Pleiotropic Locus 15q24.1 Reveals a Gender-Specific Association with Neovascular but Not Atrophic Age-Related Macular Degeneration (AMD)
Abstract
1. Introduction
2. Materials and Methods
2.1. Availability of Datasets
2.2. Description of Datasets
2.3. Association of rs2168518 with AMD
2.4. Haplotype Distribution over Different Populations
2.5. Phenome-Wide Association Analysis of rs2168518 in the UK Biobank Database
2.6. Colocalization Analysis Based on the UK Biobank GWAS Summary Statistics
2.7. Colocalization Analysis of the Genetic Signals Underlying UK Biobank Associations and AMD
2.8. eQTL Calculation
2.9. Identification of Transcription Factor Binding Sites
3. Results
3.1. Association of rs2168518 with AMD
3.1.1. Validation of a Genome-Wide Significant Association of rs2168518 with AMD
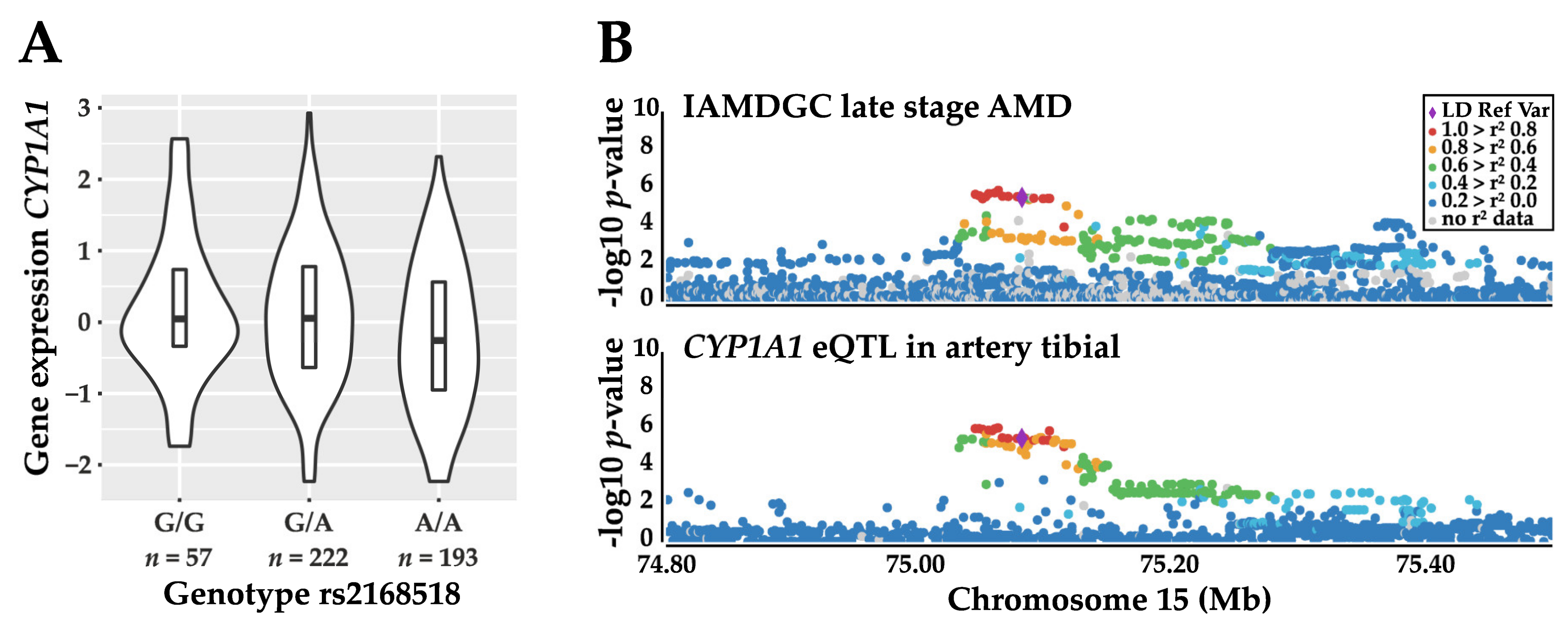
3.1.2. Refinement of the Genetic Association Signal with AMD
3.2. Haplotype Distribution in Populations
3.3. Pleiotropic Effect of rs2168518 Assessed in the UK Biobank Data
3.3.1. Association of rs2168518 with Blood Pressure Phenotypes
3.3.2. Association of rs2168518 with Metabolic Products in Urine
3.3.3. Association of rs2168518 with Other Phenotypes
3.4. Local eQTL Analysis of rs2168518 in the GTEx Dataset
3.4.1. eGenes Are Not Regulated by hsa-mir-4513
3.4.2. Colocalization of eGenes
3.4.3. Colocalization with Phenotypes
3.5. Functional Annotation of Genetic Variants at 15q24.1 with RegulomeDB 2.0
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
International AMD Genomics Consortium (IAMDGC)
References
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.-Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.; Kathryn, P.; Hebbring, S.J.; Wen, C.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2015, 48, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Strunz, T.; Kiel, C.; Sauerbeck, B.L.; Weber, B.H.F. Learning from fifteen years of genome-wide association studies in age-related macular degeneration. Cells 2020, 9, 2267. [Google Scholar] [CrossRef]
- Backes, C.; Meese, E.; Keller, A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Ooi, J.Y.; Lin, R.C.Y.; McMullen, J.R. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Futur. Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef]
- Paul, S.; Vázquez, L.A.B.; Uribe, S.P.; Reyes-Pérez, P.R.; Sharma, A. Current Status of microRNA-Based Therapeutic Approaches in Neurodegenerative Disorders. Cells 2020, 9, 1698. [Google Scholar] [CrossRef]
- De Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Chandradoss, S.D.; Schirle, N.T.; Szczepaniak, M.; Macrae, I.J.; Joo, C. A Dynamic Search Process Underlies MicroRNA Targeting. Cell 2015, 162, 96–107. [Google Scholar] [CrossRef]
