S.; Wang, H.; Sahu, S.K.; Marin, B.; Li, H.; Xu, Y.; Liang, H.; Li, Z.; Cheng, S.; et al. The genome of Prasinoderma coloniale unveils the existence of a third phylum within green plants. Nat. Ecol. Evol. 2020, 4, 1220–1231. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Sakayama, H.; De Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara Genome: Secondary Complexity and Implications for Plant Terrestrialization. Cell 2018, 174, 448–464.e24. [Google Scholar] [CrossRef]
- Papon, N.; Binder, B.M. An Evolutionary Perspective on Ethylene Sensing in Microorganisms. Trends Microbiol. 2019, 27, 193–196. [Google Scholar] [CrossRef]
- Kabbara, S.; Schmülling, T.; Papon, N. CHASEing Cytokinin Receptors in Plants, Bacteria, Fungi, and Beyond. Trends Plant Sci. 2018, 23, 179–181. [Google Scholar] [CrossRef]
- Spíchal, L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Lacey, R.F.; Binder, B.M. Ethylene Regulates the Physiology of the Cyanobacterium Synechocystis sp. PCC 6803 via an Ethylene Receptor. Plant Physiol. 2016, 171, 2798–2809. [Google Scholar] [CrossRef]
- Wang, F.-F.; Cheng, S.-T.; Wu, Y.; Ren, B.-Z.; Qian, W. A Bacterial Receptor PcrK Senses the Plant Hormone Cytokinin to Promote Adaptation to Oxidative Stress. Cell Rep. 2017, 21, 2940–2951. [Google Scholar] [CrossRef]
- Wang, F.-F.; Qian, W. The roles of histidine kinases in sensing host plant and cell–cell communication signal in a phytopathogenic bacterium. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180311. [Google Scholar] [CrossRef] [PubMed]
- Frébortová, J.; Plíhal, O.; Florová, V.; Kokáš, F.; Kubiasová, K.; Greplová, M.; Šimura, J.; Novák, O.; Frébort, I. Light influences cytokinin biosynthesis and sensing in Nostoc (cyanobacteria). J. Phycol. 2017, 53, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.J.; Lacey, R.F.; Bickford, A.B.B.; Beshears, C.P.; Gilmartin, C.J.; Binder, B.M. Cyanobacteria Respond to Low Levels of Ethylene. Front. Plant Sci. 2019, 10, 950. [Google Scholar] [CrossRef] [PubMed]
- Lacey, R.F.; Allen, C.J.; Bakshi, A.; Binder, B.M. Ethylene causes transcriptomic changes inSynechocystisduring phototaxis. Plant Direct 2018, 2, e00048. [Google Scholar] [CrossRef] [PubMed]
- Mount, S.M.; Chang, C. Evidence for a Plastid Origin of Plant Ethylene Receptor Genes. Plant Physiol. 2002, 130, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Kabbara, S.; Bidon, B.; Kilani, J.; Osman, M.; Hamze, M.; Stock, A.M.; Papon, N.; Sun, Y.; Jiang, X.; Lv, Y.; et al. Cytokinin Sensing in Bacteria. Biomolecules 2020, 10, 186. [Google Scholar] [CrossRef]
- Grant, J.R.; Katz, L.A. Building a Phylogenomic Pipeline for the Eukaryotic Tree of Life—Addressing Deep Phylogenies with Genome-Scale Data. PLoS Curr. 2014, 6. [Google Scholar] [CrossRef]
- Kabbara, S.; Hérivaux, A.; De Bernonville, T.D.; Courdavault, V.; Clastre, M.; Gastebois, A.; Osman, M.; Hamze, M.; Cock, J.M.; Schaap, P.; et al. Diversity and Evolution of Sensor Histidine Kinases in Eukaryotes. Genome Biol. Evol. 2019, 11, 86–108. [Google Scholar] [CrossRef]
