The Cynomolgus Macaque MHC Polymorphism in Experimental Medicine
Abstract
1. Introduction
2. Brief Review of Experimental MHC Polymorphism Milestones
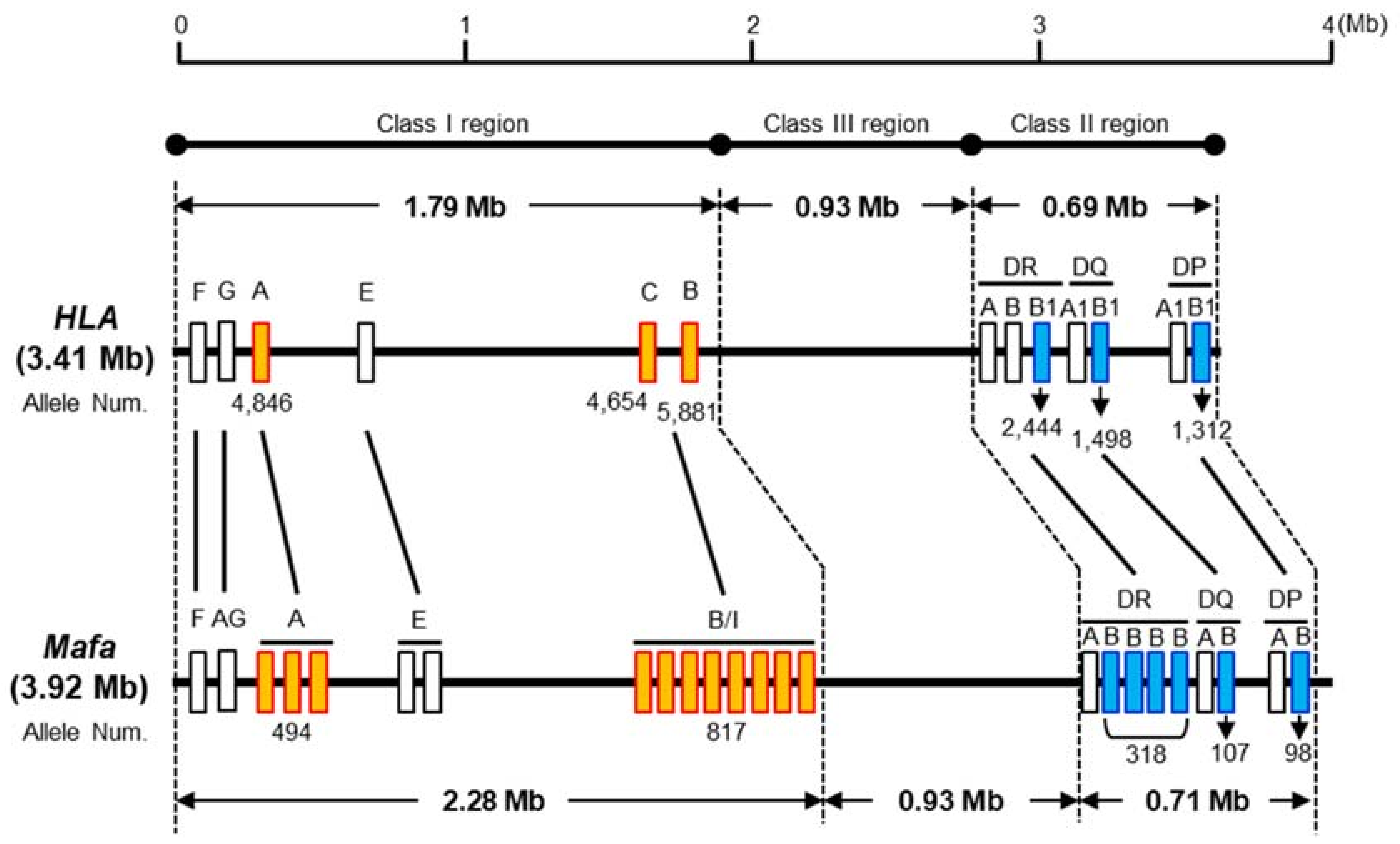
3. Genomic Structure of the Mafa MHC Region and MHC Polymorphism
3.1. Length of the Mafa Region
3.2. MHC Genes
3.3. Genes of MHC Class I Polypeptide-Related Sequences (MIC)
3.4. Non-MHC Genes
3.5. Nomenclature of Mafa Class I and Class II Alleles
3.6. Characteristics of Mafa Class I and Class II Allele Numbers
4. Diversity of Cynomolgus Macaque Populations
4.1. Study of MHC Class I Alleles in Various Populations
4.2. MHC Class II Allele Sharing
5. Comparison of MHC Polymorphism among Populations
6. Identification of MHC Region Homozygotes
7. Characterization of MHC Class I Expression Levels in Various Tissues and Conditions
8. MHC and Experimental Infectious Diseases in Cynomolgus Macaque
9. MHC and Control of SIV Infection
10. Allogenic Lymphocyte Transfer
11. Role of MHC in Induced Pluripotent (iPS) Stem Cell Allografts
12. Impact of MHC Polymorphism on Blood Counts
13. Macaque MHC and Experimental Autoimmune Diseases
14. Impact of MHC on Cynomolgus Macaque Reproduction
15. Association of MHC and Drug Related Adverse Effects
16. Interactions between MHC Proteins and Various Receptors: Killer Cell Immunoglobulin-Like Receptors (KIRs) and Leukocyte Immunoglobulin-Like Receptors (LILRs)
17. Roles of Mafa-E in Experimental Medicine
18. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Southwick, C.H.; Siddiqi, M.F. Population Status of Nonhuman-Primates in Asia, with Emphasis on Rhesus Macaques in India. Am. J. Primatol. 1994, 34, 51–59. [Google Scholar] [CrossRef]
- Sussman, R.W.; Tattersall, I. Distribution, Abundance, and Putative Ecological Strategy of Macaca-Fascicularis on the Island of Mauritius, Southwestern Indian-Ocean. Folia Primatol. 1986, 46, 28–43. [Google Scholar] [CrossRef]
- Kawamoto, Y.; Kawamoto, S.; Matsubayashi, K.; Nozawa, K.; Watanabe, T.; Stanley, M.A.; Perwitasari-Farajallah, D. Genetic diversity of longtail macaques (Macaca fascicularis) on the island of Mauritius: An assessment of nuclear and mitochondrial DNA polymorphisms. J. Med. Primatol. 2008, 37, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Tosi, A.J.; Coke, C.S. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol. Phylogenet Evol. 2007, 42, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Bonhomme, M.; Blancher, A.; Cuartero, S.; Chikhi, L.; Crouau-Roy, B. Origin and number of founders in an introduced insular primate: Estimation from nuclear genetic data. Mol. Ecol. 2008, 17, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Osada, N.; Hettiarachchi, N.; Babarinde, I.A.; Saitou, N.; Blancher, A. Whole-Genome Sequencing of Six Mauritian Cynomolgus Macaques (Macaca fascicularis) Reveals a Genome-Wide Pattern of Polymorphisms under Extreme Population Bottleneck. Genome Biol. Evol. 2015, 7, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Bonhomme, M.; Cuartero, S.; Blancher, A.; Crouau-roy, B. Assessing Natural Introgression in 2 Biomedical Model Species, the Rhesus Macaque (Macaca mulatta) and the Long-Tailed Macaque (Macaca fascicularis). J. Hered. 2009, 100, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Bunlungsup, S.; Kanthaswamy, S.; Oldt, R.F.; Smith, D.G.; Houghton, P.; Hamada, Y.; Malaivijitnond, S. Genetic analysis of samples from wild populations opens new perspectives on hybridization between long-tailed (Macaca fascicularis) and rhesus macaques (Macaca mulatta). Am. J. Primatol. 2017, 79. [Google Scholar] [CrossRef] [PubMed]
- Osada, N.; Uno, Y.; Mineta, K.; Kameoka, Y.; Takahashi, I.; Terao, K. Ancient genome-wide admixture extends beyond the current hybrid zone between Macaca fascicularis and M-mulatta. Mol. Ecol. 2010, 19, 2884–2895. [Google Scholar] [CrossRef]
- Trask, J.A.S.; Garnica, W.T.; Smith, D.G.; Houghton, P.; Lerche, N.; Kanthaswamy, S. Single-Nucleotide Polymorphisms Reveal Patterns of Allele Sharing Across the Species Boundary Between Rhesus (Macaca mulatta) and Cynomolgus (M. fascicularis) Macaques. Am. J. Primatol. 2013, 75, 135–144. [Google Scholar] [CrossRef]
- Anderson, D.J.; Kirk, A.D. Primate Models in Organ Transplantation. Csh Perspect Med. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Niu, Y.Y.; Li, Y.J.; Ai, Z.Y.; Kang, Y.; Shi, H.; Xiang, Z.; Yang, Z.H.; Tan, T.; Si, W.; et al. Generation of Cynomolgus Monkey Chimeric Fetuses using Embryonic Stem Cells. Cell Stem Cell 2015, 17, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Honda, A.; Kawano, Y.; Izu, H.; Choijookhuu, N.; Honsho, K.; Nakamura, T.; Yabuta, Y.; Yamamoto, T.; Takashima, Y.; Hirose, M.; et al. Discrimination of Stem Cell Status after Subjecting Cynomolgus Monkey Pluripotent Stem Cells to Naive Conversion. Sci. Rep. 2017, 7, 45285. [Google Scholar] [CrossRef] [PubMed]
- Hanazono, Y.; Terao, K.; Ozawa, K. Gene transfer into nonhuman primate hematopoietic stem cells: Implications for gene therapy. Stem Cells 2001, 19, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Hernandez, T.; Carnathan, D.G.; Moyle, P.M.; Toth, I.; West, N.P.; Young, P.R.; Silvestri, G.; Walker, M.J. The Contribution of Non-human Primate Models to the Development of Human Vaccines. Discov. Med. 2014, 18, 313–322. [Google Scholar] [PubMed]
- Iwasaki, K.; Uno, Y.; Utoh, M.; Yamazaki, H. Importance of cynomolgus monkeys in development of monoclonal antibody drugs. Drug Metab. Pharmacok. 2019, 34, 55–63. [Google Scholar] [CrossRef]
- Gardner, M.B.; Luciw, P.A. Macaque models of human infectious disease. ILAR J. 2008, 49, 220–255. [Google Scholar] [CrossRef]
- Verdier, J.M.; Acquatella, I.; Lautier, C.; Devau, G.; Trouche, S.; Lasbleiz, C.; Mestre-Frances, N. Lessons from the analysis of nonhuman primates for understanding human aging and neurodegenerative diseases. Front. Neurosci. 2015, 9, 64. [Google Scholar] [CrossRef]
- Eastwood, D.; Findlay, L.; Poole, S.; Bird, C.; Wadhwa, M.; Moore, M.; Burns, C.; Thorpe, R.; Stebbings, R. Monoclonal antibody TGN1412 trial failure explained by species differences in CD28 expression on CD4(+) effector memory T-cells. Brit. J. Pharmacol. 2010, 161, 512–526. [Google Scholar] [CrossRef]
- Prall, T.M.; Graham, M.E.; Karl, J.A.; Wiseman, R.W.; Ericsen, A.J.; Raveendran, M.; Alan Harris, R.; Muzny, D.M.; Gibbs, R.A.; Rogers, J.; et al. Improved full-length killer cell immunoglobulin-like receptor transcript discovery in Mauritian cynomolgus macaques. Immunogenetics 2017, 69, 325–339. [Google Scholar] [CrossRef]
