Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration
Abstract
1. Introduction
2. Materials and Methods
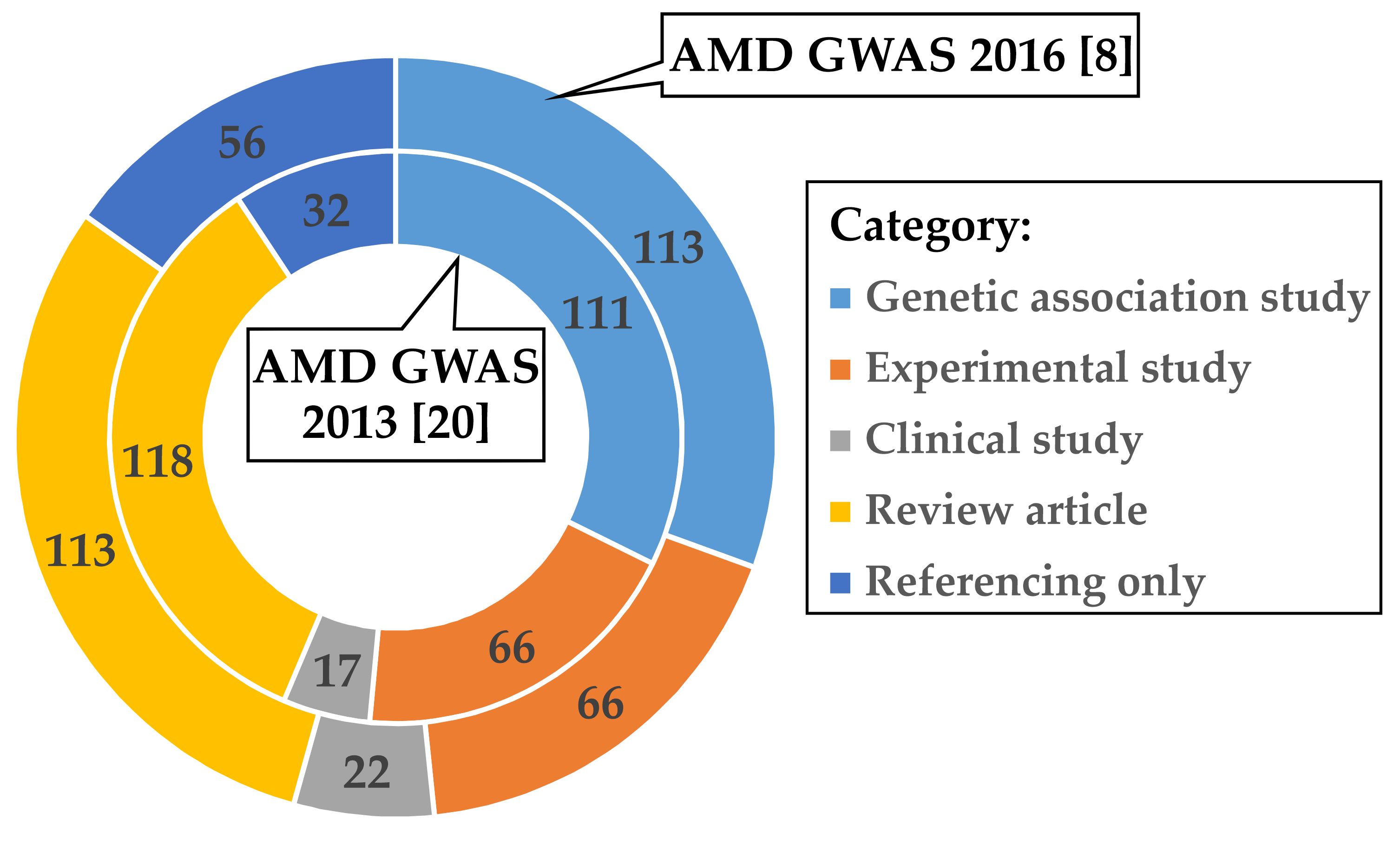
2.1. Curation of Articles Citing the Two AMD GWAS Fritsche et al. 2013 and Fritsche et al. 2016
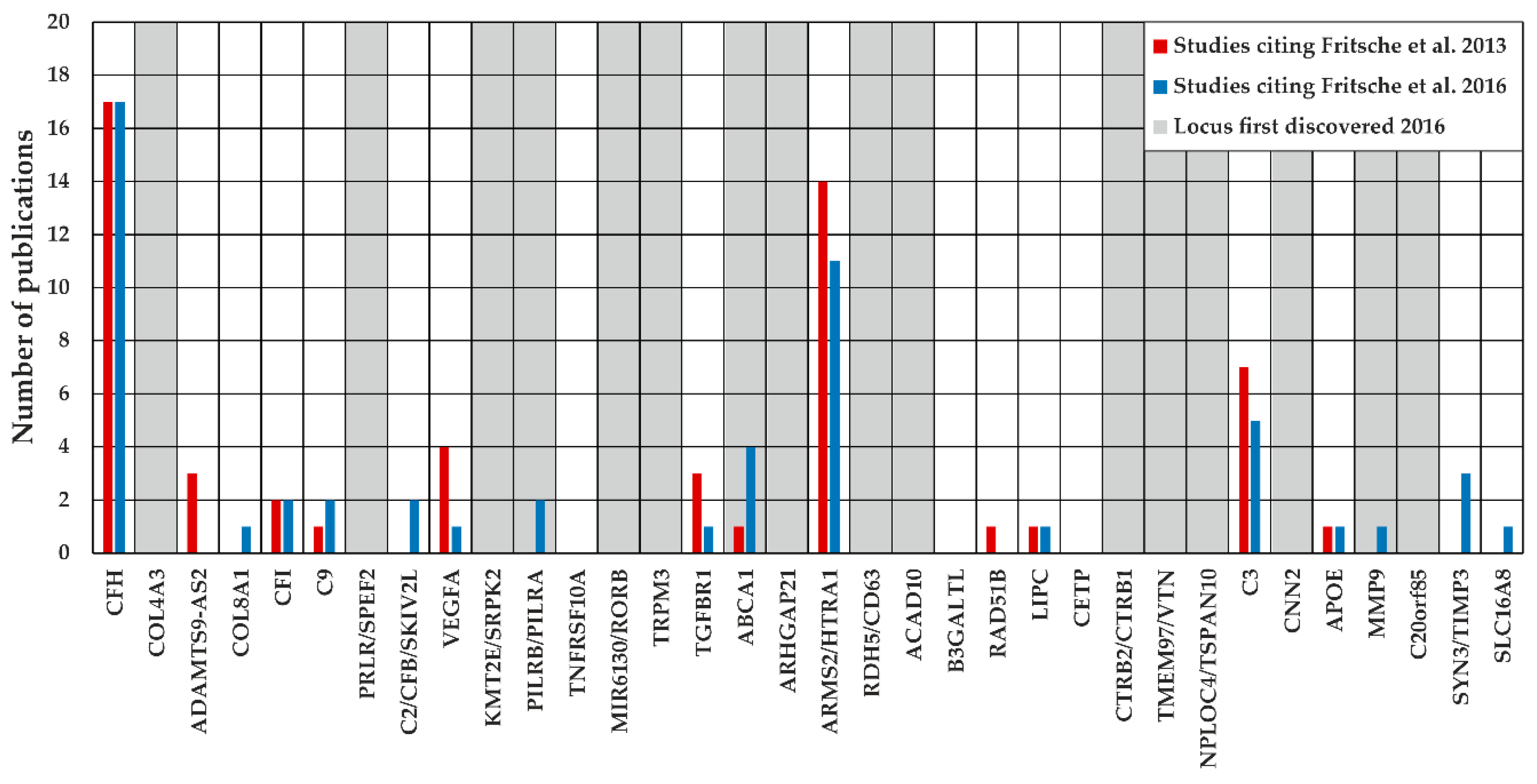
2.2. Locus Analysis of 34 Known AMD-Associated Loci
2.3. Curation and Quality Control of Studies Investigating AMD in the Context of Gene Expression Regulation
3. Results
3.1. Investigations Following the AMD GWAS of Fritsche et al. 2016
3.2. Investigation of Defined Loci Based on the Reference Data of Fritsche et al. 2016
3.3. AMD Genetics and Gene Expression Regulation
3.4. Gene Expression Regulation is Likely Associated with 15 Known AMD Loci
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.-Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Ferris, F.L.; Davis, M.D.; Clemons, T.E.; Lee, L.-Y.; Chew, E.Y.; Lindblad, A.S.; Milton, R.C.; Bressler, S.B.; Klein, R. A Simplified Severity Scale for Age-Related Macular Degeneration. Arch. Ophthalmol. 2005, 123, 1570. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, R.; Hartnett, M.E.; Atkinson, J.P.; Giclas, P.C.; Rosner, B.; Seddon, J.M. Plasma complement components and activation fragments: Associations with age-related macular degeneration genotypes and phenotypes. Investig. Ophthalmol. Vis. Sci. 2009, 50, 5818–5827. [Google Scholar] [CrossRef]
- Cougnard-Grégoire, A.; Delyfer, M.N.; Korobelnik, J.F.; Rougier, M.B.; Le Goff, M.; Dartigues, J.F.; Barberger-Gateau, P.; Delcourt, C. Elevated high-density lipoprotein cholesterol and age-related macular degeneration: The Alienor study. PLoS ONE 2014, 9, e90973. [Google Scholar] [CrossRef] [PubMed]