- Chipman, L.B.; Pasquinelli, A.E. miRNA Targeting: Growing beyond the Seed. Trends Genet. 2019, 35, 215–222. [Google Scholar] [CrossRef]
- Ghanbari, M.; Erkeland, S.J.; Xu, L.; Colijn, J.M.; Franco, O.H.; Dehghan, A.; Klaver, C.; Meester-Smoor, M.A. Genetic variants in microRNAs and their binding sites within gene 3′UTRs associate with susceptibility to age-related macular degeneration. Hum. Mutat. 2017, 38, 827–838. [Google Scholar] [CrossRef]
- Han, X.; Gharahkhani, P.; Mitchell, P.; Liew, G.; Hewitt, A.W.; MacGregor, S. Genome-wide meta-analysis identifies novel loci associated with age-related macular degeneration. J. Hum. Genet. 2020, 65, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, M.; De Vries, P.S.; De Looper, H.; Peters, M.J.; Schurmann, C.; Yaghootkar, H.D.; Frayling, T.M.; Uitterlinden, A.G.; Hofman, A.; van Meurs, J.B.J.; et al. A Genetic Variant in the Seed Region of miR-4513 Shows Pleiotropic Effects on Lipid and Glucose Homeostasis, Blood Pressure, and Coronary Artery Disease. Hum. Mutat. 2014, 35, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, L.-W.; Chen, D.; Wu, X.; Chen, M. Influence of microRNA-related polymorphisms on clinical outcomes in coronary artery disease. Am. J. Transl. Res. 2015, 7, 393–400. [Google Scholar] [PubMed]
- Mir, R.; Jha, C.K.; Elfaki, I.; Javid, J.; Rehman, S.; Khullar, N.; Banu, S.; Chahal, S.M.S. Incidence of MicroR-4513C/T Gene Variability in Coronary Artery Disease—A case-Control Study. Endocr. Metab. Immune Disord. Drug Targets 2019, 19, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, Y.; Zheng, Y.; Zhang, L.; Pan, Y.; Yu, J.; Yang, M. miR-608 and miR-4513 significantly contribute to the prognosis of lung adenocarcinoma treated with EGFR-TKIs. Lab. Investig. 2018, 99, 568–576. [Google Scholar] [CrossRef]
- Hoffmann, T.J.; Kvale, M.N.; Hesselson, S.E.; Zhan, Y.; Aquino, C.; Cao, Y.; Cawley, S.; Chung, E.; Connell, S.; Eshragh, J.; et al. Next generation genome-wide association tool: Design and coverage of a high-throughput European-optimized SNP array. Genomics 2011, 98, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Kvale, M.N.; Hesselson, S.; Hoffmann, T.J.; Cao, Y.; Chan, D.; Connell, S.; Croen, L.A.; Dispensa, B.P.; Eshragh, J.; Finn, A.; et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 2015, 200, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.J.; Eeden, S.K.V.D.; Sakoda, L.C.; Jorgenson, E.; Habel, L.A.; Graff, R.E.; Passarelli, M.N.; Cario, C.L.; Emami, N.C.; Chao, C.R.; et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015, 5, 878–891. [Google Scholar] [CrossRef]
- Neale Lab. UK Biobank. Available online: http://www.nealelab.is/uk-biobank (accessed on 30 June 2020).