- Blancher, A.; Tisseyre, P.; Dutaur, M.; Apoil, P.A.; Maurer, C.; Quesniaux, V.; Raulf, F.; Bigaud, M.; Abbal, M. Study of Cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics 2006, 58, 269–282. [Google Scholar] [CrossRef]
- Balner, H. Current knowledge of the histocompatibility complex of rhesus monkeys. Transplant. Rev. 1973, 15, 50–61. [Google Scholar]
- Balner, H.; D’Amaro, J.; Toth, E.K.; Dersjant, H.; van Vreeswijk, W. The histocompatibility complex of rhesus monkeys. Relation between RhL and the main locus controlling reactivity in mixed lymphocyte cultures. Transplant. Proc. 1973, 5, 323–329. [Google Scholar]
- Roger, J.H.; van Reeswijk, W.; D’Amaro, J.; Balner, H. The major histocompatibility complex of Rhesus monkeys IX. Current concepts of serology and genetics of Ia antigens. Tissue Antigens 1978, 11, 163–180. [Google Scholar] [CrossRef]
- van Es, A.A.; Balner, H. The major histocompatibility complex of Rhesus monkeys. XII: Cellular typing for D locus antigens in families. Tissue Antigens 1979, 13, 255–272. [Google Scholar]
- Van Rood, J.J.; Balner, R.; Gabb, B.W.; Dersjant, H.; Van Vreeswijk, W. Major histocompatibility locus of rhesus monkeys (RhL-A). Nat. New Biol. 1971, 230, 177–180. [Google Scholar]
- Balner, H.; Dorf, M.E.; de Groot, M.L.; Benacerraf, B. The histocompatibility complex of rhesus monkeys. 3. Evidence for a major MLR locus and histocompatibility-linked Ir genes. Transplant. Proc. 1973, 5, 1555–1560. [Google Scholar]
- Balner, H.; Gabb, B.W.; Toth, E.K.; Dersjant, H.; van Vreeswijk, W. The histocompatibility complex of rhesus monkeys. I. Serology and genetics of the RhL-A system. Tissue Antigens 1973, 3, 257–272. [Google Scholar] [CrossRef]
- Balner, H.; Toth, E.K. The histocompatibility complex of rhesus monkeys. II. A major locus controlling reactivity in mixed lymphocyte cultures. Tissue Antigens 1973, 3, 273–290. [Google Scholar] [CrossRef]
- Keever, C.A.; Heise, E.R. The major histocompatibility complex (CyLA) of the cynomolgus monkey. I. Serologic definition of 21 specificities. Hum. Immunol. 1983, 7, 131–149. [Google Scholar] [CrossRef]
- Keever, C.A.; Heise, E.R. The major histocompatibility complex of the cynomolgus monkey: Absorption analysis of 24 CyLA antisera. Hum. Immunol. 1985, 12, 75–90. [Google Scholar] [CrossRef]
- Keever, C.A.; Heise, E.R. The major histocompatibility complex of the cynomolgus monkey. II. Polymorphism at three serologically defined loci and correlation of haplotypes with stimulation in MLC and skin graft survival. Hum. Immunol. 1985, 12, 143–164. [Google Scholar] [CrossRef]
- Leuchte, N.; Berry, N.; Kohler, B.; Almond, N.; LeGrand, R.; Thorstensson, R.; Titti, F.; Sauermann, U. MhcDRB-sequences from cynomolgus macaques (Macaca fascicularis) of different origin. Tissue Antigens 2004, 63, 529–537. [Google Scholar] [CrossRef]
- Knapp, L.A.; Cadavid, L.F.; Eberle, M.E.; Knechtle, S.J.; Bontrop, R.E.; Watkins, D.I. Identification of new Mamu-DRB alleles using DGGE and direct sequencing. Immunogenetics 1997, 45, 171–179. [Google Scholar] [CrossRef]
- Bonhomme, M.; Blancher, A.; Crouau-Roy, B. Multiplexed microsatellites for rapid identification and characterization of individuals and populations of Cercopithecidae. Am. J. Primatol. 2005, 67, 385–391. [Google Scholar] [CrossRef]
- Watanabe, A.; Shiina, T.; Shimizu, S.; Hosomichi, K.; Yanagiya, K.; Kita, Y.F.; Kimura, T.; Soeda, E.; Torii, R.; Ogasawara, K.; et al. A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics 2007, 89, 402–412. [Google Scholar] [CrossRef]
- Wiseman, R.W.; Karl, J.A.; Bimber, B.N.; O’Leary, C.E.; Lank, S.M.; Tuscher, J.J.; Detmer, A.M.; Bouffard, P.; Levenkova, N.; Turcotte, C.L.; et al. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat. Med. 2009, 15, 1322–1326. [Google Scholar] [CrossRef]
- Aarnink, A.; Apoil, P.A.; Takahashi, I.; Osada, N.; Blancher, A. Characterization of MHC class I transcripts of a Malaysian cynomolgus macaque by high-throughput pyrosequencing and EST libraries. Immunogenetics 2011, 63, 703–713. [Google Scholar] [CrossRef]
- Westbrook, C.J.; Karl, J.A.; Wiseman, R.W.; Mate, S.; Koroleva, G.; Garcia, K.; Sanchez-Lockhart, M.; O’Connor, D.H.; Palacios, G. No assembly required: Full-length MHC class I allele discovery by PacBio circular consensus sequencing. Hum. Immunol. 2015, 76, 891–896. [Google Scholar] [CrossRef]
- Bondarenko, G.I.; Dambaeva, S.V.; Grendell, R.L.; Hughes, A.L.; Durning, M.; Garthwaite, M.A.; Golos, T.G. Characterization of cynomolgus and vervet monkey placental MHC class I expression: Diversity of the nonhuman primate AG locus. Immunogenetics 2009, 61, 431–442. [Google Scholar] [CrossRef]
- Bontrop, R.E.; Otting, N.; de Groot, N.G.; Doxiadis, G.G. Major histocompatibility complex class II polymorphisms in primates. Immunol. Rev. 1999, 167, 339–350. [Google Scholar] [CrossRef]
- Budde, M.L.; Greene, J.M.; Chin, E.N.; Ericsen, A.J.; Scarlotta, M.; Cain, B.T.; Pham, N.H.; Becker, E.A.; Harris, M.; Weinfurter, J.T.; et al. Specific CD8+ T cell responses correlate with control of simian immunodeficiency virus replication in Mauritian cynomolgus macaques. J. Virol. 2012, 86, 7596–7604. [Google Scholar] [CrossRef]
- O’Connor, S.L.; Blasky, A.J.; Pendley, C.J.; Becker, E.A.; Wiseman, R.W.; Karl, J.A.; Hughes, A.L.; O’Connor, D.H. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics 2007, 59, 449–462. [Google Scholar] [CrossRef]
- Wiseman, R.W.; Karl, J.A.; Bohn, P.S.; Nimityongskul, F.A.; Starrett, G.J.; O’Connor, D.H. Haplessly hoping: Macaque major histocompatibility complex made easy. ILAR J. 2013, 54, 196–210. [Google Scholar] [CrossRef]
- Blancher, A.; Aarnink, A.; Tanaka, K.; Ota, M.; Inoko, H.; Yamanaka, H.; Nakagawa, H.; Apoil, P.A.; Shiina, T. Study of cynomolgus monkey (Macaca fascicularis) Mhc DRB gene polymorphism in four populations. Immunogenetics 2012, 64, 605–614. [Google Scholar] [CrossRef]
- Blancher, A.; Aarnink, A.; Yamada, Y.; Tanaka, K.; Yamanaka, H.; Shiina, T. Study of MHC class II region polymorphism in the Filipino cynomolgus macaque population. Immunogenetics 2014, 66, 219–230. [Google Scholar] [CrossRef]
- Shiina, T.; Yamada, Y.; Aarnink, A.; Suzuki, S.; Masuya, A.; Ito, S.; Ido, D.; Yamanaka, H.; Iwatani, C.; Tsuchiya, H.; et al. Discovery of novel MHC-class I alleles and haplotypes in Filipino cynomolgus macaques (Macaca fascicularis) by pyrosequencing and Sanger sequencing: Mafa-class I polymorphism. Immunogenetics 2015, 67, 563–578. [Google Scholar] [CrossRef]
