Rapid Detection of Free Cancer Cells in Intraoperative Peritoneal Lavage Using One-Step Nucleic Acid Amplification (OSNA) in Gastric Cancer Patients
Abstract
1. Introduction
2. Materials and Methods
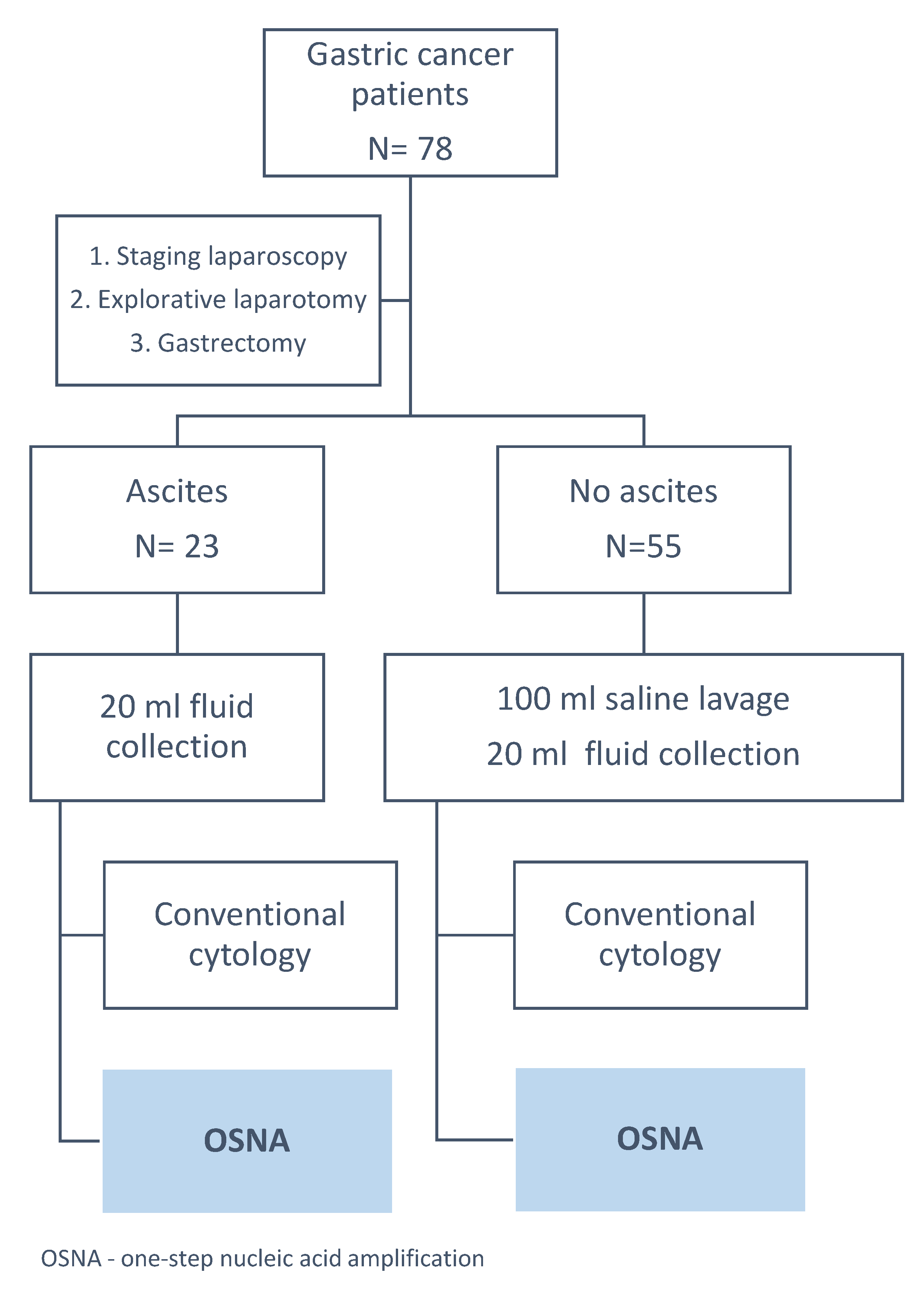
2.1. Intraoperative Peritoneal Lavage and Fluid Examination
2.2. OSNA Examination
2.3. Statistical Analysis
3. Results
3.1. Comparison of CK19 Levels in Selected Clinical Variables
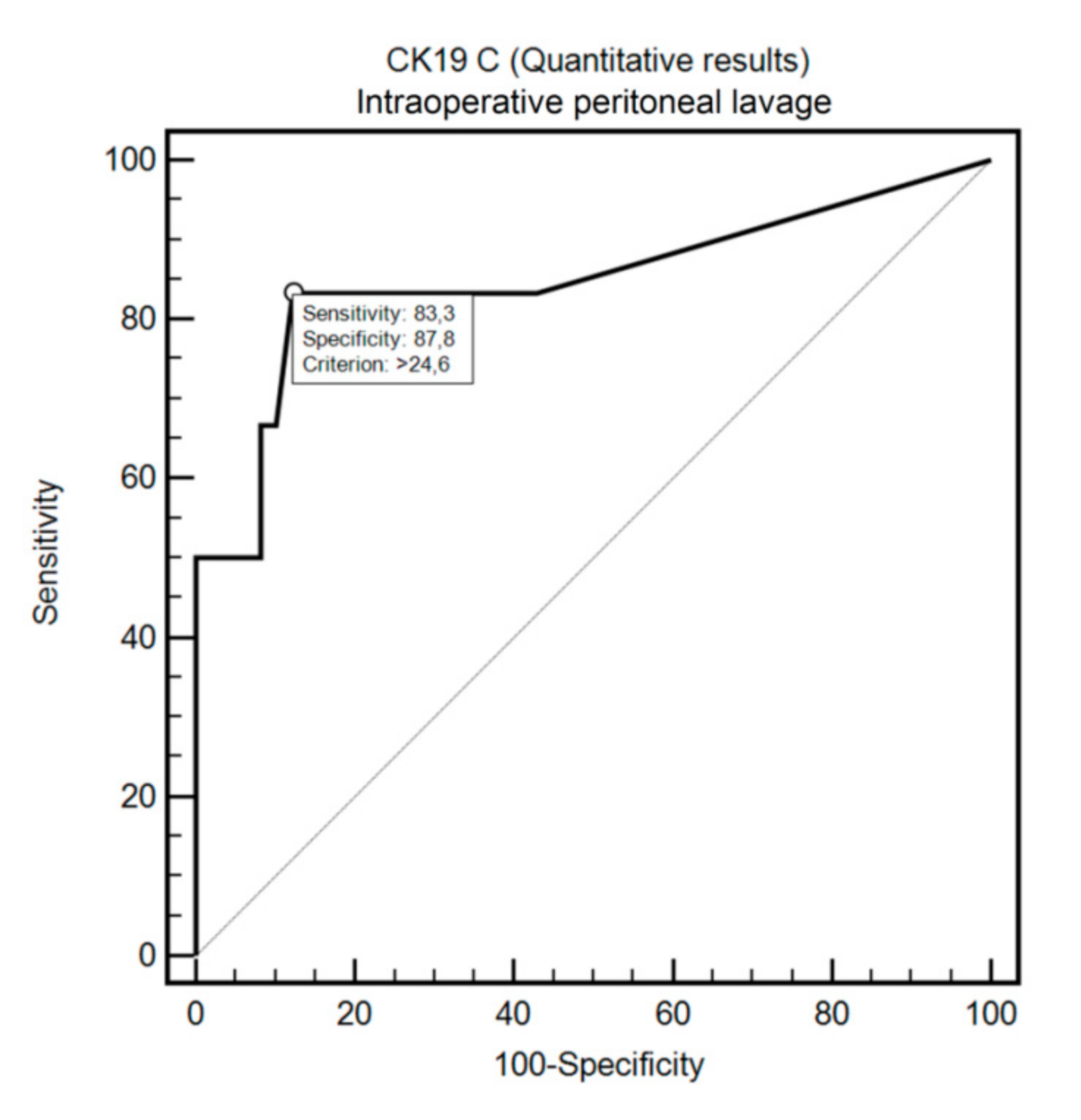
3.2. OSNA Assay in Detecting FCC in Intraoperative Peritoneal Lavage and Peritoneal Fluid with Positive Cytology Using ROC Curve Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nakabayashi, K.; Uraoka, T.; Shibuya, M.; Matsuura, N.; Tsujimoto, M. Rapid detection of CEA mRNA in peritoneal washes using One-Step Nucleic acid Amplification (OSNA) for gastric cancer patients. Clin. Chim. Acta 2015, 439, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wu, N.-D.; Liu, B. Regional but fatal: Intraperitoneal metastasis in gastric cancer. World J. Gastroenterol. 2016, 22, 7478–7485. [Google Scholar] [CrossRef] [PubMed]
- Pecqueux, M.; Fritzmann, J.; Adamu, M.; Thorlund, K.; Kahlert, C.; Reiδfelder, C.; Weitz, J.; Rahbari, N. Free intraperitoneal tumor cells and outcome in gastric cancer patients: A systematic review and meta-analysis. Oncotarget 2015, 6, 35564–35578. [Google Scholar] [CrossRef]
- Lisiecki, R.; Kruszwicka, M.; Spychała, A.; Murawa, D. Prognostic significance, diagnosis and treatment in patients with gastric cancer and positive peritoneal washings. A review of the literature. Rep. Pract. Oncol. Radiother. 2017, 22, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Kitayama, J.; Emoto, S.; Yamaguchi, H.; Ishigami, H.; Onoyama, H.; Yamashita, H.; Seto, Y.; Matsuzaki, K.; Watanabe, T. Flow cytometric quantification of intraperitoneal free tumor cells is a useful biomarker in gastric cancer patients with peritoneal metastasis. Ann. Surg. Oncol. 2015, 22, 2336–2342. [Google Scholar] [CrossRef] [PubMed]
- Yepuri, N.; Bahary, N.; Jain, A.; Dhir, M. Review and update on the role of peritoneal cytology in the treatment of gastric cancer. J. Surg. Res. 2019, 235, 607–614. [Google Scholar] [CrossRef]