- Aguet, F.; Barbeira, A.N.; Bonazzola, R.; Brown, A.E.; Castel, S.; Jo, B.; Kasela, S.; Kim-Hellmuth, S.; Liang, Y.; Oliva, M.; et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Nature 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nat. Cell Biol. 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2011; Volume 1, ISBN 3900051070. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Pollard, K.S.; Dudoit, S.; Van Der Laan, M.J. Multiple Testing Procedures: The multtest Package and Applications to Genomics. Surviv. Anal. 2005, 249–271. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- LDlink. An Interactive Web Tool for Exploring Linkage Disequilibrium in Population Groups. Available online: https://ldlink.nci.nih.gov/?tab=home (accessed on 20 July 2020).
- PheWeb. Available online: http://pheweb.sph.umich.edu (accessed on 30 June 2020).
- Neale Lab. Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank. Available online: http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank (accessed on 30 June 2020).
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- My.LocusZoom.org. Available online: https://my.locuszoom.org/ (accessed on 30 June 2020).
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- UK Biobank. Data Showcase. Available online: https://www.ukbiobank.ac.uk/data-showcase/ (accessed on 30 June 2020).
- Ongen, H.; Buil, A.; Brown, A.A.; Dermitzakis, E.T.; Delaneau, O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 2015, 32, 1479–1485. [Google Scholar] [CrossRef]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Home-miRWalk. Available online: http://mirwalk.umm.uni-heidelberg.de/ (accessed on 29 July 2020).
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Regulome. Regulome Search. Available online: https://www.regulomedb.org/regulome-search/ (accessed on 20 July 2020).
- Turley, P.; Walters, R.K.; Maghzian, O.; Okbay, A.; Lee, J.J.; Fontana, M.A.; Nguyen-Viet, T.A.; Wedow, R.; Zacher, M.; Furlotte, N.A.; et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 2018, 50, 229–237. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2017, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2019, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Tower, J. Sex-Specific Gene Expression and Life Span Regulation. Trends Endocrinol. Metab. 2017, 28, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Cotsapas, C.; Voight, B.F.; Rossin, E.; Lage, K.; Neale, B.M.; Wallace, C.; Abecasis, G.R.; Barrett, J.C.; Behrens, T.; Cho, J.; et al. Pervasive Sharing of Genetic Effects in Autoimmune Disease. PLoS Genet. 2011, 7, e1002254. [Google Scholar] [CrossRef] [PubMed]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, I.N.M.; Loh, P.-R.; Duncan, L.; Perry, J.R.; Patterson, N.; Robinson, E.B.; et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Hyman, L.; Schachat, A.P.; He, Q.; Leske, M.C. Hypertension, cardiovascular disease, and age-related macular degeneration. Age-Related Macular Degeneration Risk Factors Study Group. Arch. Ophthalmol. 2000, 118, 351–358. [Google Scholar] [CrossRef]
- Duan, Y.; Mo, J.; Klein, B.E.; Scott, I.U.; Lin, H.-M.; Caulfield, J.; Patel, M.; Liao, D. Age-Related Macular Degeneration Is Associated with Incident Myocardial Infarction among Elderly Americans. Ophthalmology 2007, 114, 732–737. [Google Scholar] [CrossRef]