- Meyer, A.; Carapito, R.; Ott, L.; Radosavljevic, M.; Georgel, P.; Adams, E.J.; Parham, P.; Bontrop, R.E.; Blancher, A.; Bahram, S. High diversity of MIC genes in non-human primates. Immunogenetics 2014, 66, 581–587. [Google Scholar] [CrossRef]
- de Groot, N.G.; Otting, N.; Robinson, J.; Blancher, A.; Lafont, B.A.; Marsh, S.G.; O’Connor, D.H.; Shiina, T.; Walter, L.; Watkins, D.I.; et al. Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics 2012, 64, 615–631. [Google Scholar] [CrossRef][Green Version]
- Bonhomme, M.; Blancher, A.; Jalil, M.F.; Crouau-Roy, B. Factors shaping genetic variation in the MHC of natural non-human primate populations. Tissue Antigens 2007, 70, 398–411. [Google Scholar] [CrossRef]
- Aarnink, A.; Bonhomme, M.; Blancher, A. Positive selection in the major histocompatibility complex class III region of cynomolgus macaques (Macaca fascicularis) of the Philippines origin. Tissue Antigens 2013, 81, 12–18. [Google Scholar] [CrossRef]
- Budde, M.L.; Wiseman, R.W.; Karl, J.A.; Hanczaruk, B.; Simen, B.B.; O’Connor, D.H. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics 2010, 62, 773–780. [Google Scholar] [CrossRef]
- Campbell, K.J.; Detmer, A.M.; Karl, J.A.; Wiseman, R.W.; Blasky, A.J.; Hughes, A.L.; Bimber, B.N.; O’Connor, S.L.; O’Connor, D.H. Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics 2009, 61, 177–187. [Google Scholar] [CrossRef]
- Kita, Y.F.; Hosomichi, K.; Kohara, S.; Itoh, Y.; Ogasawara, K.; Tsuchiya, H.; Torii, R.; Inoko, H.; Blancher, A.; Kulski, J.K.; et al. MHC class I A loci polymorphism and diversity in three Southeast Asian populations of cynomolgus macaque. Immunogenetics 2009, 61, 635–648. [Google Scholar] [CrossRef]
- Lawrence, J.; Orysiuk, D.; Prashar, T.; Pilon, R.; Fournier, J.; Rud, E.; Sandstrom, P.; Plummer, F.A.; Luo, M. Identification of 23 novel MHC class I alleles in cynomolgus macaques of Philippine and Philippine/Mauritius origins. Tissue Antigens 2012, 79, 306–307. [Google Scholar] [CrossRef]
- Ling, F.; Wei, L.Q.; Wang, T.; Wang, H.B.; Zhuo, M.; Du, H.L.; Wang, J.F.; Wang, X.N. Characterization of the major histocompatibility complex class II DOB, DPB1, and DQB1 alleles in cynomolgus macaques of Vietnamese origin. Immunogenetics 2011, 63, 155–166. [Google Scholar] [CrossRef]
- Otting, N.; de Vos-Rouweler, A.J.; Heijmans, C.M.; de Groot, N.G.; Doxiadis, G.G.; Bontrop, R.E. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 2007, 59, 367–375. [Google Scholar] [CrossRef]
- Otting, N.; Doxiadis, G.G.; Bontrop, R.E. Definition of Mafa-A and -B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis). Immunogenetics 2009, 61, 745–753. [Google Scholar] [CrossRef]
- Pendley, C.J.; Becker, E.A.; Karl, J.A.; Blasky, A.J.; Wiseman, R.W.; Hughes, A.L.; O’Connor, S.L.; O’Connor, D.H. MHC class I characterization of Indonesian cynomolgus macaques. Immunogenetics 2008, 60, 339–351. [Google Scholar] [CrossRef]
- Saito, Y.; Naruse, T.K.; Akari, H.; Matano, T.; Kimura, A. Diversity of MHC class I haplotypes in cynomolgus macaques. Immunogenetics 2012, 64, 131–141. [Google Scholar] [CrossRef]
- Uda, A.; Tanabayashi, K.; Fujita, O.; Hotta, A.; Terao, K.; Yamada, A. Identification of the MHC class I B locus in cynomolgus monkeys. Immunogenetics 2005, 57, 189–197. [Google Scholar] [CrossRef]
- Uda, A.; Tanabayashi, K.; Yamada, Y.K.; Akari, H.; Lee, Y.J.; Mukai, R.; Terao, K.; Yamada, A. Detection of 14 alleles derived from the MHC class I A locus in cynomolgus monkeys. Immunogenetics 2004, 56, 155–163. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Ni, C.; Ling, F.; Qiu, W.; Wang, H.B.; Xiao, Y.; Guo, X.J.; Huang, J.Y.; Du, H.L.; Wang, J.F.; et al. Characterization of the major histocompatibility complex class I A alleles in cynomolgus macaques of Vietnamese origin. Tissue Antigens 2012, 80, 494–501. [Google Scholar] [CrossRef]
- Karl, J.A.; Graham, M.E.; Wiseman, R.W.; Heimbruch, K.E.; Gieger, S.M.; Doxiadis, G.G.; Bontrop, R.E.; O’Connor, D.H. Major histocompatibility complex haplotyping and long-amplicon allele discovery in cynomolgus macaques from Chinese breeding facilities. Immunogenetics 2017, 69, 211–229. [Google Scholar] [CrossRef]
- Krebs, K.C.; Jin, Z.; Rudersdorf, R.; Hughes, A.L.; O’Connor, D.H. Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J. Immunol. 2005, 175, 5230–5239. [Google Scholar] [CrossRef]
- Creager, H.M.; Becker, E.A.; Sandman, K.K.; Karl, J.A.; Lank, S.M.; Bimber, B.N.; Wiseman, R.W.; Hughes, A.L.; O’Connor, S.L.; O’Connor, D.H. Characterization of full-length MHC class II sequences in Indonesian and Vietnamese cynomolgus macaques. Immunogenetics 2011, 63, 611–618. [Google Scholar] [CrossRef]
- Wang, W.; Lu, Y.E.; Zhuo, M.; Ling, F. Identification of five novel MHC class II alleles in cynomolgus macaques of Vietnamese origin. HLA 2016, 88, 61–62. [Google Scholar] [CrossRef]
- Doxiadis, G.G.M.; Rouweler, A.J.M.; de Groot, N.G.; Louwerse, A.; Otting, N.; Verschoor, E.J.; Bontrop, R.E. Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics 2006, 58, 259–268. [Google Scholar] [CrossRef]
- Aarnink, A.; Estrade, L.; Apoil, P.A.; Kita, Y.F.; Saitou, N.; Shiina, T.; Blancher, A. Study of cynomolgus monkey (Macaca fascicularis) DRA polymorphism in four populations. Immunogenetics 2010, 62, 123–136. [Google Scholar] [CrossRef]
- Boyson, J.E.; Mcadam, S.N.; Gallimore, A.; Golos, T.G.; Liu, X.M.; Gotch, F.M.; Hughes, A.L.; Watkins, D.I. The-Mhc-E Locus in Macaques Is Polymorphic and Is Conserved between Macaques and Humans. Immunogenetics 1995, 41, 59–68. [Google Scholar] [CrossRef]
- Gaur, L.K.; Nepom, G.T. Ancestral major histocompatibility complex DRB genes beget conserved patterns of localized polymorphisms. Proc. Natl. Acad. Sci. USA 1996, 93, 5380–5383. [Google Scholar] [CrossRef]
- Alvarez, M.; MartinezLaso, J.; Varela, P.; DiazCampos, N.; GomezCasado, E.; VargasAlarcon, G.; GarciaTorre, C.; ArnaizVillena, A. High polymorphism of Mhc-E locus in non-human primates: Alleles with identical exon 2 and 3 are found in two different species. Tissue Antigens 1997, 49, 160–167. [Google Scholar] [CrossRef]
- Otting, N.; de Groot, N.G.; Doxiadis, G.G.; Bontrop, R.E. Extensive Mhc-DQB variation in humans and non-human primate species. Immunogenetics 2002, 54, 230–239. [Google Scholar] [CrossRef]
- Sano, K.; Shiina, T.; Kohara, S.; Yanagiya, K.; Hosomichi, K.; Shimizu, S.; Anzai, T.; Watanabe, A.; Ogasawara, K.; Torii, R.; et al. Novel cynomolgus macaque MHC-DPB1 polymorphisms in three South-East Asian populations. Tissue Antigens 2006, 67, 297–306. [Google Scholar] [CrossRef]