- Seddon, J.M.; Cote, J.; Page, W.F.; Aggen, S.H.; Neale, M.C. The US twin study of age-related macular degeneration: Relative roles of genetic and environmental influences. Arch. Ophthalmol. 2005, 123, 321–327. [Google Scholar] [CrossRef]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement factor H polymorphism in age-related macular degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 2012, 30, 1095–1106. [Google Scholar] [CrossRef]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Spain, S.L.; Barrett, J.C. Strategies for fine-mapping complex traits. Hum. Mol. Genet. 2015, 24, R111–R119. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, F.; Heid, I.M.; Weber, B.H.F.; Fritsche, L.G.; Igl, W.; Bailey, J.N.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; et al. Recombinant haplotypes narrow the ARMS2/HTRA1 association signal for age-related macular degeneration. Genetics 2017, 205, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, F.; Kiel, C.; Zimmermann, M.E.; Gorski, M.; Grassmann, V.; Stark, K.; Heid, I.M.; Weber, B.H.F. Genetic pleiotropy between age-related macular degeneration and 16 complex diseases and traits. Genome Med. 2017, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Westra, H.J.; Franke, L. From genome to function by studying eQTLs. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1896–1902. [Google Scholar] [CrossRef]
- Schadt, E.E.; Molony, C.; Chudin, E.; Hao, K.; Yang, X.; Lum, P.Y.; Kasarskis, A.; Zhang, B.; Wang, S.; Suver, C.; et al. Mapping the Genetic Architecture of Gene Expression in Human Liver. PLoS Biol. 2008, 6, e107. [Google Scholar] [CrossRef]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.J.H.; Jansen, R.; de Geus, E.J.C.; Boomsma, D.I.; Wright, F.A.; et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef]