- Grassmann, F.; International AMD Genomics Consortium (IAMDGC); Kiel, C.; Zimmermann, M.E.; Gorski, M.; Grassmann, V.; Stark, K.J.; Heid, I.M.; Weber, B.H.F.; Fritsche, L.G. Genetic pleiotropy between age-related macular degeneration and 16 complex diseases and traits. Genome Med. 2017, 9, 29. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A.; Zwaan, R.A.; Etz, A.; Lucas, R.E.; Donnellan, M.B.; Howe, P.D.L.; Perfors, A.; Holcombe, A.O.; Gershman, S.J.; Hardwicke, T.E.; et al. Why replication has more scientific value than original discovery. Behav. Brain Sci. 2018, 41, e137. [Google Scholar] [CrossRef]
- Strunz, T.; Kiel, C.; Grassmann, F.; Ratnapriya, R.; Kwicklis, M.; Karlstetter, M.; Fauser, S.; Arend, N.; Swaroop, A.; Langmann, T.; et al. A mega-analysis of expression quantitative trait loci in retinal tissue. PLoS Genet. 2020, 16, e1008934. [Google Scholar] [CrossRef] [PubMed]
- Orozco, L.D.; Chen, H.-H.; Cox, C.; Katschke, K.J.; Arceo, R.; Espiritu, C.; Caplazi, P.; Nghiem, S.S.; Chen, Y.-J.; Modrusan, Z.; et al. Integration of eQTL and a Single-Cell Atlas in the Human Eye Identifies Causal Genes for Age-Related Macular Degeneration. Cell Rep. 2020, 30, 1246–1259.e6. [Google Scholar] [CrossRef] [PubMed]
- Ratnapriya, R.; Sosina, O.A.; Starostik, M.R.; Kwicklis, M.; Kapphahn, R.J.; Fritsche, L.G.; Walton, A.; Arvanitis, M.; Gieser, L.; Pietraszkiewicz, A.; et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 2019, 51, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Masland, R.H. The Neuronal Organization of the Retina. Neuron 2012, 76, 266–280. [Google Scholar] [CrossRef]
- Menon, M.; Mohammadi, S.; Davila-Velderrain, J.; Goods, B.A.; Cadwell, T.D.; Xing, Y.; Stemmer-Rachamimov, A.; Shalek, A.K.; Love, J.C.; Kellis, M.; et al. Single-cell transcriptomic atlas of the human retina identifies cell types associated with age-related macular degeneration. Nat. Commun. 2019, 10, 4902–4909. [Google Scholar] [CrossRef]
- Liang, Q.; Dharmat, R.; Owen, L.; Shakoor, A.; Li, Y.; Kim, S.; Vitale, A.; Kim, I.; Morgan, D.; Liang, S.; et al. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun. 2019, 10, 5743. [Google Scholar] [CrossRef]
- Rheaume, B.A.; Jereen, A.; Bolisetty, M.; Sajid, M.S.; Yang, Y.; Renna, K.; Sun, L.; Robson, P.; Trakhtenberg, E.F. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat. Commun. 2018, 9, 2759. [Google Scholar] [CrossRef]
- Li, M.; Jia, C.; Kazmierkiewicz, K.L.; Bowman, A.S.; Tian, L.; Liu, Y.; Gupta, N.A.; Gudiseva, H.V.; Yee, S.S.; Kim, M.; et al. Comprehensive analysis of gene expression in human retina and supporting tissues. Hum. Mol. Genet. 2014, 23, 4001–4014. [Google Scholar] [CrossRef]
- Gonzalez, F.J. Molecular genetics of the P-450 superfamily. Pharmacol. Ther. 1990, 45, 1–38. [Google Scholar] [CrossRef]
- Shi, Z.; Dragin, N.; Gálvez-Peralta, M.; Jorge-Nebert, L.F.; Miller, M.L.; Wang, B.; Nebert, D.W. Organ-specific roles of CYP1A1 during detoxication of dietary benzo[a]pyrene. Mol. Pharmacol. 2010, 78, 46–57. [Google Scholar] [CrossRef]