- Wiseman, R.W.; Wojcechowskyj, J.A.; Greene, J.M.; Blasky, A.J.; Gopon, T.; Soma, T.; Friedrich, T.C.; O’Connor, S.L.; O’Connor, D.H. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from mauritius. J. Virol. 2007, 81, 349–361. [Google Scholar] [CrossRef]
- Ling, F.; Zhuo, M.; Ni, C.; Zhang, G.Q.; Wang, T.; Li, W.; Wei, L.Q.; Du, H.L.; Wang, J.F.; Wang, X.N. Comprehensive identification of high-frequency and co-occurring Mafa-B, Mafa-DQB1, and Mafa-DRB alleles in cynomolgus macaques of Vietnamese origin. Hum. Immunol. 2012, 73, 547–553. [Google Scholar] [CrossRef]
- Mitchell, J.L.; Mee, E.T.; Almond, N.M.; Cutler, K.; Rose, N.J. Characterisation of MHC haplotypes in a breeding colony of Indonesian cynomolgus macaques reveals a high level of diversity. Immunogenetics 2012, 64, 123–129. [Google Scholar] [CrossRef]
- de Groot, N.; Doxiadis, G.G.M.; Otting, N.; de Vos-Rouweler, A.J.M.; Bontrop, R.E. Differential recombination dynamics within the MHC of macaque species. Immunogenetics 2014, 66, 535–544. [Google Scholar] [CrossRef]
- Klimanskaya, I.; Chung, Y.; Becker, S.; Lu, S.J.; Lanza, R. Human embryonic stem cell lines derived from single blastomeres. Nature 2006, 444, 481–485. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Wiseman, R.W.; Wojcechowskyj, J.A.; Greene, J.M.; Blasky, A.J.; O’Connor, D.H. MHC identical cynomolgus macaques from Mauritius. J. Med. Primatol. 2007, 36, 296. [Google Scholar]
- Greene, J.M.; Burwitz, B.J.; Blasky, A.J.; Mattila, T.L.; Hong, J.J.; Rakasz, E.G.; Wiseman, R.W.; Hasenkrug, K.J.; Skinner, P.J.; O’Connor, S.L.; et al. Allogeneic lymphocytes persist and traffic in feral MHC-matched mauritian cynomolgus macaques. PLoS ONE 2008, 3, e2384. [Google Scholar] [CrossRef]
- Mee, E.T.; Stebbings, R.; Hall, J.; Giles, E.; Almond, N.; Rose, N.J. Allogeneic lymphocyte transfer in MHC-identical siblings and MHC-identical unrelated Mauritian cynomolgus macaques. Plos ONE 2014, 9, e88670. [Google Scholar] [CrossRef][Green Version]
- Shiba, Y.; Gomibuchi, T.; Seto, T.; Wada, Y.; Ichimura, H.; Tanaka, Y.; Ogasawara, T.; Okada, K.; Shiba, N.; Sakamoto, K.; et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016, 538, 388–391. [Google Scholar] [CrossRef]
- Osada, N.; Hirata, M.; Tanuma, R.; Suzuki, Y.; Sugano, S.; Terao, K.; Kusuda, J.; Kameoka, Y.; Hashimoto, K.; Takahashi, I. Collection of Macaca fascicularis cDNAs derived from bone marrow, kidney, liver, pancreas, spleen, and thymus. BMC Res. Notes 2009, 2, 199. [Google Scholar] [CrossRef]
- Takahata, N. MHC diversity and selection. Immunol. Rev. 1995, 143, 225–247. [Google Scholar] [CrossRef]
- Greene, J.M.; Wiseman, R.W.; Lank, S.M.; Bimber, B.N.; Karl, J.A.; Burwitz, B.J.; Lhost, J.J.; Hawkins, O.E.; Kunstman, K.J.; Broman, K.W.; et al. Differential MHC class I expression in distinct leukocyte subsets. BMC Immunol. 2011, 12. [Google Scholar] [CrossRef]
- Koterski, J.; Twenhafel, N.; Porter, A.; Reed, D.S.; Martino-Catt, S.; Sobral, B.; Crasta, O.; Downey, T.; DaSilva, L. Gene expression profiling of nonhuman primates exposed to aerosolized Venezuelan equine encephalitis virus. FEMS Immunol. Med. Microbiol. 2007, 51, 462–472. [Google Scholar] [CrossRef]
- Mothe, B.R.; Lindestam Arlehamn, C.S.; Dow, C.; Dillon, M.B.C.; Wiseman, R.W.; Bohn, P.; Karl, J.; Golden, N.A.; Gilpin, T.; Foreman, T.W.; et al. The TB-specific CD4(+) T cell immune repertoire in both cynomolgus and rhesus macaques largely overlap with humans. Tuberculosis (Edinb) 2015, 95, 722–735. [Google Scholar] [CrossRef]
- Martin, M.P.; Carrington, M. Immunogenetics of HIV disease. Immunol. Rev. 2013, 254, 245–264. [Google Scholar] [CrossRef]
- Kawashima, Y.; Pfafferott, K.; Frater, J.; Matthews, P.; Payne, R.; Addo, M.; Gatanaga, H.; Fujiwara, M.; Hachiya, A.; Koizumi, H.; et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 2009, 458, 641–645. [Google Scholar] [CrossRef]
- Avila-Rios, S.; Carlson, J.M.; John, M.; Mallal, S.; Brumme, Z.L. Clinical and evolutionary consequences of HIV adaptation to HLA: Implications for vaccine and cure. Curr. Opin. HIV AIDS 2019, 14, 194–204. [Google Scholar] [CrossRef]
- Chikata, T.; Carlson, J.M.; Tamura, Y.; Borghan, M.A.; Naruto, T.; Hashimoto, M.; Murakoshi, H.; Le, A.Q.; Mallal, S.; John, M.; et al. Host-specific adaptation of HIV-1 subtype B in the Japanese population. J. Virol. 2014, 88, 4764–4775. [Google Scholar] [CrossRef]
- Kloverpris, H.N.; Leslie, A.; Goulder, P. Role of HLA Adaptation in HIV Evolution. Front. Immunol. 2016, 6. [Google Scholar] [CrossRef]
- Loffredo, J.T.; Maxwell, J.; Qi, Y.; Glidden, C.E.; Borchardt, G.J.; Soma, T.; Bean, A.T.; Beal, D.R.; Wilson, N.A.; Rehrauer, W.M.; et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 2007, 81, 8827–8832. [Google Scholar] [CrossRef]
- Mothe, B.R.; Weinfurter, J.; Wang, C.; Rehrauer, W.; Wilson, N.; Allen, T.M.; Allison, D.B.; Watkins, D.I. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2003, 77, 2736–2740. [Google Scholar] [CrossRef]
- Muhl, T.; Krawczak, M.; Ten Haaft, P.; Hunsmann, G.; Sauermann, U. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J. Immunol. 2002, 169, 3438–3446. [Google Scholar] [CrossRef]
- Yant, L.J.; Friedrich, T.C.; Johnson, R.C.; May, G.E.; Maness, N.J.; Enz, A.M.; Lifson, J.D.; O’Connor, D.H.; Carrington, M.; Watkins, D.I. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 2006, 80, 5074–5077. [Google Scholar] [CrossRef]
- Marcilla, M.; Alvarez, I.; Ramos-Fernandez, A.; Lombardia, M.; Paradela, A.; Albar, J.P. Comparative Analysis of the Endogenous Peptidomes Displayed by HLA-B*27 and Mamu-B*08: Two MHC Class I Alleles Associated with Elite Control of HIV/SIV Infection. J. Proteome Res. 2016, 15, 1059–1069. [Google Scholar] [CrossRef]
- Loffredo, J.T.; Sidney, J.; Bean, A.T.; Beal, D.R.; Bardet, W.; Wahl, A.; Hawkins, O.E.; Piaskowski, S.; Wilson, N.A.; Hildebrand, W.H.; et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 2009, 182, 7763–7775. [Google Scholar] [CrossRef]
- Valentine, L.E.; Loffredo, J.T.; Bean, A.T.; Leon, E.J.; MacNair, C.E.; Beal, D.R.; Piaskowski, S.M.; Klimentidis, Y.C.; Lank, S.M.; Wiseman, R.W.; et al. Infection with “escaped” virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J. Virol. 2009, 83, 11514–11527. [Google Scholar] [CrossRef]
- Blancher, A.; Aarnink, A.; Savy, N.; Takahata, N. Use of Cumulative Poisson Probability Distribution as an Estimator of the Recombination Rate in an Expanding Population: Example of the Macaca fascicularis Major Histocompatibility Complex. G3 (Bethesda) 2012, 2, 123–130. [Google Scholar] [CrossRef]