- Gamazon, E.R.; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Nicolae, D.L.; Cox, N.J.; et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef]
- Web of Science Core Collection—Web of Science Group. Available online: https://clarivate.com/webofsciencegroup/solutions/web-of-science-core-collection/ (accessed on 7 July 2020).
- Fritsche, L.G.; Chen, W.; Schu, M.; Yaspan, B.L.; Yu, Y.; Thorleifsson, G.; Zack, D.J.; Arakawa, S.; Cipriani, V.; Ripke, S.; et al. Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013, 45, 433–439. [Google Scholar] [CrossRef]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 6 August 2020).
- Wang, J.; Zheng, J.; Wang, Z.; Li, H.; Deng, M. Inferring Gene-Disease Association by an Integrative Analysis of eQTL Genome-Wide Association Study and Protein-Protein Interaction Data. Hum. Hered. 2019, 83, 117–129. [Google Scholar] [CrossRef]
- Aguet, F.; Brown, A.A.; Castel, S.E.; Davis, J.R.; He, Y.; Jo, B.; Mohammadi, P.; Park, Y.S.; Parsana, P.; Segrè, A.V.; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef]
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Brown, A.; Castel, S.E.; Jo, B.; Kasela, S.; Kim-Hellmuth, S.; Liang, Y.; Oliva, M.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv 2019, 787903. [Google Scholar] [CrossRef]
- Ratnapriya, R.; Sosina, O.A.; Starostik, M.R.; Kwicklis, M.; Kapphahn, R.J.; Fritsche, L.G.; Walton, A.; Arvanitis, M.; Gieser, L.; Pietraszkiewicz, A.; et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 2019, 51, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- GTEx Portal Datasets. Available online: https://www.gtexportal.org/home/datasets (accessed on 7 July 2020).
