The NAE Pathway: Autobahn to the Nucleus for Cell Surface Receptors
Abstract
1. Introduction: Numerous Cell Surface Receptors Traffic to The Nucleus
2. Proposed Nuclear Functions of Cell Surface Receptors and Functional Significance
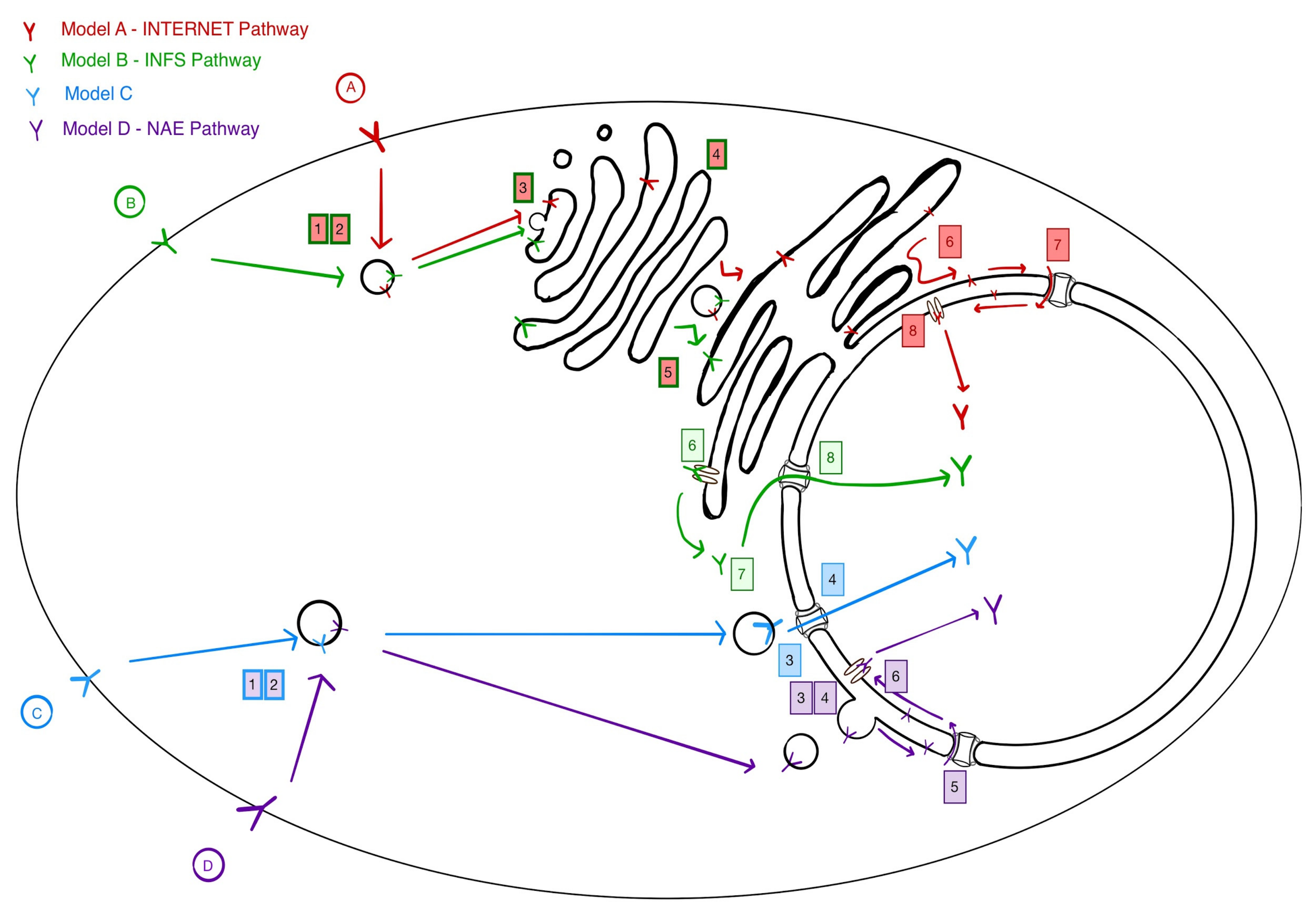
3. Four Models for Receptor Trafficking to the Nucleus
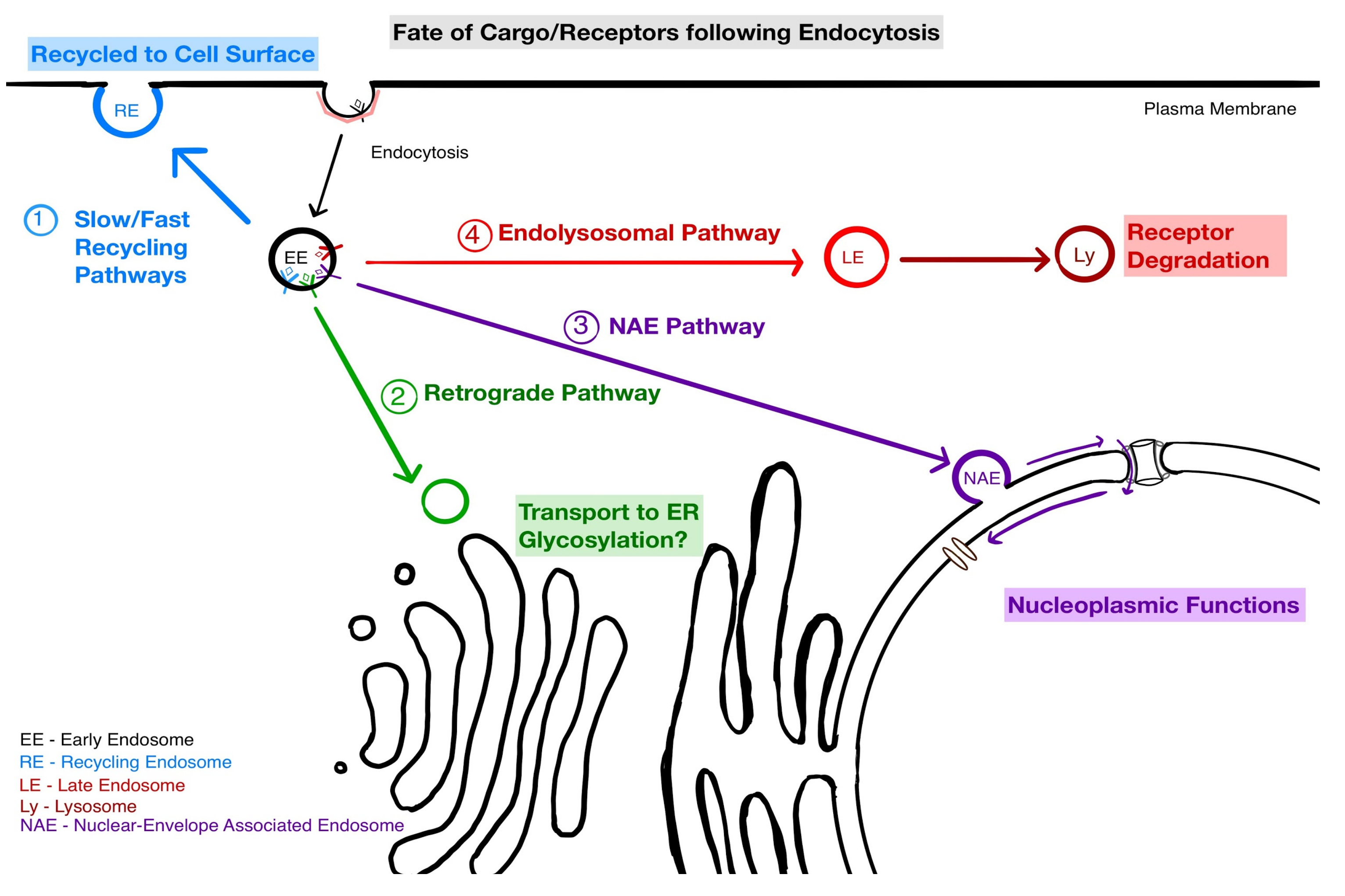
4. Implications of NAE for Endosomal Sorting
5. Mechanisms at Play in the NAE Pathway
5.1. The Docking of NAE
5.2. Fusion of NAE With the Nuclear Envelope: A Hug-and-Kiss Process?
5.3. Translocation of Receptors to the INM: A Role for the NPC
6. Extraction of Receptors from the INM
7. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Podlecki, D.A.; Smith, R.M.; Kao, M.; Tsai, P.; Huecksteadt, T.; Brandenburg, D.; Lasher, R.S.; Jarett, L.; Olefsky, J.M. Nuclear translocation of the insulin receptor. A possible mediator of insulin’s long term effects. J. Biol. Chem. 1987, 262, 3362–3368. [Google Scholar] [PubMed]
- Kamio, T.; Shigematsu, K.; Sou, H.; Kawai, K.; Tsuchiyama, H. Immunohistochemical expression of epidermal growth factor receptors in human adrenocortical carcinoma. Hum. Pathol. 1990, 21, 277–282. [Google Scholar] [CrossRef]
- Marti, U.; Burwen, S.J.; Wells, A.; Barker, M.E.; Huling, S.; Feren, A.M.; Jones, A.L. Localization of epidermal growth factor receptor in hepatocyte nuclei. Hepatology 1991, 13, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005, 1, 2005.0010. [Google Scholar] [CrossRef] [PubMed]
- Maher, P.A. Nuclear Translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J. Cell Biol. 1996, 134, 529–536. [Google Scholar] [CrossRef]
- Shay-Salit, A.; Shushy, M.; Wolfovitz, E.; Yahav, H.; Breviario, F.; Dejana, E.; Resnick, N. VEGF receptor 2 and the adherens junction as a mechanical transducer in vascular endothelial cells. Proc. Natl. Acad. Sci. USA 2002, 99, 9462–9467. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Lennartsson, J.; Heldin, C.H. PDGFRβ translocates to the nucleus and regulates chromatin remodeling via TATA element–modifying factor 1. J. Cell Biol. 2018, 217, 1701–1717. [Google Scholar] [CrossRef]
- De Angelis Campos, A.C.; Rodrigues, M.A.; de Andrade, C.; de Goes, A.M.; Nathanson, M.H.; Gomes, D.A. Epidermal growth factor receptors destined for the nucleus are internalized via a clathrin-dependent pathway. Biochem. Biophys. Res. Commun. 2011, 412, 341–346. [Google Scholar] [CrossRef]
- Boivin, B.; Vaniotis, G.; Allen, B.G.; Hébert, T.E. G protein-coupled receptors in and on the cell nucleus: A new signaling paradigm? J. Recept. Signal Transduct. Res. 2008, 28, 15–28. [Google Scholar] [CrossRef]
- Cohentannoudji, J. G Protein-Coupled Receptors: New insights into signaling and regulation. Biol. Cell 2004, 96, 325–326. [Google Scholar] [CrossRef]
- Janiszewska, M.; De Vito, C.; Le Bitoux, M.-A.; Fusco, C.; Stamenkovic, I. Transportin regulates nuclear import of CD44. J. Biol. Chem. 2010, 285, 30548–30557. [Google Scholar] [CrossRef]
- Lee, J.-L.; Wang, M.-J.; Chen, J.-Y. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J. Cell Biol. 2009, 185, 949–957. [Google Scholar] [CrossRef]
- Chaumet, A.; Wright, G.D.; Seet, S.H.; Tham, K.M.; Gounko, N.V.; Bard, F. Nuclear envelope-associated endosomes deliver surface proteins to the nucleus. Nat. Commun. 2015, 6, 8218. [Google Scholar] [CrossRef]