- Cain, B.T.; Pham, N.H.; Budde, M.L.; Greene, J.M.; Weinfurter, J.T.; Scarlotta, M.; Harris, M.; Chin, E.; O’Connor, S.L.; Friedrich, T.C.; et al. T cell response specificity and magnitude against SIVmac239 are not concordant in major histocompatibility complex-matched animals. Retrovirology 2013, 10, 116. [Google Scholar] [CrossRef]
- Florese, R.H.; Wiseman, R.W.; Venzon, D.; Karl, J.A.; Demberg, T.; Larsen, K.; Flanary, L.; Kalyanaraman, V.S.; Pal, R.; Titti, F.; et al. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: Influence of Mauritian MHC haplotypes on susceptibility/resistance to SHIV89.6P infection. Vaccine 2008, 26, 3312–3321. [Google Scholar] [CrossRef]
- Burwitz, B.J.; Pendley, C.J.; Greene, J.M.; Detmer, A.M.; Lhost, J.J.; Karl, J.A.; Piaskowski, S.M.; Rudersdorf, R.A.; Wallace, L.T.; Bimber, B.N.; et al. Mauritian Cynomolgus Macaques Share Two Exceptionally Common Major Histocompatibility Complex Class I Alleles That Restrict Simian Immunodeficiency Virus-Specific CD8(+) T Cells. J. Virol. 2009, 83, 6011–6019. [Google Scholar] [CrossRef]
- Mee, E.T.; Berry, N.; Ham, C.; Sauermann, U.; Maggiorella, M.T.; Martinon, F.; Verschoor, E.; Heeney, J.L.; Le Grand, R.; Titti, F.; et al. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics 2009, 61, 327–339. [Google Scholar] [CrossRef]
- Mee, E.T.; Berry, N.; Ham, C.; Aubertin, A.; Lines, J.; Hall, J.; Stebbings, R.; Page, M.; Almond, N.; Rose, N.J. Mhc haplotype M3 is associated with early control of SHIVsbg infection in Mauritian cynomolgus macaques. Tissue Antigens 2010, 76, 223–229. [Google Scholar] [CrossRef]
- Aarnink, A.; Dereuddre-Bosquet, N.; Vaslin, B.; Le Grand, R.; Winterton, P.; Apoil, P.A.; Blancher, A. Influence of the MHC genotype on the progression of experimental SIV infection in the Mauritian cynomolgus macaque. Immunogenetics 2011, 63, 267–274. [Google Scholar] [CrossRef]
- de Manuel, M.; Shiina, T.; Suzuki, S.; Dereuddre-Bosquet, N.; Garchon, H.J.; Tanaka, M.; Congy-Jolivet, N.; Aarnink, A.; Le Grand, R.; Marques-Bonet, T.; et al. Whole genome sequencing in the search for genes associated with the control of SIV infection in the Mauritian macaque model. Sci. Rep. 2018, 8, 7131. [Google Scholar] [CrossRef]
- Shiina, T.; Suzuki, S.; Congy-Jolivet, N.; Aarnink, A.; Garchon, H.J.; Dereuddre-Bosquet, N.; Vaslin, B.; Tchitchek, N.; Desjardins, D.; Autran, B.; et al. Cynomolgus macaque IL37 polymorphism and control of SIV infection. Sci. Rep. 2019, 9, 7981. [Google Scholar] [CrossRef]
- Borsetti, A.; Ferrantelli, F.; Maggiorella, M.T.; Sernicola, L.; Bellino, S.; Gallinaro, A.; Farcomeni, S.; Mee, E.T.; Rose, N.J.; Cafaro, A.; et al. Effect of MHC Haplotype on Immune Response upon Experimental SHIVSF162P4cy Infection of Mauritian Cynomolgus Macaques. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Antony, J.M.; MacDonald, K.S. A critical analysis of the cynomolgus macaque, Macaca fascicularis, as a model to test HIV-1/SIV vaccine efficacy. Vaccine 2015, 33, 3073–3083. [Google Scholar] [CrossRef]
- O’Connor, S.L.; Lhost, J.J.; Becker, E.A.; Detmer, A.M.; Johnson, R.C.; MacNair, C.E.; Wiseman, R.W.; Karl, J.A.; Greene, J.M.; Burwitz, B.J.; et al. MHC Heterozygote Advantage in Simian Immunodeficiency Virus-Infected Mauritian Cynomolgus Macaques. Sci. Transl. Med. 2010, 2. [Google Scholar] [CrossRef]
- Greene, J.M.; Chin, E.N.; Budde, M.L.; Lhost, J.J.; Hines, P.J.; Burwitz, B.J.; Broman, K.W.; Nelson, J.E.; Friedrich, T.C.; O’Connor, D.H. Ex Vivo SIV-Specific CD8 T Cell Responses in Heterozygous Animals Are Primarily Directed against Peptides Presented by a Single MHC Haplotype. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Gellerup, D.D.; Balgeman, A.J.; Nelson, C.W.; Ericsen, A.J.; Scarlotta, M.; Hughes, A.L.; O’Connor, S.L. Conditional Immune Escape during Chronic Simian Immunodeficiency Virus Infection. J. Virol. 2016, 90, 545–552. [Google Scholar] [CrossRef]
- Li, H.Z.; Omange, R.W.; Czarnecki, C.; Correia-Pinto, J.F.; Crecente-Campo, J.; Richmond, M.; Li, L.; Schultz-Darken, N.; Alonso, M.J.; Whitney, J.B.; et al. Mauritian cynomolgus macaques with M3M4 MHC genotype control SIVmac251 infection. J. Med. Primatol. 2017, 46, 137–143. [Google Scholar] [CrossRef]
- Bruel, T.; Hamimi, C.; Dereuddre-Bosquet, N.; Cosma, A.; Shin, S.Y.; Corneau, A.; Versmisse, P.; Karlsson, I.; Malleret, B.; Targat, B.; et al. Long-term control of simian immunodeficiency virus (SIV) in cynomolgus macaques not associated with efficient SIV-specific CD8+ T-cell responses. J. Virol. 2015, 89, 3542–3556. [Google Scholar] [CrossRef]
- Kaeuferle, T.; Krauss, R.; Blaeschke, F.; Willier, S.; Feuchtinger, T. Strategies of adoptive T -cell transfer to treat refractory viral infections post allogeneic stem cell transplantation. J. Hematol. Oncol. 2019, 12, 13. [Google Scholar] [CrossRef]
- Ottaviano, G.; Chiesa, R.; Feuchtinger, T.; Vickers, M.A.; Dickinson, A.; Gennery, A.R.; Veys, P.; Todryk, S. Adoptive T Cell Therapy Strategies for Viral Infections in Patients Receiving Haematopoietic Stem Cell Transplantation. Cells 2019, 8. [Google Scholar] [CrossRef]
- Greene, J.M.; Lhost, J.J.; Hines, P.J.; Scarlotta, M.; Harris, M.; Burwitz, B.J.; Budde, M.L.; Dudley, D.M.; Pham, N.; Cain, B.; et al. Adoptive transfer of lymphocytes isolated from simian immunodeficiency virus SIVmac239Deltanef-vaccinated macaques does not affect acute-phase viral loads but may reduce chronic-phase viral loads in major histocompatibility complex-matched recipients. J. Virol. 2013, 87, 7382–7392. [Google Scholar] [CrossRef]
- Mohns, M.S.; Greene, J.M.; Cain, B.T.; Pham, N.H.; Gostick, E.; Price, D.A.; O’Connor, D.H. Expansion of Simian Immunodeficiency Virus (SIV)-Specific CD8 T Cell Lines from SIV-Naive Mauritian Cynomolgus Macaques for Adoptive Transfer. J. Virol. 2015, 89, 9748–9757. [Google Scholar] [CrossRef]
- Chapuis, A.G.; Desmarais, C.; Emerson, R.; Schmitt, T.M.; Shibuya, K.C.; Lai, I.P.; Wagener, F.; Chou, J.; Roberts, I.M.; Coffey, D.G.; et al. Tracking the fate and origin of clinically relevant adoptively transferred CD8(+) T cells in vivo. Science Immunol. 2017, 2. [Google Scholar] [CrossRef]
- O’Neil, R.T.; Saha, S.; Veach, R.A.; Welch, R.C.; Woodard, L.E.; Rooney, C.M.; Wilson, M.H. Transposon-modified antigen-specific T lymphocytes for sustained therapeutic protein delivery in vivo. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Center for iPS Cell Research and Application, Kyoto University. Available online: https://www.cira.kyoto-u.ac.jp/e/research/stock.html (accessed on 23 August 2019).