- Defosse, T.A.; Sharma, A.; Mondal, A.K.; De Bernonville, T.D.; Latgé, J.-P.; Calderone, R.; Giglioli-Guivarc’H, N.; Courdavault, V.; Clastre, M.; Papon, N. Hybrid histidine kinases in pathogenic fungi. Mol. Microbiol. 2015, 95, 914–924. [Google Scholar] [CrossRef]
- Hérivaux, A.; So, Y.-S.; Gastebois, A.; Latgé, J.-P.; Bouchara, J.-P.; Bahn, Y.-S.; Papon, N. Major Sensing Proteins in Pathogenic Fungi: The Hybrid Histidine Kinase Family. PLOS Pathog. 2016, 12, e1005683. [Google Scholar] [CrossRef]
- Foo, E.; McAdam, E.L.; Weller, J.L.; Reid, J.B. Interactions between ethylene, gibberellins, and brassinosteroids in the development of rhizobial and mycorrhizal symbioses of pea. J. Exp. Bot. 2016, 67, 2413–2424. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.T.D.L.; Vierheilig, H.; Ocampo, J.A.; Garcia-Garrido, J.M. Altered pattern of arbuscular mycorrhizal formation in tomato ethylene mutants. Plant Signal. Behav. 2011, 6, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Chanclud, E.; Kisiala, A.; Emery, N.R.J.; Chalvon, V.; Ducasse, A.; Romiti-Michel, C.; Gravot, A.; Kroj, T.; Morel, J.-B. Cytokinin Production by the Rice Blast Fungus Is a Pivotal Requirement for Full Virulence. PLoS Pathog. 2016, 12, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Cosme, M.; Ramireddy, E.; Franken, P.; Schmülling, T.; Wurst, S. Shootand root-borne cytokinin influences arbuscular mycorrhizal symbiosis. Mycorrhiza 2016, 26, 709–720. [Google Scholar] [CrossRef]
- Laffont, C.; Rey, T.; André, O.; Novero, M.; Kazmierczak, T.; Debelle, F.; Bonfante, P.; Jacquet, C.; Frugier, F. The CRE1 Cytokinin Pathway Is Differentially Recruited Depending on Medicago truncatula Root Environments and Negatively Regulates Resistance to a Pathogen. PLoS ONE 2015, 10, e0116819. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, J.; Galuszka, P.; Tudzynski, P. Functional characterization of the first filamentous fungal tRNA -isopentenyltransferase and its role in the virulence of Claviceps purpurea. New Phytol. 2016, 211, 980–992. [Google Scholar] [CrossRef]
- Goh, D.M.; Cosme, M.; Kisiala, A.; Mulholland, S.; Said, Z.M.F.; Spíchal, L.; Emery, R.N.; Declerck, S.; Guinel, F. A Stimulatory Role for Cytokinin in the Arbuscular Mycorrhizal Symbiosis of Pea. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Hérivaux, A.; De Bernonville, T.D.; Le Roux, C.; Clastre, M.; Courdavault, V.; Gastebois, A.; Bouchara, J.-P.; James, T.Y.; Latgé, J.-P.; Martin, F.; et al. The Identification of Phytohormone Receptor Homologs in Early Diverging Fungi Suggests a Role for Plant Sensing in Land Colonization by Fungi. mBio 2017, 8, e01739-16. [Google Scholar] [CrossRef]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Genet. 2020, 1–12. [Google Scholar] [CrossRef]
- Pons, S.; Fournier, S.; Chervin, C.; Bécard, G.; Rochange, S.; Frey, N.F.D.; Pagès, V.P. Phytohormone production by the arbuscular mycorrhizal fungus Rhizophagus irregularis. PLoS ONE 2020, 15, e0240886. [Google Scholar] [CrossRef]
- Aoki, M.; Kisiala, A.; Li, S.; Stock, N.L.; Brunetti, C.R.; Huber, R.J.; Emery, R.N. Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination. Biomolecules 2019, 9, 702. [Google Scholar] [CrossRef] [PubMed]