- Wells, A.; Marti, U. Signalling shortcuts: Cell-surface receptors in the nucleus? Nat. Rev. Mol. Cell Biol. 2002, 3, 697–702. [Google Scholar] [CrossRef]
- Bryant, D.M.; Stow, J.L. Nuclear translocation of cell-surface receptors: Lessons from fibroblast growth factor. Traffic 2005, 6, 947–954. [Google Scholar] [CrossRef]
- Hsu, J.L.; Hung, M.-C. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016, 35, 575–588. [Google Scholar] [CrossRef]
- Raper, S.E.; Burwen, S.J.; Barker, M.E.; Jones, A.L. Translocation of epidermal growth factor to the hepatocyte nucleus during rat liver regeneration. Gastroenterology 1987, 92, 1243–1250. [Google Scholar] [CrossRef]
- Liccardi, G.; Hartley, J.A.; Hochhauser, D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 2011, 71, 1103–1114. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Li, C.; Wheeler, D.L. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov. Med. 2011, 12, 419–432. [Google Scholar]
- Rakowicz-Szulczynska, E.M.; Rodeck, U.; Herlyn, M.; Koprowski, H. Chromatin binding of epidermal growth factor, nerve growth factor, and platelet-derived growth factor in cells bearing the appropriate surface receptors. Proc. Natl. Acad. Sci. USA 1986, 83, 3728–3732. [Google Scholar] [CrossRef]
- Lin, S.Y.; Makino, K.; Xia, W.; Matin, A.; Wen, Y.; Kwong, K.Y.; Bourguignon, L.; Hung, M.C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001, 3, 802–808. [Google Scholar] [CrossRef]
- Hung, L.-Y.; Tseng, J.T.; Lee, Y.-C.; Xia, W.; Wang, Y.-N.; Wu, M.-L.; Chuang, Y.-H.; Lai, C.-H.; Chang, W.-C. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008, 36, 4337–4351. [Google Scholar] [CrossRef]
- Lo, H.-W.; Hsu, S.-C.; Ali-Seyed, M.; Gunduz, M.; Xia, W.; Wei, Y.; Bartholomeusz, G.; Shih, J.-Y.; Hung, M.-C. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 2005, 7, 575–589. [Google Scholar] [CrossRef]
- Hanada, N.; Lo, H.-W.; Day, C.-P.; Pan, Y.; Nakajima, Y.; Hung, M.-C. Co-regulation of B-Myb expression by E2F1 and EGF receptor. In Molecular Carcinogenesis; University of Texas MD Anderson Cancer Center: Houston, TX, USA, 2006; pp. 10–17. [Google Scholar]
- Hancock, M.L.; Meyer, R.C.; Mistry, M.; Khetani, R.S.; Wagschal, A.; Shin, T.; Ho Sui, S.J.; Näär, A.M.; Flanagan, J.G. Insulin Receptor Associates with Promoters Genome-wide and Regulates Gene Expression. Cell 2019, 177, 722–736.e22. [Google Scholar] [CrossRef]
- Li, L.-Y.; Chen, H.; Hsieh, Y.-H.; Wang, Y.-N.; Chu, H.-J.; Chen, Y.-H.; Chen, H.-Y.; Chien, P.-J.; Ma, H.-T.; Tsai, H.-C.; et al. Nuclear ErbB2 enhances translation and cell growth by activating transcription of ribosomal RNA genes. Cancer Res. 2011, 71, 4269–4279. [Google Scholar] [CrossRef]
- Domingues, I.; Rino, J.; Demmers, J.A.A.; de Lanerolle, P.; Santos, S.C.R. VEGFR2 translocates to the nucleus to regulate its own transcription. PLoS ONE 2011, 6, e25668. [Google Scholar] [CrossRef]
- Bischoff, J.R.; Anderson, L.; Zhu, Y.; Mossie, K.; Ng, L.; Souza, B.; Schryver, B.; Flanagan, P.; Clairvoyant, F.; Ginther, C.; et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998, 17, 3052–3065. [Google Scholar] [CrossRef]
- Wang, S.-C.; Nakajima, Y.; Yu, Y.-L.; Xia, W.; Chen, C.-T.; Yang, C.-C.; McIntush, E.W.; Li, L.-Y.; Hawke, D.H.; Kobayashi, R.; et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 2006, 8, 1359–1368. [Google Scholar] [CrossRef]