- Sugita, S.; Iwasaki, Y.; Makabe, K.; Kamao, H.; Mandai, M.; Shiina, T.; Ogasawara, K.; Hirami, Y.; Kurimoto, Y.; Takahashi, M. Successful Transplantation of Retinal Pigment Epithelial Cells from MHC Homozygote iPSCs in MHC-Matched Models. Stem Cell Rep. 2016, 7, 635–648. [Google Scholar] [CrossRef]
- Sugita, S.; Makabe, K.; Fujii, S.; Iwasaki, Y.; Kamao, H.; Shiina, T.; Ogasawara, K.; Takahashi, M. Detection of Retinal Pigment Epithelium-Specific Antibody in iPSC-Derived Retinal Pigment Epithelium Transplantation Models. Stem Cell Rep. 2017, 9, 1501–1515. [Google Scholar] [CrossRef]
- Morizane, A.; Doi, D.; Kikuchi, T.; Okita, K.; Hotta, A.; Kawasaki, T.; Hayashi, T.; Onoe, H.; Shiina, T.; Yamanaka, S.; et al. Direct comparison of autologous and allogeneic transplantation of iPSC-derived neural cells in the brain of a non-human primate. Stem Cell Rep. 2013, 1, 283–292. [Google Scholar] [CrossRef]
- Morizane, A.; Kikuchi, T.; Hayashi, T.; Mizuma, H.; Takara, S.; Doi, H.; Mawatari, A.; Glasser, M.F.; Shiina, T.; Ishigaki, H.; et al. MHC matching improves engraftment of iPSC-derived neurons in non-human primates. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Kawamura, T.; Miyagawa, S.; Fukushima, S.; Maeda, A.; Kashiyama, N.; Kawamura, A.; Miki, K.; Okita, K.; Yoshida, Y.; Shiina, T.; et al. Cardiomyocytes Derived from MHC-Homozygous Induced Pluripotent Stem Cells Exhibit Reduced Allogeneic Immunogenicity in MHC-Matched Non-human Primates. Stem Cell Rep. 2016, 6, 312–320. [Google Scholar] [CrossRef]
- Sugita, S.; Iwasaki, Y.; Makabe, K.; Kimura, T.; Futagami, T.; Suegami, S.; Takahashi, M. Lack of T Cell Response to iPSC-Derived Retinal Pigment Epithelial Cells from HLA Homozygous Donors. Stem Cell Rep. 2016, 7, 619–634. [Google Scholar] [CrossRef]
- Yamasaki, J.; Iwatani, C.; Tsuchiya, H.; Okahara, J.; Sankai, T.; Torii, R. Vitrification and transfer of cynomolgus monkey (Macaca fascicularis) embryos fertilized by intracytoplasmic sperm injection. Theriogenology 2011, 76, 33–38. [Google Scholar] [CrossRef]
- Aarnink, A.; Garchon, H.J.; Puissant-Lubrano, B.; Blancher-Sardou, M.; Apoil, P.A.; Blancher, A. Impact of MHC class II polymorphism on blood counts of CD4+T lymphocytes in macaque. Immunogenetics 2011, 63, 95–102. [Google Scholar] [CrossRef]
- Aarnink, A.; Garchon, H.J.; Okada, Y.; Takahashi, A.; Matsuda, K.; Kubo, M.; Nakamura, Y.; Blancher, A. Comparative analysis in cynomolgus macaque identifies a novel human MHC locus controlling platelet blood counts independently of BAK1. J. Thromb. Haemost. 2013, 11, 384–386. [Google Scholar] [CrossRef]
- Bagavant, H.; Sharp, C.; Kurth, B.; Tung, K.S. Induction and immunohistology of autoimmune ovarian disease in cynomolgus macaques (Macaca fascicularis). Am. J. Pathol. 2002, 160, 141–149. [Google Scholar] [CrossRef]
- Massacesi, L.; Joshi, N.; Lee-Parritz, D.; Rombos, A.; Letvin, N.L.; Hauser, S.L. Experimental allergic encephalomyelitis in cynomolgus monkeys. Quantitation of T cell responses in peripheral blood. J. Clin. Invest. 1992, 90, 399–404. [Google Scholar] [CrossRef]
- Shimozuru, Y.; Yamane, S.; Fujimoto, K.; Terao, K.; Honjo, S.; Nagai, Y.; Sawitzke, A.D.; Terato, K. Collagen-induced arthritis in nonhuman primates: Multiple epitopes of type II collagen can induce autoimmune-mediated arthritis in outbred cynomolgus monkeys. Arthritis Rheum. 1998, 41, 507–514. [Google Scholar] [CrossRef]
- Bitoun, S.; Roques, P.; Maillere, B.; Le Grand, R.; Mariette, X. Valine 11 and phenylalanine 13 have a greater impact on the T-cell response to citrullinated peptides than the 70-74 shared epitope of the DRB1 molecule in macaques. Ann. Rheum. Dis. 2019. [Google Scholar] [CrossRef]
- Bitoun, S.; Roques, P.; Larcher, T.; Nocturne, G.; Serguera, C.; Chretien, P.; Serre, G.; Grand, R.L.; Mariette, X. Both Systemic and Intra-articular Immunization with Citrullinated Peptides Are Needed to Induce Arthritis in the Macaque. Front. Immunol. 2017, 8, 1816. [Google Scholar] [CrossRef]
- Choi, E.W.; Lee, K.W.; Park, H.; Kim, H.; Lee, J.H.; Song, J.W.; Yang, J.; Kwon, Y.; Kim, T.M.; Park, J.B.; et al. Therapeutic effects of anti-CD154 antibody in cynomolgus monkeys with advanced rheumatoid arthritis. Sci. Rep. 2018, 8, 2135. [Google Scholar] [CrossRef]
- Ho, H.N.; Yang, Y.S.; Hsieh, R.P.; Lin, H.R.; Chen, S.U.; Chen, H.F.; Huang, S.C.; Lee, T.Y.; Gill, T.J., 3rd. Sharing of human leukocyte antigens in couples with unexplained infertility affects the success of in vitro fertilization and tubal embryo transfer. Am. J. Obstet. Gynecol. 1994, 170, 63–71. [Google Scholar] [CrossRef]
- Jin, K.; Ho, H.N.; Speed, T.P.; Gill, T.J., 3rd. Reproductive failure and the major histocompatibility complex. Am. J. Hum. Genet. 1995, 56, 1456–1467. [Google Scholar]
- Kirby, D.R. The egg and immunology. Proc. R. Soc. Med. 1970, 63, 59–61. [Google Scholar]
- Mee, E.T.; Badhan, A.; Karl, J.A.; Wiseman, R.W.; Cutler, K.; Knapp, L.A.; Almond, N.; O’Connor, D.H.; Rose, N.J. MHC haplotype frequencies in a UK breeding colony of Mauritian cynomolgus macaques mirror those found in a distinct population from the same geographic origin. J. Med. Primatol. 2009, 38, 1–14. [Google Scholar] [CrossRef]
- Aarnink, A.; Mee, E.T.; Savy, N.; Congy-Jolivet, N.; Rose, N.J.; Blancher, A. Deleterious impact of feto-maternal MHC compatibility on the success of pregnancy in a macaque model. Immunogenetics 2014, 66, 105–113. [Google Scholar] [CrossRef]
- Ober, C. Current Topic—Hla and Reproduction—Lessons from Studies in the Hutterites. Placenta 1995, 16, 569–577. [Google Scholar] [CrossRef]
- Parimi, N.; Tromp, G.; Kuivaniemi, H.; Nien, J.K.; Gomez, R.; Romero, R.; Goddard, K.A. Analytical approaches to detect maternal/fetal genotype incompatibilities that increase risk of pre-eclampsia. BMC Med. Genet. 2008, 9, 60. [Google Scholar] [CrossRef]
- Ziegler, A.; Santos, P.S.; Kellermann, T.; Uchanska-Ziegler, B. Self/nonself perception, reproduction and the extended MHC. Self Nonself 2010, 1, 176–191. [Google Scholar] [CrossRef]
- Wu, H.; Whritenour, J.; Sanford, J.C.; Houle, C.; Adkins, K.K. Identification of MHC Haplotypes Associated with Drug-induced Hypersensitivity Reactions in Cynomolgus Monkeys. Toxicol. Pathol. 2017, 45, 127–133. [Google Scholar] [CrossRef]
- Pompeu, Y.A.; Stewart, J.D.; Mallal, S.; Phillips, E.; Peters, B.; Ostrov, D.A. The structural basis of HLA-associated drug hypersensitivity syndromes. Immunol. Rev. 2012, 250, 158–166. [Google Scholar] [CrossRef]
- Campbell, K.S.; Purdy, A.K. Structure/function of human killer cell immunoglobulin-like receptors: Lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 2011, 132, 315–325. [Google Scholar] [CrossRef]
- Hudson, L.E.; Allen, R.L. Leukocyte Ig-Like Receptors—A Model for MHC Class I Disease Associations. Front. Immunol. 2016, 7, 281. [Google Scholar] [CrossRef]