- Aoki, M.M.; Emery, R.J.N.; Anjard, C.; Brunetti, C.R.; Huber, R.J. Cytokinins in Dictyostelia—A Unique Model for Studying the Functions of Signaling Agents From Species to Kingdoms. Front. Cell Dev. Biol. 2020, 8, 511. [Google Scholar] [CrossRef] [PubMed]
- Anjard, C.; Loomis, W.F. Cytokinins induce sporulation in Dictyostelium. Development 2008, 135, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Loomis, W.F. Cell signaling during development of Dictyostelium. Dev. Biol. 2014, 391, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Shaulsky, G.; Escalante, R.; Loomis, W.F. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 1996, 15, 3890–3898. [Google Scholar] [CrossRef] [PubMed]
- Kabbara, S.; Bidon, B.; Kilani, J.; De Bernonville, T.D.; Clastre, M.; Courdavault, V.; Cock, J.M.; Papon, N. Megaviruses: An involvement in phytohormone receptor gene transfer in brown algae? Gene 2019, 704, 149–151. [Google Scholar] [CrossRef]
- North, J.A.; Narrowe, A.B.; Xiong, W.; Byerly, K.M.; Zhao, G.; Young, S.J.; Murali, S.; Wildenthal, J.A.; Cannon, W.R.; Wrighton, K.C.; et al. A nitrogenase-like enzyme system catalyzes methionine, ethylene, and methane biogenesis. Science 2020, 369, 1094–1098. [Google Scholar] [CrossRef]
- North, J.A.; Miller, A.R.; Wildenthal, J.A.; Young, S.J.; Tabita, F.R. Microbial pathway for anaerobic 5’-methylthioadenosine metabolism coupled to ethylene formation. Proc. Natl. Acad. Sci. USA 2017, 114, E10455–E10464. [Google Scholar] [CrossRef]
- Amagai, A. Ethylene as a potent inducer of sexual development. Dev. Growth Differ. 2011, 53, 617–623. [Google Scholar] [CrossRef]
- Daudu, D.; Kisiala, A.; Ribeiro, C.W.; Mélin, C.; Perrot, L.; Clastre, M.; Courdavault, V.; Papon, N.; Oudin, A.; Courtois, M.; et al. Setting-up a fast and reliable cytokinin biosensor based on a plant histidine kinase receptor expressed in Saccharomyces cerevisiae. J. Biotechnol. 2019, 289, 103–111. [Google Scholar] [CrossRef]
- Andreas, P.; Kisiala, A.; Emery, R.N.; De Clerck-Floate, R.; Tooker, J.F.; Price, P.W.; Miller, D.G.; Chen, M.-S.; Connor, E.F. Cytokinins Are Abundant and Widespread among Insect Species. Plants 2020, 9, 208. [Google Scholar] [CrossRef] [PubMed]
- Kisiala, A.; Kambhampati, S.; Stock, N.L.; Aoki, M.; Emery, R.N. Quantification of Cytokinins Using High-Resolution Accurate-Mass Orbitrap Mass Spectrometry and Parallel Reaction Monitoring (PRM). Anal. Chem. 2019, 91, 15049–15056. [Google Scholar] [CrossRef] [PubMed]
- Seegobin, M.; Kisiala, A.; Noble, A.; Kaplan, D.; Brunetti, C.; Emery, R.J.N. Canis familiaris tissues are characterized by different profiles of cytokinins typical of the tRNA degradation pathway. FASEB J. 2018, 32, 6575–6581. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Yamashino, T. Biochemical Characterization of Plant Hormone Cytokinin-Receptor Histidine Kinases Using Microorganisms. Methods Enzymol. 2010, 471, 335–356. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.E.; Binder, B.M. Biochemical Characterization of Plant Ethylene Receptors Following Transgenic Expression in Yeast. Methods Enzymol. 2007, 422, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Daudu, D.; Allion, E.; Liesecke, F.; Papon, N.; Courdavault, V.; De Bernonville, T.D.; Mélin, C.; Oudin, A.; Clastre, M.; LaNoue, A.; et al. CHASE-Containing Histidine Kinase Receptors in Apple Tree: From a Common Receptor Structure to Divergent Cytokinin Binding Properties and Specific Functions. Front. Plant Sci. 2017, 8, 1614. [Google Scholar] [CrossRef]