- Koganti, S.B.; Boddepalli, S.; Nambada, M.; Thumma, V.M.; Nagari, B.; Sastry, R.A. Positive peritoneal lavage cytology-implications for staging and management of gastric cancer. Indian J. Surg. Oncol. 2016, 7, 430–435. [Google Scholar] [CrossRef][Green Version]
- Yaguchi, Y.; Sugasawa, H.; Tsujimoto, H.; Takata, H.; Nakabayashi, K.; Ichikura, T.; Ono, S.; Hiraki, S.; Sakamoto, N.; Horio, T.; et al. One-step nucleic acid amplification (OSNA) for the application of sentinel node concept in gastric cancer. Ann. Surg. Oncol. 2011, 18, 2289–2296. [Google Scholar] [CrossRef]
- Kumagai, K.; Yamamoto, N.; Miyashiro, I.; Tomita, Y.; Katai, H.; Kushima, R.; Tsuda, H.; Kitagawa, Y.; Takeuchi, H.; Mukai, M.; et al. Multicenter study evaluating the clinical performance of the OSNA assay for the molecular detection of lymph node metastases in gastric cancer patients. Gastric Cancer 2014, 17, 273–280. [Google Scholar] [CrossRef]
- Bizzarri, N.; Anchora, L.P.; Zannoni, G.F.; Santoro, A.; Valente, M.; Inzani, F.; Gallotta, V.; Conte, C.; Chiantera, V.; Fanfani, F.; et al. Role of one-step nucleic acid amplification (OSNA) to detect sentinel lymph node low-volume metastasis in early-stage cervical cancer. Int. J. Gynecol. Cancer 2020, 30, 364–371. [Google Scholar] [CrossRef]
- Fanfani, F.; Monterossi, G.; Ghizzoni, V.; Rossi, E.D.; Dinoi, G.; Inzani, F.; Fagotti, A.; Alletti, S.G.; Scarpellini, F.; Nero, C.; et al. One-Step Nucleic Acid Amplification (OSNA): A fast molecular test based on CK19 mRNA concentration for assessment of lymph-nodes metastases in early stage endometrial cancer. PLoS ONE 2018, 13, e0195877. [Google Scholar] [CrossRef] [PubMed]
- Kostun, J.; Pesta, M.; Sláma, J.; Slunéčko, R.; Vlasák, P.; Bouda, J.; Novotný, Z.; Topolčan, O.; Kucera, R.; Kulda, V.; et al. One-step nucleic acid amplification vs ultrastaging in the detection of sentinel lymph node metastasis in endometrial cancer patients. J. Surg. Oncol. 2019, 119, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Engels, S.; Goos, P.; Süykers, M.-C.; Henke, R.-P.; Gerullis, H.; Wawroschek, F. Detection of CK19 mRNA Using One-step Nucleic Acid Amplification (OSNA) in Prostate Cancer: Preliminary Results. J. Cancer 2018, 9, 4611–4617. [Google Scholar] [CrossRef] [PubMed]