- Orozco, L.D.; Chen, H.-H.; Cox, C.; Katschke, K.J.; Arceo, R.; Espiritu, C.; Caplazi, P.; Nghiem, S.S.; Chen, Y.-J.; Modrusan, Z.; et al. Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration. Cell Rep. 2020, 30, 1246–1259.e6. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Eye eQTL Browser. Available online: http://eye-eqtl.com/ (accessed on 20 July 2020).
- Strunz, T.; Grassmann, F.; Gayán, J.; Nahkuri, S.; Souza-Costa, D.; Maugeais, C.; Fauser, S.; Nogoceke, E.; Weber, B.H.F. A mega-analysis of expression quantitative trait loci (eQTL) provides insight into the regulatory architecture of gene expression variation in liver. Sci. Rep. 2018, 8, 5865. [Google Scholar] [CrossRef]
- Strunz, T.; Kiel, C.; Grassmann, F.; Ratnapriya, R.; Kwicklis, M.; Karlstetter, M.; Fauser, S.; Swaroop, A.; Arend, N.; Langmann, T.; et al. A mega-analysis of expression quantitative trait loci in retinal tissue. PLoS Genet. 2020. [Google Scholar] [CrossRef]
- Strunz, T.; Lauwen, S.; Kiel, C.; Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; et al. A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration. Sci. Rep. 2020, 10, 1584. [Google Scholar] [CrossRef]
- Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; Gabriel, S.B.; et al. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Liutkeviciene, R.; Vilkeviciute, A.; Gedvilaite, G.; Kaikaryte, K.; Kriauciuniene, L. Haplotypes of HTRA1 rs1120638, TIMP3 rs9621532, VEGFA rs833068, CFI rs10033900, ERCC6 rs3793784, and KCTD10 rs56209061 Gene Polymorphisms in Age-Related Macular Degeneration. Dis. Markers 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Popp, N.A.; Agrón, E.; Hageman, G.S.; Tuo, J.; Chew, E.Y.; Chan, C.C. No Sex Differences in the Frequencies of Common Single Nucleotide Polymorphisms Associated with Age-Related Macular Degeneration. Curr. Eye Res. 2017, 42, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Heesterbeek, T.J.; Lechanteur, Y.T.E.; Lorés-Motta, L.; Schick, T.; Daha, M.R.; Altay, L.; Liakopoulos, S.; Smailhodzic, D.; den Hollander, A.I.; Hoyng, C.B.; et al. Complement activation levels are related to disease stage in AMD. Investig. Ophthalmol. Vis. Sci. 2020, 61, 18. [Google Scholar] [CrossRef]
- Roh, M.; Shin, H.J.; Laíns, I.; Providência, J.; Caseiro-Alves, M.; Barreto, P.; Vavvas, D.G.; Miller, J.B.; Kim, I.K.; Gaziano, J.M.; et al. Higher intake of polyunsaturated fatty acid and monounsaturated fatty acid is inversely associated with AMD. Investig. Ophthalmol. Vis. Sci. 2020, 61, 20. [Google Scholar] [CrossRef]
- Treister, A.D.; Nesper, P.L.; Fayed, A.E.; Gill, M.K.; Mirza, R.G.; Fawzi, A.A. Prevalence of subclinical CNV and choriocapillaris nonperfusion in fellow eyes of unilateral exudative AMD on OCT angiography. Transl. Vis. Sci. Technol. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Haeussler, M.; Zweig, A.S.; Tyner, C.; Speir, M.L.; Rosenbloom, K.R.; Raney, B.J.; Lee, C.M.; Lee, B.T.; Hinrichs, A.S.; Gonzalez, J.N.; et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019, 47, D853–D858. [Google Scholar] [CrossRef]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- UCSC Genome Browser Home. Available online: https://genome.ucsc.edu/ (accessed on 6 August 2020).