- Yu, Y.-L.; Chou, R.-H.; Liang, J.-H.; Chang, W.-J.; Su, K.-J.; Tseng, Y.-J.; Huang, W.-C.; Wang, S.-C.; Hung, M.-C. Targeting the EGFR/PCNA signaling suppresses tumor growth of triple-negative breast cancer cells with cell-penetrating PCNA peptides. PLoS ONE 2013, 8, e61362. [Google Scholar] [CrossRef]
- Dittmann, K.; Mayer, C.; Fehrenbacher, B.; Schaller, M.; Raju, U.; Milas, L.; Chen, D.J.; Kehlbach, R.; Rodemann, H.P. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J. Biol. Chem. 2005, 280, 31182–31189. [Google Scholar] [CrossRef]
- Chou, R.-H.; Wang, Y.-N.; Hsieh, Y.-H.; Li, L.-Y.; Xia, W.; Chang, W.-C.; Chang, L.-C.; Cheng, C.-C.; Lai, C.-C.; Hsu, J.L.; et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell 2014, 30, 224–237. [Google Scholar] [CrossRef]
- Dittmann, K.; Mayer, C.; Fehrenbacher, B.; Schaller, M.; Kehlbach, R.; Rodemann, H.P. Nuclear EGFR shuttling induced by ionizing radiation is regulated by phosphorylation at residue Thr654. FEBS Lett. 2010, 584, 3878–3884. [Google Scholar] [CrossRef]
- Xia, W.; Wei, Y.; Du, Y.; Liu, J.; Chang, B.; Yu, Y.-L.; Huo, L.-F.; Miller, S.; Hung, M.-C. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol. Carcinog. 2009, 48, 610–617. [Google Scholar] [CrossRef]
- Traynor, A.M.; Weigel, T.L.; Oettel, K.R.; Yang, D.T.; Zhang, C.; Kim, K.; Salgia, R.; Iida, M.; Brand, T.M.; Hoang, T.; et al. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer 2013, 81, 138–141. [Google Scholar] [CrossRef]
- Brand, T.M.; Iida, M.; Luthar, N.; Starr, M.M.; Huppert, E.J.; Wheeler, D.L. Nuclear EGFR as a molecular target in cancer. Radiother. Oncol. 2013, 108, 370–377. [Google Scholar] [CrossRef]
- Li, C.; Iida, M.; Dunn, E.F.; Ghia, A.J.; Wheeler, D.L. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene 2009, 28, 3801–3813. [Google Scholar] [CrossRef]
- Huang, W.-C.; Chen, Y.-J.; Li, L.-Y.; Wei, Y.-L.; Hsu, S.-C.; Tsai, S.-L.; Chiu, P.-C.; Huang, W.-P.; Wang, Y.-N.; Chen, C.-H.; et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J. Biol. Chem. 2011, 286, 20558–20568. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Miller, S.A.; Wang, Y.; Hung, M.-C. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am. J. Transl. Res. 2009, 1, 249–258. [Google Scholar]
- Schillaci, R.; Guzmán, P.; Cayrol, F.; Beguelin, W.; Díaz Flaqué, M.C.; Proietti, C.J.; Pineda, V.; Palazzi, J.; Frahm, I.; Charreau, E.H.; et al. Clinical relevance of ErbB-2/HER2 nuclear expression in breast cancer. BMC Cancer 2012, 12, 74. [Google Scholar] [CrossRef]
- Cordo Russo, R.I.; Béguelin, W.; Díaz Flaqué, M.C.; Proietti, C.J.; Venturutti, L.; Galigniana, N.; Tkach, M.; Guzmán, P.; Roa, J.C.; O’Brien, N.A.; et al. Targeting ErbB-2 nuclear localization and function inhibits breast cancer growth and overcomes trastuzumab resistance. Oncogene 2014, 34, 3413. [Google Scholar] [CrossRef]
- Moreau, D.; Kumar, P.; Wang, S.C.; Chaumet, A.; Chew, S.Y.; Chevalley, H.; Bard, F. Genome-wide RNAi screens identify genes required for Ricin and PE intoxications. Dev. Cell 2011, 21, 231–244. [Google Scholar] [CrossRef]
- Wernick, N.L.B.; Chinnapen, D.J.-F.; Cho, J.A.; Lencer, W.I. Cholera toxin: An intracellular journey into the cytosol by way of the endoplasmic reticulum. Toxins 2010, 2, 310–325. [Google Scholar] [CrossRef]
- Spooner, R.A.; Lord, J.M. Ricin trafficking in cells. Toxins 2015, 7, 49–65. [Google Scholar] [CrossRef]
- Lee, M.-S.; Cherla, R.P.; Tesh, V.L. Shiga toxins: Intracellular trafficking to the ER leading to activation of host cell stress responses. Toxins 2010, 2, 1515–1535. [Google Scholar] [CrossRef]