- Jarrett, S.G.; Boulton, M.E. Consequences of oxidative stress in age-related macular degeneration. Mol. Asp. Med. 2012, 33, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Esfandiary, H.; Chakravarthy, U.; Patterson, C.; Young, I.S.E.; Hughes, A. Association study of detoxification genes in age related macular degeneration. Br. J. Ophthalmol. 2005, 89, 470–474. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perepechaeva, M.L.; Grishanova, A.Y.; Rudnitskaya, E.A.; Kolosova, N.G. The Mitochondria-Targeted Antioxidant SkQ1 Downregulates Aryl Hydrocarbon Receptor-Dependent Genes in the Retina of OXYS Rats with AMD-Like Retinopathy. J. Ophthalmol. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kopf, P.G.; Scott, J.A.; Agbor, L.N.; Boberg, J.R.; Elased, K.M.; Huwe, J.K.; Walker, M.K. Cytochrome P4501A1 Is Required for Vascular Dysfunction and Hypertension Induced by 2,3,7,8-Tetrachlorodibenzo-p-Dioxin. Toxicol. Sci. 2010, 117, 537–546. [Google Scholar] [CrossRef] [PubMed]




| Sample Size | OR [95% CI] | p-Value | Q-Value | |
|---|---|---|---|---|
| Late-stage AMD | 33,976 | 1.089 [1.052–1.127] | 8.74 × 10−7 | 4.00 × 10−6 |
| Geographic atrophy (GA) | 21,067 | 1.077 [1.016–1.142] | 0.013 | 0.024 |
| Choroidal neovascularization (CNV) | 28,581 | 1.091 [1.050–1.133] | 8.29E-06 | 1.87 × 10−5 |
| GA & CNV | 19,992 | 1.073 [1.001–1.150] | 0.048 | 0.054 |
| Early stage AMD | 24,489 | 1.036 [0.992–1.081] | 0.112 | 0.112 |
| <75 years | 17,326 | 1.121 [1.066–1.177] | 7.41 × 10−6 | 1.87 × 10−5 |
| >75 years | 15,825 | 1.055 [1.005–1.108] | 0.030 | 0.039 |
| Male | 14,352 | 1.139 [1.081–1.200] | 8.89 × 10−7 | 4.00 × 10−6 |
| Female | 19,624 | 1.053 [1.007–1.102] | 0.025 | 0.038 |
| Haplotype | rs2168518 Allele | EUR | EAS | SAS | AFR |
|---|---|---|---|---|---|
| H1 | A | 0.5577 | 0.1508 | 0.1483 | 0.0257 |
| H2 | G | 0.3429 | 0.5149 | 0.7321 | 0.7973 |
| H3 | G | 0.0268 | 0.2788 | 0.0828 | 0.0408 |
| H4 | A | 0.0109 | - | - | - |
| H5 | A | - | 0.0169 | - | - |
| H6 | G | - | - | - | 0.0983 |
| H7 | G | - | - | - | 0.0106 |
| PheWeb Association | Summary Statistics Association | Coloc Probability with IAMDGC AMD | ||||
|---|---|---|---|---|---|---|
| UK Biobank Phenotype | p-Value | Gender | p-Value | Gender Specificity | Same Signal | Two Signals |
| Diastolic blood pressure, automated reading | 1.50 × 10−23 | both sexes | 2.27 × 10−27 | no | 0.604 | 0.385 |
| female | 7.27 × 10−18 | 0.964 | 0.035 | |||
| male | 2.58 × 10−11 | 0.610 | 0.379 | |||
| Vascular/heart problems diagnosed by doctor: high blood pressure | 7.20 × 10−23 | both sexes | 2.24 × 10−26 | no | 0.976 | 0.024 |
| female | 2.89 × 10−14 | 0.918 | 0.079 | |||
| male | 9.90 × 10−14 | 0.970 | 0.029 | |||
| Vascular/heart problems diagnosed by doctor: none of the above | 2.60 × 10−21 | both sexes | 2.79 × 10−25 | no | 0.970 | 0.029 |
| female | 9.10 × 10−14 | 0.860 | 0.136 | |||
| male | 4.11 × 10−13 | 0.969 | 0.030 | |||
| Non-cancer illness code, self-reported: hypertension | 2.70 × 10−21 | both sexes | 2.15 × 10−24 | no | 0.975 | 0.024 |
| female | 3.15 × 10−13 | 0.936 | 0.062 | |||
| male | 9.12 × 10−13 | 0.965 | 0.034 | |||
| Systolic blood pressure, automated reading | 1.60 × 10−11 | both sexes | 9.33 × 10−14 | no | 0.735 | 0.258 |
| female | 1.12 × 10−10 | 0.969 | 0.030 | |||
| male | 7.66 × 10−5 | 0.428 | 0.523 | |||