- Schem, C.; Maass, N.; Bauerschlag, D.O.; Carstensen, M.H.; Löning, T.; Röder, C.; Batic, O.; Jonat, W.; Tiemann, K. One-step nucleic acid amplification-a molecular method for the detection of lymph node metastases in breast cancer patients; results of the German study group. Virchows Arch. 2009, 454, 203–210. [Google Scholar] [CrossRef]
- Safavieh, M.; Kanakasabapathy, M.K.; Tarlan, F.; Ahmed, M.U.; Zourob, M.; Asghar, W.; Shafiee, H. Emerging Loop-Mediated Isothermal Amplification-Based Microchip and Microdevice Technologies for Nucleic Acid Detection. ACS Biomater. Sci. Eng. 2016, 2, 278–294. [Google Scholar] [CrossRef]
- Tsujimoto, M.; Nakabayashi, K.; Yoshidome, K.; Kaneko, T.; Iwase, T.; Akiyama, F.; Kato, Y.; Tsuda, H.; Ueda, S.; Sato, K.; et al. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin. Cancer Res. 2007, 13, 4807–4816. [Google Scholar] [CrossRef]
- Shimada, A.; Takeuchi, H.; Nishi, T.; Mayanagi, S.; Fukuda, K.; Suda, K.; Nakamura, R.; Wada, N.; Kawakubo, H.; Nakahara, T.; et al. Utility of the one-step nucleic acid amplification assay in sentinel node mapping for early gastric cancer patients. Gastric Cancer 2020, 23, 418–425. [Google Scholar] [CrossRef]
- Visser, M.; Jiwa, M.; Horstman, A.; Brink, A.A.; Pol, R.P.; Van Diest, P.; Snijders, P.J.; Meijer, C.J.L.M. Intra-operative rapid diagnostic method based on CK19 mRNA expression for the detection of lymph node metastases in breast cancer. Int. J. Cancer 2008, 122, 2562–2567. [Google Scholar] [CrossRef]
- Yoneda, A.; Taniguchi, K.; Torashima, Y.; Susumu, S.; Kanetaka, K.; Kuroki, T.; Eguchi, S. The detection of gastric cancer cells in intraoperative peritoneal lavage using the reverse transcription--loop-mediated isothermal amplification method. J. Surg. Res. 2014, 187, e1–e6. [Google Scholar] [CrossRef]
- Kowalewska, M.; Chechlinska, M.; Nowak, R. Carcinoembryonic antigen and cytokeratin 20 in peritoneal cells of cancer patients: Are we aware of what we are detecting by mRNA examination? Br. J. Cancer 2008, 98, 512–513. [Google Scholar] [CrossRef]
- Sibio, S.; Fiorani, C.; Stolfi, C.; Divizia, A.; Pezzuto, R.; Montagnese, F.; Bagaglini, G.; Sammartino, P.; Sica, G.S. Detection methods and clinical significance of free peritoneal tumor cells found during colorectal cancer surgery. World J. Gastrointest. Surg. 2015, 7, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Ruud, P.; Hovig, E.; Fodstad, O. Identification of a novel cytokeratin 19 pseudogene that may interfere with reverse transcriptase-polymerase chain reaction assays used to detect micrometastatic tumor cells. Int. J. Cancer 1999, 80, 119–125. [Google Scholar] [CrossRef]