- Michalska, M.; Wolf, P. Pseudomonas Exotoxin A: Optimized by evolution for effective killing. Front. Microbiol. 2015, 6, 963. [Google Scholar] [CrossRef]
- Stachowiak, M.K.; Maher, P.A.; Stachowiak, E.K. Integrative Nuclear Signaling in Cell Development—A Role for FGF Receptor-1. DNA Cell Biol. 2007, 26, 811–826. [Google Scholar] [CrossRef]
- Giri, D.K.; Ali-Seyed, M.; Li, L.Y.; Lee, D.F.; Ling, P.; Bartholomeusz, G.; Wang, S.C.; Hung, M.C. Endosomal Transport of ErbB-2: Mechanism for Nuclear Entry of the Cell Surface Receptor. Mol. Cell. Biol. 2005, 25, 11005–11018. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Wang, H.; Yamaguchi, H.; Lee, H.-J.; Lee, H.-H.; Hung, M.-C. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem. Biophys. Res. Commun. 2010, 399, 498–504. [Google Scholar] [CrossRef]
- Yamane, J.; Kubo, A.; Nakayama, K.; Yuba-Kubo, A.; Katsuno, T.; Tsukita, S.; Tsukita, S. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp. Cell Res. 2007, 313, 3472–3485. [Google Scholar] [CrossRef]
- Cherfils, J. Arf GTPases and their effectors: Assembling multivalent membrane-binding platforms. Curr. Opin. Struct. Biol. 2014, 29, 67–76. [Google Scholar] [CrossRef]
- Nakai, W.; Kondo, Y.; Saitoh, A.; Naito, T.; Nakayama, K.; Shin, H.-W. ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol. Biol. Cell 2013, 24, 2570–2581. [Google Scholar] [CrossRef]
- Gu, F.; Gruenberg, J. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J. Biol. Chem. 2000, 275, 8154–8160. [Google Scholar] [CrossRef]
- Gabriely, G.; Kama, R.; Gerst, J.E. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol. Cell. Biol. 2007, 27, 526–540. [Google Scholar] [CrossRef]
- Sciaky, N.; Presley, J.; Smith, C.; Zaal, K.J.; Cole, N.; Moreira, J.E.; Terasaki, M.; Siggia, E.; Lippincott-Schwartz, J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 1997, 139, 1137–1155. [Google Scholar] [CrossRef]
- Lee, H.-H.; Wang, Y.-N.; Hung, M.-C. Non-canonical signaling mode of the epidermal growth factor receptor family. Am. J. Cancer Res. 2015, 5, 2944–2958. [Google Scholar]
- Gonias, S.L.; Campana, W.M. LDL receptor-related protein-1: A regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am. J. Pathol. 2014, 184, 18–27. [Google Scholar] [CrossRef]
- Lillis, A.P.; Van Duyn, L.B.; Murphy-Ullrich, J.E.; Strickland, D.K. LDL receptor-related protein 1: Unique tissue-specific functions revealed by selective gene knockout studies. Physiol. Rev. 2008, 88, 887–918. [Google Scholar] [CrossRef]
- Amaya, M.J.; Oliveira, A.G.; Guimarães, E.S.; Casteluber, M.C.F.; Carvalho, S.M.; Andrade, L.M.; Pinto, M.C.X.; Mennone, A.; Oliveira, C.A.; Resende, R.R.; et al. The insulin receptor translocates to the nucleus to regulate cell proliferation in liver. Hepatology 2014, 59, 274–283. [Google Scholar] [CrossRef]
- Hirschberg, K.; Miller, C.M.; Ellenberg, J.; Presley, J.F.; Siggia, E.D.; Phair, R.D.; Lippincott-Schwartz, J. Kinetic analysis of secretory protein traffic and characterization of golgi to plasma membrane transport intermediates in living cells. J. Cell Biol. 1998, 143, 1485–1503. [Google Scholar] [CrossRef]
- Braham, K.; Junqua, S.; Tursz, T.; Le Pecq, J.B.; Lipinski, M. Kinetic analysis of choriocarcinoma cell intoxication induced by ricin and ricin A chain immunotoxin. Cancer Res. 1988, 48, 806–811. [Google Scholar]