| Medication for cholesterol, blood pressure or diabetes: blood pressure medication | 6.30 × 10−11 | male | 3.63 × 10−12 | 0.938 | 0.061 | |
| Medication for cholesterol, blood pressure, diabetes, or take exogenous hormones: blood pressure medication | 3.90 × 10−10 | female | 8.58 × 10−12 | 0.890 | 0.106 | |
| Creatinine (enzymatic) in urine | 5.30 × 10−9 | both sexes | 2.85 × 10−7 | no | 0.037 | 0.936 |
| female | 8.92 × 10−8 | 0.038 | 0.934 | |||
| male | 0.031 | 0.037 | 0.936 | |||
| Treatment/medication code: ramipril | 7.40 × 10−8 | both sexes | 8.01 × 10−8 | yes, only in male | 0.964 | 0.035 |
| female | 0.103 | 0.034 | 0.067 | |||
| male | 4.03 × 10−8 | 0.898 | 0.099 | |||
| Treatment/medication code: bendroflumethiazide | 1.20 × 10−7 | both sexes | 5.29 × 10−8 | no | 0.948 | 0.051 |
| female | 9.58 × 10−6 | 0.901 | 0.080 | |||
| male | 1.37 × 10−3 | 0.486 | 0.085 | |||
| Medication for cholesterol, blood pressure or diabetes: none of the above | 1.30 × 10−7 | male | 1.23 × 10−8 | 0.830 | 0.165 | |
| Hearing difficulty/problems with background noise | 4.10 × 10−6 | both sexes | 1.43 × 10−6 | yes, only in female | 0.917 | 0.079 |
| female | 6.01 × 10−9 | 0.966 | 0.033 | |||
| male | 0.345 | 0.007 | 0.062 | |||
| Birth weight of first child | 1.20 × 10−5 | female | 2.48 × 10−5 | 0.785 | 0.175 | |
| Mineral and other dietary supplements: glucosamine | 2.50 × 10−5 | both sexes | 3.59 × 10−5 | no | 0.877 | 0.068 |
| female | 9.41 × 10−3 | 0.118 | 0.098 | |||
| male | 8.17 × 10−4 | 0.469 | 0.056 | |||
| Sodium in urine | 7.20 × 10−5 | both sexes | 8.38 × 10−4 | no | 0.037 | 0.935 |
| female | 1.58 × 10−3 | 0.037 | 0.935 | |||
| male | 0.106 | 0.010 | 0.300 | |||
| Eye problems/disorders: macular degeneration | 5.10 × 10−3 | both sexes | 2.66 × 10−3 | |||
| female | 0.157 | |||||
| male | 2.82 × 10−3 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiel, C.; Strunz, T.; International AMD Genomics Consortium (Project Manager Susan Blanton) IAMDGC; Grassmann, F.; Weber, B.H.F. Pleiotropic Locus 15q24.1 Reveals a Gender-Specific Association with Neovascular but Not Atrophic Age-Related Macular Degeneration (AMD). Cells 2020, 9, 2257. https://doi.org/10.3390/cells9102257
Kiel C, Strunz T, International AMD Genomics Consortium (Project Manager Susan Blanton) IAMDGC, Grassmann F, Weber BHF. Pleiotropic Locus 15q24.1 Reveals a Gender-Specific Association with Neovascular but Not Atrophic Age-Related Macular Degeneration (AMD). Cells. 2020; 9(10):2257. https://doi.org/10.3390/cells9102257
Chicago/Turabian StyleKiel, Christina, Tobias Strunz, International AMD Genomics Consortium (Project Manager Susan Blanton) IAMDGC, Felix Grassmann, and Bernhard H. F. Weber. 2020. "Pleiotropic Locus 15q24.1 Reveals a Gender-Specific Association with Neovascular but Not Atrophic Age-Related Macular Degeneration (AMD)" Cells 9, no. 10: 2257. https://doi.org/10.3390/cells9102257
APA StyleKiel, C., Strunz, T., International AMD Genomics Consortium (Project Manager Susan Blanton) IAMDGC, Grassmann, F., & Weber, B. H. F. (2020). Pleiotropic Locus 15q24.1 Reveals a Gender-Specific Association with Neovascular but Not Atrophic Age-Related Macular Degeneration (AMD). Cells, 9(10), 2257. https://doi.org/10.3390/cells9102257