- Majima, T.; Ichikura, T.; Takayama, E.; Chochi, K.; Mochizuki, H. Detecting circulating cancer cells using reverse transcriptase-polymerase chain reaction for cytokeratin mRNA in peripheral blood from patients with gastric cancer. Jpn. J. Clin. Oncol. 2000, 30, 499–503. [Google Scholar] [CrossRef][Green Version]
- Shim, H.-J.; Kim, H.-J.; Lee, S.H.; Bae, W.-K.; Hwang, E.-C.; Cho, S.-H.; Chung, I.-J.; Bang, H.-J.; Hwang, J.E. Observational Study of Peritoneal Washing Cytology-Positive Gastric Cancer without Gross Peritoneal Metastasis in Patients who Underwent Radical D2 Gastrectomy. Sci. Rep. 2020, 10, 9549. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xu, A.; Sun, X.; Zhao, X.; Xia, Y.; Rao, H.; Zhang, Y.; Zhang, R.; Chen, L.; Zhang, T.; et al. Combined Surgery and Extensive Intraoperative Peritoneal Lavage vs Surgery Alone for Treatment of Locally Advanced Gastric Cancer: The SEIPLUS Randomized Clinical Trial. JAMA Surg. 2019, 154, 610–616. [Google Scholar] [CrossRef]
- Misawa, K.; Mochizuki, Y.; Sakai, M.; Teramoto, H.; Morimoto, D.; Nakayama, H.; Tanaka, N.; Matsui, T.; Ito, Y.; Ito, S.; et al. Randomized clinical trial of extensive intraoperative peritoneal lavage versus standard treatment for resectable advanced gastric cancer (CCOG 1102 trial). BJS 2019, 106, 1602–1610. [Google Scholar] [CrossRef] [PubMed]

| Variable | Peritoneal Fluid | Intraoperative Peritoneal Lavage |
|---|---|---|
| n (%) or Median ± SD. Median (Range) | ||
| Sex | ||
| Males | 11 (47.8%) | 32 (58.2%) |
| Females | 12 (52.2%) | 23 (41.8%) |
| Age | 60.2 + 12.6. | 62.4 + 11.4 |
| 60 (36–86) | 63 (37–87) | |
| Group | ||
| Explorative laparotomy (M1) | 12 (52.2%) | 6 (10.9%) |
| Staging Laparoscopy | 3 (13.0%) | 16 (29.1%) |
| Surgery after CTH | 7 (30.4%) | 25 (45.4%) |
| Upfront Surgery | 1 (4.4%) | 8 (14.5%) |
| Lauren type | ||
| Intestinal | 4 (17.4%) | 25 (45.5%) |
| Mixed | 6 (26.1%) | 5 (9.1%) |
| Diffused | 12 (52.2%) | 22 (40.0%) |
| Unclassified | 1 (4.3%) | 3 (5.4%) |
| pT | ||