- Bashirova, A.A.; Martin-Gayo, E.; Jones, D.C.; Qi, Y.; Apps, R.; Gao, X.; Burke, P.S.; Taylor, C.J.; Rogich, J.; Wolinsky, S.; et al. LILRB2 interaction with HLA class I correlates with control of HIV-1 infection. PLoS Genet. 2014, 10, e1004196. [Google Scholar] [CrossRef]
- Alaoui, L.; Palomino, G.; Zurawski, S.; Zurawski, G.; Coindre, S.; Dereuddre-Bosquet, N.; Lecuroux, C.; Goujard, C.; Vaslin, B.; Bourgeois, C.; et al. Early SIV and HIV infection promotes the LILRB2/MHC-I inhibitory axis in cDCs. Cell Mol. Life Sci. 2018, 75, 1871–1887. [Google Scholar] [CrossRef]
- Chazara, O.; Xiong, S.; Moffett, A. Maternal KIR and fetal HLA-C: A fine balance. J. Leukoc. Biol. 2011, 90, 703–716. [Google Scholar] [CrossRef]
- Colucci, F. The role of KIR and HLA interactions in pregnancy complications. Immunogenetics 2017, 69, 557–565. [Google Scholar] [CrossRef]
- Parham, P.; Moffett, A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol. 2013, 13, 133–144. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Holzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.E.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef]
- Lin, A.; Yan, W.H. The Emerging Roles of Human Leukocyte Antigen-F in Immune Modulation and Viral Infection. Front. Immunol. 2019, 10, 964. [Google Scholar] [CrossRef]
- Morandi, F.; Rizzo, R.; Fainardi, E.; Rouas-Freiss, N.; Pistoia, V. Recent Advances in Our Understanding of HLA-G Biology: Lessons from a Wide Spectrum of Human Diseases. J. Immunol. Res. 2016, 2016, 4326495. [Google Scholar] [CrossRef]
- Sharpe, H.R.; Bowyer, G.; Brackenridge, S.; Lambe, T. HLA-E: Exploiting pathogen-host interactions for vaccine development. Clin. Exp. Immunol. 2019, 196, 167–177. [Google Scholar] [CrossRef]
- Bimber, B.N.; Moreland, A.J.; Wiseman, R.W.; Hughes, A.L.; O’Connor, D.H. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: Novel insights into nonhuman primate KIR gene content and organization. J. Immunol. 2008, 181, 6301–6308. [Google Scholar] [CrossRef]
- Walter, L.; Ansari, A.A. MHC and KIR Polymorphisms in Rhesus Macaque SIV Infection. Front. Immunol. 2015, 6, 540. [Google Scholar] [CrossRef]
- Grimsley, C.; Kawasaki, A.; Gassner, C.; Sageshima, N.; Nose, Y.; Hatake, K.; Geraghty, D.E.; Ishitani, A. Definitive high resolution typing of HLA-E allelic polymorphisms: Identifying potential errors in existing allele data. Tissue Antigens 2002, 60, 206–212. [Google Scholar] [CrossRef]
- Braud, V.; Jones, E.Y.; McMichael, A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 1997, 27, 1164–1169. [Google Scholar] [CrossRef]
- Lee, N.; Goodlett, D.R.; Ishitani, A.; Marquardt, H.; Geraghty, D.E. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 1998, 160, 4951–4960. [Google Scholar]
- Borrego, F.; Ulbrecht, M.; Weiss, E.H.; Coligan, J.E.; Brooks, A.G. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J. Exp. Med. 1998, 187, 813–818. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.J.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Lee, N.; Llano, M.; Carretero, M.; Ishitani, A.; Navarro, F.; Lopez-Botet, M.; Geraghty, D.E. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 1998, 95, 5199–5204. [Google Scholar] [CrossRef]
- Caccamo, N.; Pietra, G.; Sullivan, L.C.; Brooks, A.G.; Prezzemolo, T.; La Manna, M.P.; Di Liberto, D.; Joosten, S.A.; van Meijgaarden, K.E.; Di Carlo, P.; et al. Human CD8 T lymphocytes recognize Mycobacterium tuberculosis antigens presented by HLA-E during active tuberculosis and express type 2 cytokines. Eur. J. Immunol. 2015, 45, 1069–1081. [Google Scholar] [CrossRef]
- Heinzel, A.S.; Grotzke, J.E.; Lines, R.A.; Lewinsohn, D.A.; McNabb, A.L.; Streblow, D.N.; Brand, V.M.; Grieser, H.J.; Belisle, J.T.; Lewinsohn, D.M. HLA-E-dependent presentation of Mtb-derived antigen to human CD8(+) T cells. J. Exp. Med. 2002, 196, 1473–1481. [Google Scholar] [CrossRef]
- Joosten, S.A.; van Meijgaarden, K.E.; van Weeren, P.C.; Kazi, F.; Geluk, A.; Savage, N.D.L.; Drijfhout, J.W.; Flower, D.R.; Hanekom, W.A.; Klein, M.R.; et al. Mycobacterium tuberculosis Peptides Presented by HLA-E Molecules Are Targets for Human CD8(+) T-Cells with Cytotoxic as well as Regulatory Activity. PLoS Pathogens 2010, 6. [Google Scholar] [CrossRef]
- van Meijgaarden, K.E.; Haks, M.C.; Caccamo, N.; Dieli, F.; Ottenhoff, T.H.M.; Joosten, S.A. Human CD8(+) T-cells Recognizing Peptides from Mycobacterium tuberculosis (Mtb) Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Mtb Inhibitory Phenotype and Represent a Novel Human T-cell Subset. PLoS Pathogens 2015, 11. [Google Scholar] [CrossRef]
- Salerno-Goncalves, R.; Fernandez-Vina, M.; Lewinsohn, D.M.; Sztein, M.B. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 2004, 173, 5852–5862. [Google Scholar] [CrossRef]
- Jorgensen, P.B.; Livbjerg, A.H.; Hansen, H.J.; Petersen, T.; Hollsberg, P. Epstein-Barr virus Peptide Presented by HLA-E is Predominantly Recognized by CD8(bright) Cells in multiple Sclerosis Patients. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Mazzarino, P.; Pietra, G.; Vacca, P.; Falco, M.; Colau, D.; Coulie, P.; Moretta, L.; Mingari, M.C. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur. J. Immunol. 2005, 35, 3240–3247. [Google Scholar] [CrossRef]
- Pietra, G.; Romagnani, C.; Mazzarino, P.; Falco, M.; Millo, E.; Moretta, A.; Moretta, L.; Mingari, M.C. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8(+) cytolytic T lymphocytes. Proc. Natl. Acad. Sci. USA 2003, 100, 10896–10901. [Google Scholar] [CrossRef]
- Nattermann, J.; Nischalke, H.D.; Hofmeister, V.; Ahlenstiel, G.; Zimmermann, H.; Leifeld, L.; Weiss, E.H.; Sauerbruch, T.; Spengler, U. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am. J. Pathol. 2005, 166, 443–453. [Google Scholar] [CrossRef]
- Schulte, D.; Vogel, M.; Langhans, B.; Kramer, B.; Korner, C.; Nischalke, H.D.; Steinberg, V.; Michalk, M.; Berg, T.; Rockstroh, J.K.; et al. The HLA-E-R/HLA-E-R Genotype Affects the Natural Course of Hepatitis C Virus (HCV) Infection and Is Associated with HLA-E-Restricted Recognition of an HCV-Derived Peptide by Interferon-gamma-Secreting Human CD8(+) T Cells. J. Infect. Dis. 2009, 200, 1397–1401. [Google Scholar] [CrossRef][Green Version]
- Hansen, S.G.; Wu, H.L.; Burwitz, B.J.; Hughes, C.M.; Hammond, K.B.; Ventura, A.B.; Reed, J.S.; Gilbride, R.M.; Ainslie, E.; Morrow, D.W.; et al. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science 2016, 351, 714–720. [Google Scholar] [CrossRef]
- Hansen, S.G.; Piatak, M., Jr.; Ventura, A.B.; Hughes, C.M.; Gilbride, R.M.; Ford, J.C.; Oswald, K.; Shoemaker, R.; Li, Y.; Lewis, M.S.; et al. Immune clearance of highly pathogenic SIV infection. Nature 2013, 502, 100–104. [Google Scholar] [CrossRef]