- Cotsapas, C.; Voight, B.F.; Rossin, E.; Lage, K.; Neale, B.M.; Wallace, C.; Abecasis, G.R.; Barrett, J.C.; Behrens, T.; Cho, J.; et al. Pervasive Sharing of Genetic Effects in Autoimmune Disease. PLoS Genet. 2011, 7, e1002254. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.-R.; Duncan, L.; Perry, J.R.B.; Patterson, N.; Robinson, E.B.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015. [Google Scholar] [CrossRef]
- Shiratori, I.; Ogasawara, K.; Saito, T.; Lanier, L.L.; Arase, H. Activation of Natural Killer Cells and Dendritic Cells upon Recognition of a Novel CD99-like Ligand by Paired Immunoglobulin-like Type 2 Receptor. J. Exp. Med. 2004, 199, 525–533. [Google Scholar] [CrossRef]
- Mousseau, D.D.; Banville, D.; L’Abbé, D.; Bouchard, P.; Shen, S.-H. PILRα, a Novel Immunoreceptor Tyrosine-based Inhibitory Motif-bearing Protein, Recruits SHP-1 upon Tyrosine Phosphorylation and Is Paired with the Truncated Counterpart PILRβ. J. Biol. Chem. 2000, 275, 4467–4474. [Google Scholar] [CrossRef] [PubMed]
- Bowman, S.L.; Bi-Karchin, J.; Le, L.; Marks, M.S. The road to lysosome-related organelles: Insights from Hermansky-Pudlak syndrome and other rare diseases. Traffic 2019, 20, 404–435. [Google Scholar] [CrossRef] [PubMed]
- Huizing, M.; Malicdan, M.C.V.; Wang, J.A.; Pri-Chen, H.; Hess, R.A.; Fischer, R.; O’Brien, K.J.; Merideth, M.A.; Gahl, W.A.; Gochuico, B.R. Hermansky–Pudlak syndrome: Mutation update. Hum. Mutat. 2020, 41, 543–580. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Webster, B.R.; Li, J.H.; Sack, M.N. Identification of a molecular component of the mitochondrial acetyltransferase programme: A novel role for GCN5L1. Biochem. J. 2012, 443, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.R.; Scott, I.; Han, K.; Li, J.H.; Lu, Z.; Stevens, M.V.; Malide, D.; Chen, Y.; Samsel, L.; Connelly, P.S.; et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J. Cell Sci. 2013, 126, 4843–4849. [Google Scholar] [CrossRef]
- Karunadharma, P.P.; Nordgaard, C.L.; Olsen, T.W.; Ferrington, D.A. Mitochondrial DNA damage as a potential mechanism for Age-Related macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2010, 51, 5470–5479. [Google Scholar] [CrossRef]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K.; et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef]
- Cano-Gamez, E.; Trynka, G. From GWAS to Function: Using Functional Genomics to Identify the Mechanisms Underlying Complex Diseases. Front. Genet. 2020, 11, 424. [Google Scholar] [CrossRef]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef]
- Uyar, B.; Palmer, D.; Kowald, A.; Escobar, H.M.; Barrantes, I.; Möller, S.; Akalin, A.; Fuellen, G. Single-cell analyses of aging, inflammation and senescence. Ageing Res. Rev. 2020, 101156. [Google Scholar] [CrossRef]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Jiang, S.; Musayeva, A.; Gericke, A. Oxidative Stress and Vascular Dysfunction in the Retina: Therapeutic Strategies. Antioxidants 2020, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hanus, J.; Abu-Asab, M.; Shen, D.; Ogilvy, A.; Ou, J.; Chu, X.; Shi, G.; Li, W.; Wang, S.; et al. NLRP3 Upregulation in Retinal Pigment Epithelium in Age-Related Macular Degeneration. Int. J. Mol. Sci. 2016, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Ulirsch, J.C.; Nandakumar, S.K.; Wang, L.; Giani, F.C.; Zhang, X.; Rogov, P.; Melnikov, A.; McDonel, P.; Do, R.; Mikkelsen, T.S.; et al. Systematic Functional Dissection of Common Genetic Variation Affecting Red Blood Cell Traits. Cell 2016, 165, 1530–1545. [Google Scholar] [CrossRef]