- Gruenberg, J.; Howell, K.E. Membrane traffic in endocytosis: Insights from cell-free assays. Annu. Rev. Cell Biol. 1989, 5, 453–481. [Google Scholar] [CrossRef]
- Naslavsky, N.; Caplan, S. The enigmatic endosome–sorting the ins and outs of endocytic trafficking. J. Cell Sci. 2018, 131, jcs216499. [Google Scholar] [CrossRef]
- Progida, C.; Bakke, O. Bidirectional traffic between the Golgi and the endosomes—Machineries and regulation. J. Cell Sci. 2016, 129, 3971–3982. [Google Scholar] [CrossRef]
- Brodsky, F.M.; Chen, C.Y.; Knuehl, C.; Towler, M.C.; Wakeham, D.E. Biological basket weaving: Formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 2001, 17, 517–568. [Google Scholar] [CrossRef]
- Goh, L.K.; Huang, F.; Kim, W.; Gygi, S.; Sorkin, A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J. Cell Biol. 2010, 189, 871–883. [Google Scholar] [CrossRef]
- Sigismund, S.; Algisi, V.; Nappo, G.; Conte, A.; Pascolutti, R.; Cuomo, A.; Bonaldi, T.; Argenzio, E.; Verhoef, L.G.G.C.; Maspero, E.; et al. Threshold-controlled ubiquitination of the EGFR directs receptor fate. EMBO J. 2013, 32, 2140–2157. [Google Scholar] [CrossRef]
- Motley, A.; Bright, N.A.; Seaman, M.N.J.; Robinson, M.S. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 2003, 162, 909–918. [Google Scholar] [CrossRef]
- Huang, F.; Khvorova, A.; Marshall, W.; Sorkin, A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J. Biol. Chem. 2004, 279, 16657–16661. [Google Scholar] [CrossRef]
- Pascolutti, R.; Algisi, V.; Conte, A.; Raimondi, A.; Pasham, M.; Upadhyayula, S.; Gaudin, R.; Maritzen, T.; Barbieri, E.; Caldieri, G.; et al. Molecularly Distinct Clathrin-Coated Pits Differentially Impact EGFR Fate and Signaling. Cell Rep. 2019, 27, 3049–3061. [Google Scholar] [CrossRef]
- Christoforidis, S.; McBride, H.M.; Burgoyne, R.D.; Zerial, M. The Rab5 effector EEA1 is a core component of endosome docking. Nature 1999, 397, 621–625. [Google Scholar] [CrossRef]
- Mills, I.G.; Jones, A.T.; Clague, M.J. Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr. Biol. 1998, 8, 881–884. [Google Scholar] [CrossRef]
- Hainan, L.; Huilin, L.; Khan, M.A.; Xin, Z.; YuJiang, Y.; Hui, Z.; Naiquan, Y. The basic route of the nuclear translocation porcine growth hormone (GH)-growth hormone receptor (GHR) complex (pGH/GHR) in porcine hepatocytes. Gen. Comp. Endocrinol. 2018, 266, 101–109. [Google Scholar] [CrossRef]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef]
- Batzer, A.G.; Rotin, D.; Ureña, J.M.; Skolnik, E.Y.; Schlessinger, J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol. Cell. Biol. 1994, 14, 5192–5201. [Google Scholar] [CrossRef]
- Tomas, A.; Futter, C.E.; Eden, E.R. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014, 24, 26–34. [Google Scholar] [CrossRef]
- Mohapatra, B.; Ahmad, G.; Nadeau, S.; Zutshi, N.; An, W.; Scheffe, S.; Dong, L.; Feng, D.; Goetz, B.; Arya, P.; et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of Cbl-family ubiquitin ligases. Biochim. Biophys. Acta 2013, 1833, 122–139. [Google Scholar] [CrossRef]
- Huber, L.A.; Teis, D. Lysosomal signaling in control of degradation pathways. Curr. Opin. Cell Biol. 2016, 39, 8–14. [Google Scholar] [CrossRef]
- Clague, M.J.; Liu, H.; Urbé, S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell 2012, 23, 457–467. [Google Scholar] [CrossRef]
- Komada, M. Controlling receptor downregulation by ubiquitination and deubiquitination. Curr. Drug Discov. Technol. 2008, 5, 78–84. [Google Scholar] [CrossRef]