| 1a | 1 (10%) | 6 (19.3%) |
| 1b | 1 (10%) | 3 (9.7%) |
| 2 | 2 (20%) | 4 (12.9%) |
| 3 | 2 (20%) | 10 (32.3%) |
| 4a | 1 (10%) | 6 (19.3%) |
| 4b | 3 (30%) | 2 (6.4%) |
| pN | ||
| 0 | 4 (40%) | 17 (54.8%) |
| 1 | 1 (10%) | 5 (16.1%) |
| 2 | 1 (10%) | 4 (12.9%) |
| 3, 3a, 3b | 4 (40%) | 5 (16.1%) |
| pM | ||
| 0 | 7 (70%) | 28 (90.3%) |
| 1 | 3 (30%) | 3 (9.7%) |
| Cytology | ||
| Positive | 13 (56.5%) | 6 (10.9%) |
| Negative | 10 (43.5%) | 49 (89.1%) |
| Variable | Peritoneal Fluid | Intraoperative Peritoneal Lavage | ||
|---|---|---|---|---|
| Median (copies/µL) | p | Median (copies/µL) | p | |
| pT | 0.0335 | 0.6778 | ||
| 1a/b, 2 | 2.48 | 0 | ||
| 3 | 100 | 0.03 | ||
| 4a, 4b | 415.7 | 0 | ||
| pN | 0.0478 | 0.6264 | ||
| 0 | 2.48 | 0 | ||
| 1–3 | 334.8 | 0 | ||
| pM | 0.1056 | 0.0125 | ||
| 0 (C−) | 5 | 0 | ||
| 0 (C+) | 38508.2 | 2200 | ||
| 1 (C−) | 25.1 | 0 | ||
| 1 (C+) | 960 | 54.9 | ||
| Cytology | 0.0099 | 0.0027 | ||
| Positive | 960 | 577.5 | ||
| Negative | 6.9 | 0 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gęca, K.; Rawicz-Pruszyński, K.; Mielko, J.; Mlak, R.; Sędłak, K.; Polkowski, W.P. Rapid Detection of Free Cancer Cells in Intraoperative Peritoneal Lavage Using One-Step Nucleic Acid Amplification (OSNA) in Gastric Cancer Patients. Cells 2020, 9, 2168. https://doi.org/10.3390/cells9102168
Gęca K, Rawicz-Pruszyński K, Mielko J, Mlak R, Sędłak K, Polkowski WP. Rapid Detection of Free Cancer Cells in Intraoperative Peritoneal Lavage Using One-Step Nucleic Acid Amplification (OSNA) in Gastric Cancer Patients. Cells. 2020; 9(10):2168. https://doi.org/10.3390/cells9102168
Chicago/Turabian StyleGęca, Katarzyna, Karol Rawicz-Pruszyński, Jerzy Mielko, Radosław Mlak, Katarzyna Sędłak, and Wojciech P. Polkowski. 2020. "Rapid Detection of Free Cancer Cells in Intraoperative Peritoneal Lavage Using One-Step Nucleic Acid Amplification (OSNA) in Gastric Cancer Patients" Cells 9, no. 10: 2168. https://doi.org/10.3390/cells9102168
APA StyleGęca, K., Rawicz-Pruszyński, K., Mielko, J., Mlak, R., Sędłak, K., & Polkowski, W. P. (2020). Rapid Detection of Free Cancer Cells in Intraoperative Peritoneal Lavage Using One-Step Nucleic Acid Amplification (OSNA) in Gastric Cancer Patients. Cells, 9(10), 2168. https://doi.org/10.3390/cells9102168