- Menon, M.; Mohammadi, S.; Davila-Velderrain, J.; Goods, B.A.; Cadwell, T.D.; Xing, Y.; Stemmer-Rachamimov, A.; Shalek, A.K.; Love, J.C.; Kellis, M.; et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun. 2019, 10, 4902. [Google Scholar] [CrossRef]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 24100–24107. [Google Scholar] [CrossRef]
- Voigt, A.P.; Whitmore, S.S.; Flamme-Wiese, M.J.; Riker, M.J.; Wiley, L.A.; Tucker, B.A.; Stone, E.M.; Mullins, R.F.; Scheetz, T.E. Molecular characterization of foveal versus peripheral human retina by single-cell RNA sequencing. Exp. Eye Res. 2019, 184, 234–242. [Google Scholar] [CrossRef]
| Study | Study Type | Category | Tissue | Sample Size | AMD Status of Tissue Donors | AMD Loci [8] 1 with Findings |
|---|---|---|---|---|---|---|
| Ratnapriya et al. (2019) [25] | eQTL, TWAS | Single study | Retina | 406 | Non-AMD (94), AMD (312) | eQTL: 9, TWAS: 10 |
| Orozco et al. (2020) [28] | eQTL | Single study | Retina, RPE/choroid | 121 | Non-AMD (98), AMD (23) | 11 in retina, 9 in RPE/choroid |
| Strunz et al. (2020) [32] | eQTL | Mega-analysis | Retina | 311 | Non-AMD | 4 |
| Strunz et al. (2018) [31] | eQTL | Mega-analysis | Liver | 588 | Unknown | 5 |
| Strunz et al. (2020) [33] | TWAS | - | 27 tissues | 134–421 | Unknown | 25 |
| Locus ID1 | Category 1 (Retina + Other Tissues) | Category 2 (Retina) | Category 3 (Predominantly Other Tissues) |
|---|---|---|---|
| CFH | - | - | KCNT2 (−), CFH (+/−), CFHR1 (+), CFHR3 (+), ZBTB41(+) |
| COL8A1 | - | - | NIT2 (−), TBC1D23 (−) |
| CFI | - | CFI (−) | PLA2G12A (+), CASP6 (+) |
| C2/CFB/SKIV2L | - | HLA-DQB1 (−) 2 | - |
| PILRB/PILRA | PILRA (+), PILRB (+), STAG3L5P (+) | - | PMS2P1 (−), TSC22D4 (+), ZCWPW1 (+), NYAP1 (−) |
| TNFRSF10A | - | - | TNFRSF10A (−) |
| ARMS2/HTRA1 | HTRA1 (+/−) | - | PLEKHA1 (+/−), ARMS2 (−), BTBD16 (+/−), DMBT1 (−) |
| RDH5/CD63 | - | BLOC1S1 (+), AC009779.3 (−) | RDH5 (−) |
| B3GALTL | B3GLCT (+/−) | - | - |
| CETP | - | - | CETP (−) |
| CTRB2/CTRB1 | - | - | CFDP1 (−) |
| TMEM97/VTN | TMEM199 (+) | - | POLDIP2 (+) |
| C3 | - | - | GPR108 (+) |
| CNN2 | - | - | MED16 (+) |
| MMP9 | - | - | PLTP (+), SLC12A5 (+/−) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strunz, T.; Kiel, C.; Sauerbeck, B.L.; Weber, B.H.F. Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration. Cells 2020, 9, 2267. https://doi.org/10.3390/cells9102267
Strunz T, Kiel C, Sauerbeck BL, Weber BHF. Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration. Cells. 2020; 9(10):2267. https://doi.org/10.3390/cells9102267
Chicago/Turabian StyleStrunz, Tobias, Christina Kiel, Bastian L. Sauerbeck, and Bernhard H. F. Weber. 2020. "Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration" Cells 9, no. 10: 2267. https://doi.org/10.3390/cells9102267
APA StyleStrunz, T., Kiel, C., Sauerbeck, B. L., & Weber, B. H. F. (2020). Learning from Fifteen Years of Genome-Wide Association Studies in Age-Related Macular Degeneration. Cells, 9(10), 2267. https://doi.org/10.3390/cells9102267