- Pareja, F.; Ferraro, D.A.; Rubin, C.; Cohen-Dvashi, H.; Zhang, F.; Aulmann, S.; Ben-Chetrit, N.; Pines, G.; Navon, R.; Crosetto, N.; et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene 2012, 31, 4599–4608. [Google Scholar] [CrossRef]
- Suarez-Quian, C.A.; Dai, M.Z.; Onoda, M.; Kriss, R.M.; Dym, M. Epidermal growth factor receptor localization in the rat and monkey testes. Biol. Reprod. 1989, 41, 921–932. [Google Scholar] [CrossRef]
- Faria, J.A.Q.A.; de Andrade, C.; Goes, A.M.; Rodrigues, M.A.; Gomes, D.A. Effects of different ligands on epidermal growth factor receptor (EGFR) nuclear translocation. Biochem. Biophys. Res. Commun. 2016, 478, 39–45. [Google Scholar] [CrossRef]
- Xu, Y.; Shao, Y.; Zhou, J.; Voorhees, J.J.; Fisher, G.J. Ultraviolet irradiation-induces epidermal growth factor receptor (EGFR) nuclear translocation in human keratinocytes. J. Cell. Biochem. 2009, 107, 873–880. [Google Scholar] [CrossRef]
- Granger, E.; McNee, G.; Allan, V.; Woodman, P. The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin. Cell Dev. Biol. 2014, 31, 20–29. [Google Scholar] [CrossRef]
- Gundersen, G.G.; Worman, H.J. Nuclear positioning. Cell 2013, 152, 1376–1389. [Google Scholar] [CrossRef]
- Starr, D.A. KASH and SUN proteins. Curr. Biol. 2011, 21, R414–R415. [Google Scholar] [CrossRef]
- Sosa, B.A.; Rothballer, A.; Kutay, U.; Schwartz, T.U. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 2012, 149, 1035–1047. [Google Scholar] [CrossRef]
- Rajgor, D.; Shanahan, C.M. Nesprins: From the nuclear envelope and beyond. Expert Rev. Mol. Med. 2013, 15, e5. [Google Scholar] [CrossRef]
- Padmakumar, V.C.; Libotte, T.; Lu, W.; Zaim, H.; Abraham, S.; Noegel, A.A.; Gotzmann, J.; Foisner, R.; Karakesisoglou, I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 2005, 118, 3419–3430. [Google Scholar] [CrossRef]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef]
- Gautreau, A.; Oguievetskaia, K.; Ungermann, C. Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb. Perspect. Biol. 2014, 6, a016832. [Google Scholar] [CrossRef]
- Brandhorst, D.; Zwilling, D.; Rizzoli, S.O.; Lippert, U.; Lang, T.; Jahn, R. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. USA 2006, 103, 2701–2706. [Google Scholar] [CrossRef]
- Ohya, T.; Miaczynska, M.; Coskun, U.; Lommer, B.; Runge, A.; Drechsel, D.; Kalaidzidis, Y.; Zerial, M. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature 2009, 459, 1091–1097. [Google Scholar] [CrossRef]
- Spang, A. Membrane Tethering Complexes in the Endosomal System. Front. Cell Dev. Biol. 2016, 4, 35. [Google Scholar] [CrossRef]
- Fesce, R.; Grohovaz, F.; Valtorta, F.; Meldolesi, J. Neurotransmitter release: Fusion or “kiss-and-run”? Trends Cell Biol. 1994, 4, 1–4. [Google Scholar] [CrossRef]
- Storrie, B.; Desjardins, M. The biogenesis of lysosomes: Is it a kiss and run, continuous fusion and fission process? Bioessays 1996, 18, 895–903. [Google Scholar] [CrossRef]
- Klein, O.; Sagi-Eisenberg, R. Anaphylactic Degranulation of Mast Cells: Focus on Compound Exocytosis. J. Immunol. Res. 2019, 2019, 9542656. [Google Scholar] [CrossRef]
- Mironov, A.A.; Sesorova, I.V.; Beznoussenko, G.V. Golgi’s way: A long path toward the new paradigm of the intra-Golgi transport. Histochem. Cell Biol. 2013, 140, 383–393. [Google Scholar] [CrossRef]
- Kurokawa, K.; Okamoto, M.; Nakano, A. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 2014, 5, 3653. [Google Scholar] [CrossRef]