- Kubiasová, K.; Montesinos, J.C.; Šamajová, O.; Nisler, J.; Mik, V.; Semerádová, H.; Plíhalová, L.; Novak, O.; Marhavý, P.; Cavallari, N.; et al. Cytokinin fluoroprobe reveals multiple sites of cytokinin perception at plasma membrane and endoplasmic reticulum. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Brütting, C.; Crava, C.M.; Schäfer, M.; Schuman, M.C.; Meldau, S.; Adam, N.; Baldwin, I.T. Cytokinin transfer by a free-living mirid to Nicotiana attenuata recapitulates a strategy of endophytic insects. eLife 2018, 7. [Google Scholar] [CrossRef]
- Body, M.J.A.; Appel, H.M.; Edger, P.P.; Schultz, J.C. A gall-forming insect manipulates hostplant phytohormone synthesis, concentrations, and signaling. bioRxiv 2019, 658823. [Google Scholar] [CrossRef]
- Siddique, S.; Radakovic, Z.S.; De La Torre, C.M.; Chronis, D.; Novák, O.; Ramireddy, E.; Holbein, J.; Matera, C.; Hütten, M.; Gutbrod, P.; et al. A parasitic nematode releases cytokinin that controls cell division and orchestrates feeding site formation in host plants. Proc. Natl. Acad. Sci. USA 2015, 112, 12669–12674. [Google Scholar] [CrossRef]
- Siddique, S.; Grundler, F.M. Parasitic nematodes manipulate plant development to establish feeding sites. Curr. Opin. Microbiol. 2018, 46, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.-S.; Sintaha, M.; Cheung, M.-Y.; Lam, H.-M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed]
- Bedini, A.; Mercy, L.; Schneider, C.; Franken, P.; Lucic-Mercy, E. Unraveling the Initial Plant Hormone Signaling, Metabolic Mechanisms and Plant Defense Triggering the Endomycorrhizal Symbiosis Behavior. Front. Plant Sci. 2018, 9, 1800. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bidon, B.; Kabbara, S.; Courdavault, V.; Glévarec, G.; Oudin, A.; Héricourt, F.; Carpin, S.; Spíchal, L.; Binder, B.M.; Cock, J.M.; et al. Cytokinin and Ethylene Cell Signaling Pathways from Prokaryotes to Eukaryotes. Cells 2020, 9, 2526. https://doi.org/10.3390/cells9112526
Bidon B, Kabbara S, Courdavault V, Glévarec G, Oudin A, Héricourt F, Carpin S, Spíchal L, Binder BM, Cock JM, et al. Cytokinin and Ethylene Cell Signaling Pathways from Prokaryotes to Eukaryotes. Cells. 2020; 9(11):2526. https://doi.org/10.3390/cells9112526
Chicago/Turabian StyleBidon, Baptiste, Samar Kabbara, Vincent Courdavault, Gaëlle Glévarec, Audrey Oudin, François Héricourt, Sabine Carpin, Lukáš Spíchal, Brad M. Binder, J. Mark Cock, and et al. 2020. "Cytokinin and Ethylene Cell Signaling Pathways from Prokaryotes to Eukaryotes" Cells 9, no. 11: 2526. https://doi.org/10.3390/cells9112526
APA StyleBidon, B., Kabbara, S., Courdavault, V., Glévarec, G., Oudin, A., Héricourt, F., Carpin, S., Spíchal, L., Binder, B. M., Cock, J. M., & Papon, N. (2020). Cytokinin and Ethylene Cell Signaling Pathways from Prokaryotes to Eukaryotes. Cells, 9(11), 2526. https://doi.org/10.3390/cells9112526