- Wu, H.L.; Wiseman, R.W.; Hughes, C.M.; Webb, G.M.; Abdulhaqq, S.A.; Bimber, B.N.; Hammond, K.B.; Reed, J.S.; Gao, L.; Burwitz, B.J.; et al. The Role of MHC-E in T Cell Immunity Is Conserved among Humans, Rhesus Macaques, and Cynomolgus Macaques. J. Immunol. 2018, 200, 49–60. [Google Scholar] [CrossRef]
- Andre, P.; Denis, C.; Soulas, C.; Bourbon-Caillet, C.; Lopez, J.; Arnoux, T.; Blery, M.; Bonnafous, C.; Gauthier, L.; Morel, A.; et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell 2018, 175, 1731–1743.e13. [Google Scholar] [CrossRef]
- Walter, L.; Petersen, B. Diversification of both KIR and NKG2 natural killer cell receptor genes in macaques—Implications for highly complex MHC-dependent regulation of natural killer cells. Immunology 2017, 150, 139–145. [Google Scholar] [CrossRef]
- ’t Hart, B.A.; Gran, B.; Weissert, R. EAE: Imperfect but useful models of multiple sclerosis. Trends Mol. Med. 2011, 17, 119–125. [Google Scholar] [CrossRef]
- Jagessar, S.A.; Holtman, I.R.; Hofman, S.; Morandi, E.; Heijmans, N.; Laman, J.D.; Gran, B.; Faber, B.W.; van Kasteren, S.I.; Eggen, B.J.L.; et al. Lymphocryptovirus Infection of Nonhuman Primate B Cells Converts Destructive into Productive Processing of the Pathogenic CD8 T Cell Epitope in Myelin Oligodendrocyte Glycoprotein. J. Immunol. 2016, 197, 1074–1088. [Google Scholar] [CrossRef]
- Haanstra, K.G.; Wubben, J.A.M.; Jonker, M.; ’t Hart, B.A. Induction of Encephalitis in Rhesus Monkeys Infused with Lymphocryptovirus-Infected B-Cells Presenting MOG(34-56) Peptide. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Jagessar, S.A.; Heijmans, N.; Bauer, J.; Blezer, E.L.A.; Laman, J.D.; Hellings, N.; ’t Hart, B.A. B-Cell Depletion Abrogates T Cell-Mediated Demyelination in an Antibody-Nondependent Common Marmoset Experimental Autoimmune Encephalomyelitis Model. J. Neuropath. Exp. Neur. 2012, 71, 716–728. [Google Scholar] [CrossRef]
- Shen, S.; Pyo, C.W.; Vu, Q.; Wang, R.; Geraghty, D.E. The essential detail: The genetics and genomics of the primate immune response. ILAR J. 2013, 54, 181–195. [Google Scholar] [CrossRef][Green Version]
- Liu, Z.; Cai, Y.; Wang, Y.; Nie, Y.; Zhang, C.; Xu, Y.; Zhang, X.; Lu, Y.; Wang, Z.; Poo, M.; et al. Cloning of Macaque Monkeys by Somatic Cell Nuclear Transfer. Cell 2018, 172, 881–887.e7. [Google Scholar] [CrossRef]



| Region | Allele Group | HLA Locus | Allele Num. | Mafa Locus | Allele Num. |
|---|---|---|---|---|---|
| Class I | MHC-F | HLA-F | 44 | Mafa-F | 8 |
| MHC-G | HLA-G | 68 | Mafa-G | 10 | |
| MHC-AG | X | X | Mafa-AG | 36 | |
| MHC-A | HLA-A | 5,018 | Mafa-A1 | 320 | |
| Mafa-A2 | 83 | ||||
| Mafa-A3 | 30 | ||||
| Mafa-A4 | 38 | ||||
| Mafa-A5 | 8 | ||||
| Mafa-A6 | 14 | ||||
| Mafa-A8 | 1 | ||||
| MHC-E | HLA-E | 30 | Mafa-E | 16 | |
| MHC-C | HLA-C | 4,852 | X | X | |
| MHC-B | HLA-B | 6,096 | Mafa-B | 717 | |
| Mafa-B11L | 5 | ||||
| Mafa-B16 | 11 | ||||
| Mafa-B17 | 4 | ||||
| Mafa-B20 | 2 | ||||
| Mafa-B21 | 2 | ||||
| MHC-I | X | X | Mafa-I | 76 | |
| Class II | MHC-DR | HLA-DRA | 7 | Mafa-DRA | 52 |
| HLA-DRB1 | 2,403 | Mafa-DRB*W | 160 | ||
| HLA-DRB3 | 217 | Mafa-DRB1 | 82 | ||
| HLA-DRB4 | 108 | Mafa-DRB3 | 13 | ||
| HLA-DRB5 | 77 | Mafa-DRB4 | 10 | ||
| Mafa-DRB5 | 18 | ||||
| Mafa-DRB6 | 35 | ||||
| MHC-DQ | HLA-DQA1 | 149 | Mafa-DQA1 | 100 | |
| HLA-DQB1 | 1,560 | Mafa-DQB1 | 107 | ||
| MHC-DO | HLA-DOA | 12 | Mafa-DOA | 15 | |
| HLA-DOB | 13 | Mafa-DOB | 16 | ||
| MHC-DM | HLA-DMA | 7 | Mafa-DMA | 11 | |
| HLA-DMB | 13 | Mafa-DMB | 7 | ||
| MHC-DP | HLA-DPA1 | 106 | Mafa-DPA1 | 91 | |
| HLA-DPB1 | 1,360 | Mafa-DPB1 | 98 | ||
| Total | 22,140 | 2196 |
| Reference | Year | Population of Macaques | Class I E | Class I F | Class I A | Class I B | Class I I | DRA | DRB Exon 2 | DRB cDNA | Class II Others | Microsat. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Boyson et al. [70] | 1995 | Not specified | X | |||||||||
| Gaur and Nepom [71] | 1996 | Not specified | X | |||||||||
| Alvarez et al. [72] | 1997 | Not specified | X | |||||||||
| Otting et al. [73] | 2002 | Not specified | DQB | |||||||||
| Leuchte et al. [33] | 2004 | Mauritius | X | |||||||||
| Uda et al. [62] | 2004 | Not specified | X | |||||||||
| Krebs et al. [65] | 2005 | Vietnam, Mauritius, Chinese breeding facilities | X (RSCA) (1) | X (RSCA) | ||||||||
| Uda et al. [61] | 2005 | Not specified | X | X | ||||||||
| Blancher et al. [21] | 2006 | Mauritius, The Philippines | X | |||||||||
| Sano K et al. [74] | 2006 | Indonesia, Vietnam, The Philippines | DPB1 | |||||||||
| Doxiadis et al. [68] | 2006 | Indonesia Malaysia Thailand Java Sumatra Vietnam, Chinese breeding facilities | X | DPB1, DQA1, DQB1 | ||||||||
| O’Connor et al. [43] | 2007 | Mauritius | X | X | X | DPA, DPB, DQA, DQB, | X | |||||
| Wiseman et al. [75] | 2007 | Mauritius | X (RSCA) | X (RSCA) | X | X | ||||||
| Pendley et al. [59] | 2008 | Indonesia | X | X | X | |||||||
| Wiseman et al. [37] | 2009 | Mauritius | X | X | ||||||||
| Campbell et al. [53] | 2009 | The Philippines | X | X | X | |||||||
| Kita et al. [54] | 2009 | Indonesia, Vietnam, The Philippines | X | |||||||||
| Aarnink et al. [69] | 2010 | Indonesia, Mauritius, The Philippines, Vietnam | X | X | ||||||||
| Craeger et al. [66] | 2011 | Indonesia and Vietnam | X | X | DPA, DPB, DQA, DQB, cDNA | |||||||
| Ling et al. [56] | 2011 | Vietnam | DOB, DPB1, DQB1 | |||||||||
| Blancher et al. [45] | 2012 | The Philippines Java, Vietnam, Mauritius | X | X | X | |||||||
| Ling et al. [76] | 2012 | Vietnam | X | X | DQB1 | |||||||
| Mitchell et al. [77] | 2012 | Indonesia | X (RSCA) | X (RSCA) | X | |||||||
| de Groot et al. [78] | 2014 | Indonesia, Indochina | X | X | X | DQA, DQB | X | |||||
| Shiina et al. [47] | 2015 | The Philippines | X | X | X | X | X | |||||
| Karl et al. [64] | 2017 | Chinese breeding facilities | X | X | X |
| Locus | Total Allele Num. | Vietnamese | Indonesian | Filipino |
|---|---|---|---|---|
| Mafa-A1 | 76 | 34 | 32 | 13 |
| Mafa-DRA | 22 | 17 | _ | 12 |
| Mafa-DRB | 134 | 84 | 60 | 35 |
| Mafa-DPB1 | 40 | 26 | 23 | 12 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiina, T.; Blancher, A. The Cynomolgus Macaque MHC Polymorphism in Experimental Medicine. Cells 2019, 8, 978. https://doi.org/10.3390/cells8090978
Shiina T, Blancher A. The Cynomolgus Macaque MHC Polymorphism in Experimental Medicine. Cells. 2019; 8(9):978. https://doi.org/10.3390/cells8090978
Chicago/Turabian StyleShiina, Takashi, and Antoine Blancher. 2019. "The Cynomolgus Macaque MHC Polymorphism in Experimental Medicine" Cells 8, no. 9: 978. https://doi.org/10.3390/cells8090978
APA StyleShiina, T., & Blancher, A. (2019). The Cynomolgus Macaque MHC Polymorphism in Experimental Medicine. Cells, 8(9), 978. https://doi.org/10.3390/cells8090978