- Spang, A. Retrograde traffic from the Golgi to the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2013, 5, a013391. [Google Scholar] [CrossRef]
- Spang, A. The DSL1 complex: The smallest but not the least CATCHR. Traffic 2012, 13, 908–913. [Google Scholar] [CrossRef]
- Meiringer, C.T.A.; Rethmeier, R.; Auffarth, K.; Wilson, J.; Perz, A.; Barlowe, C.; Schmitt, H.D.; Ungermann, C. The Dsl1 protein tethering complex is a resident endoplasmic reticulum complex, which interacts with five soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors (SNAREs): Implications for fusion and fusion regulation. J. Biol. Chem. 2011, 286, 25039–25046. [Google Scholar] [CrossRef]
- Eden, E.R. The formation and function of ER-endosome membrane contact sites. Biochim. Biophys. Acta 2016, 1861, 874–879. [Google Scholar] [CrossRef]
- Katta, S.S.; Smoyer, C.J.; Jaspersen, S.L. Destination: Inner nuclear membrane. Trends Cell Biol. 2014, 24, 221–229. [Google Scholar] [CrossRef]
- Maimon, T.; Elad, N.; Dahan, I.; Medalia, O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012, 20, 998–1006. [Google Scholar] [CrossRef]
- Turgay, Y.; Ungricht, R.; Rothballer, A.; Kiss, A.; Csucs, G.; Horvath, P.; Kutay, U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010, 29, 2262–2275. [Google Scholar] [CrossRef]
- Soullam, B.; Worman, H.J. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 1995, 130, 15–27. [Google Scholar] [CrossRef]
- Lusk, C.P.; Blobel, G.; King, M.C. Highway to the inner nuclear membrane: Rules for the road. Nat. Rev. Mol. Cell Biol. 2007, 8, 414–420. [Google Scholar] [CrossRef]
- Meinema, A.C.; Poolman, B.; Veenhoff, L.M. The transport of integral membrane proteins across the nuclear pore complex. Nucleus 2012, 3, 322–329. [Google Scholar] [CrossRef][Green Version]
- Lo, H.-W.; Ali-Seyed, M.; Wu, Y.; Bartholomeusz, G.; Hsu, S.-C.; Hung, M.-C. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J. Cell. Biochem. 2006, 98, 1570–1583. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Yamaguchi, H.; Hsu, J.-M.; Hung, M.-C. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 2010, 29, 3997–4006. [Google Scholar] [CrossRef]
- Reilly, J.F.; Maher, P.A. Importin beta-mediated nuclear import of fibroblast growth factor receptor: Role in cell proliferation. J. Cell Biol. 2001, 152, 1307–1312. [Google Scholar] [CrossRef]
- Hsu, S.-C.; Hung, M.-C. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J. Biol. Chem. 2007, 282, 10432–10440. [Google Scholar] [CrossRef]
- Chen, Q.Q.; Chen, X.Y.; Jiang, Y.Y.; Liu, J. Identification of novel nuclear localization signal within the ErbB-2 protein. Cell Res. 2005, 15, 504–510. [Google Scholar] [CrossRef]
- Tseng, H.-C.; Lyu, P.-C.; Lin, W.-C. Nuclear localization of orphan receptor protein kinase (Ror1) is mediated through the juxtamembrane domain. BMC Cell Biol. 2010, 11, 48. [Google Scholar] [CrossRef]
- Rapoport, T.A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007, 450, 663–669. [Google Scholar] [CrossRef]
- Lakkaraju, A.K.; Abrami, L.; Lemmin, T.; Blaskovic, S.; Kunz, B.; Kihara, A.; Dal Peraro, M.; van der Goot, F.G. Palmitoylated calnexin is a key component of the ribosome-translocon complex. EMBO J. 2012, 31, 1823–1835. [Google Scholar] [CrossRef]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninghausen, O.; Mandon, E.C.; Becker, T.; Förster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef]
- Johnson, A.E.; Haigh, N.G. The ER Translocon and Retrotranslocation: Is the Shift into Reverse Manual or Automatic? Cell 2000, 102, 709–712. [Google Scholar] [CrossRef][Green Version]
- Wiertz, E.J.; Tortorella, D.; Bogyo, M.; Yu, J.; Mothes, W.; Jones, T.R.; Rapoport, T.A.; Ploegh, H.L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 1996, 384, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Pilon, M.; Schekman, R.; Römisch, K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997, 16, 4540–4548. [Google Scholar] [CrossRef] [PubMed]
- Plemper, R.K.; Böhmler, S.; Bordallo, J.; Sommer, T.; Wolf, D.H. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 1997, 388, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Nakatsukasa, K.; Brodsky, J.L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 2008, 9, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, J.; Huang, E.Y.; Olzmann, J.A. Endoplasmic Reticulum-Associated Degradation and Lipid Homeostasis. Annu. Rev. Nutr. 2016, 36, 511–542. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Herrgen, H.; Winkeler, A.; Herzog, V. Cholera toxin is exported from microsomes by the Sec61p complex. J. Cell Biol. 2000, 148, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Yamaguchi, H.; Huo, L.; Du, Y.; Lee, H.J. The translocon Sec61β localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. 2010, 285, 38720–38729. [Google Scholar] [CrossRef] [PubMed]
- Smoyer, C.J.; Katta, S.S.; Gardner, J.M.; Stoltz, L.; McCroskey, S.; Bradford, W.D.; McClain, M.; Smith, S.E.; Slaughter, B.D.; Unruh, J.R.; et al. Analysis of membrane proteins localizing to the inner nuclear envelope in living cells. J. Cell Biol. 2016, 215, 575–590. [Google Scholar] [CrossRef]
- Lang, S.; Pfeffer, S.; Lee, P.-H.; Cavalié, A.; Helms, V.; Förster, F.; Zimmermann, R. An Update on Sec61 Channel Functions, Mechanisms, and Related Diseases. Front. Physiol. 2017, 8, 887. [Google Scholar] [CrossRef]
- Geiger, R.; Andritschke, D.; Friebe, S.; Herzog, F.; Luisoni, S.; Heger, T.; Helenius, A. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat. Cell Biol. 2011, 13, 1305–1314. [Google Scholar] [CrossRef]
- Petris, G.; Casini, A.; Sasset, L.; Cesaratto, F.; Bestagno, M.; Cereseto, A.; Burrone, O.R. CD4 and BST-2/tetherin proteins retro-translocate from endoplasmic reticulum to cytosol as partially folded and multimeric molecules. J. Biol. Chem. 2014, 289, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Courgeon, M.; He, D.Q.; Liu, H.H.; Legent, K.; Treisman, J.E. The Drosophila Epidermal Growth Factor Receptor does not act in the nucleus. J. Cell Sci. 2018, 131, jcs220251. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, P.; Chaumet, A.; Royle, S.J.; Bard, F.A. The NAE Pathway: Autobahn to the Nucleus for Cell Surface Receptors. Cells 2019, 8, 915. https://doi.org/10.3390/cells8080915
Shah P, Chaumet A, Royle SJ, Bard FA. The NAE Pathway: Autobahn to the Nucleus for Cell Surface Receptors. Cells. 2019; 8(8):915. https://doi.org/10.3390/cells8080915
Chicago/Turabian StyleShah, Poonam, Alexandre Chaumet, Stephen J. Royle, and Frederic A. Bard. 2019. "The NAE Pathway: Autobahn to the Nucleus for Cell Surface Receptors" Cells 8, no. 8: 915. https://doi.org/10.3390/cells8080915
APA StyleShah, P., Chaumet, A., Royle, S. J., & Bard, F. A. (2019). The NAE Pathway: Autobahn to the Nucleus for Cell Surface Receptors. Cells, 8(8), 915. https://doi.org/10.3390/cells8080915




