An Omics Approach to Extracellular Vesicles from HIV-1 Infected Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Treatment of PBMCs
2.2. Reagents and Antibodies
2.3. Nanotrap® Particle Pulldown
2.4. EV Isolation
2.5. EV Purification
2.6. ZetaView Nanoparticle Tracking Analysis
2.7. Mass Spectrometry
2.8. [3H] Thymidine DNA Synthesis Assay
2.9. Protein-Protein Interactions (PPIs) Analysis
2.10. Western Blot Analysis
2.11. RNA and DNA Isolation
2.12. RNA Sequencing Library Construction
2.13. Bioinformatics
2.14. Mitochondrial DNA PCR
3. Results
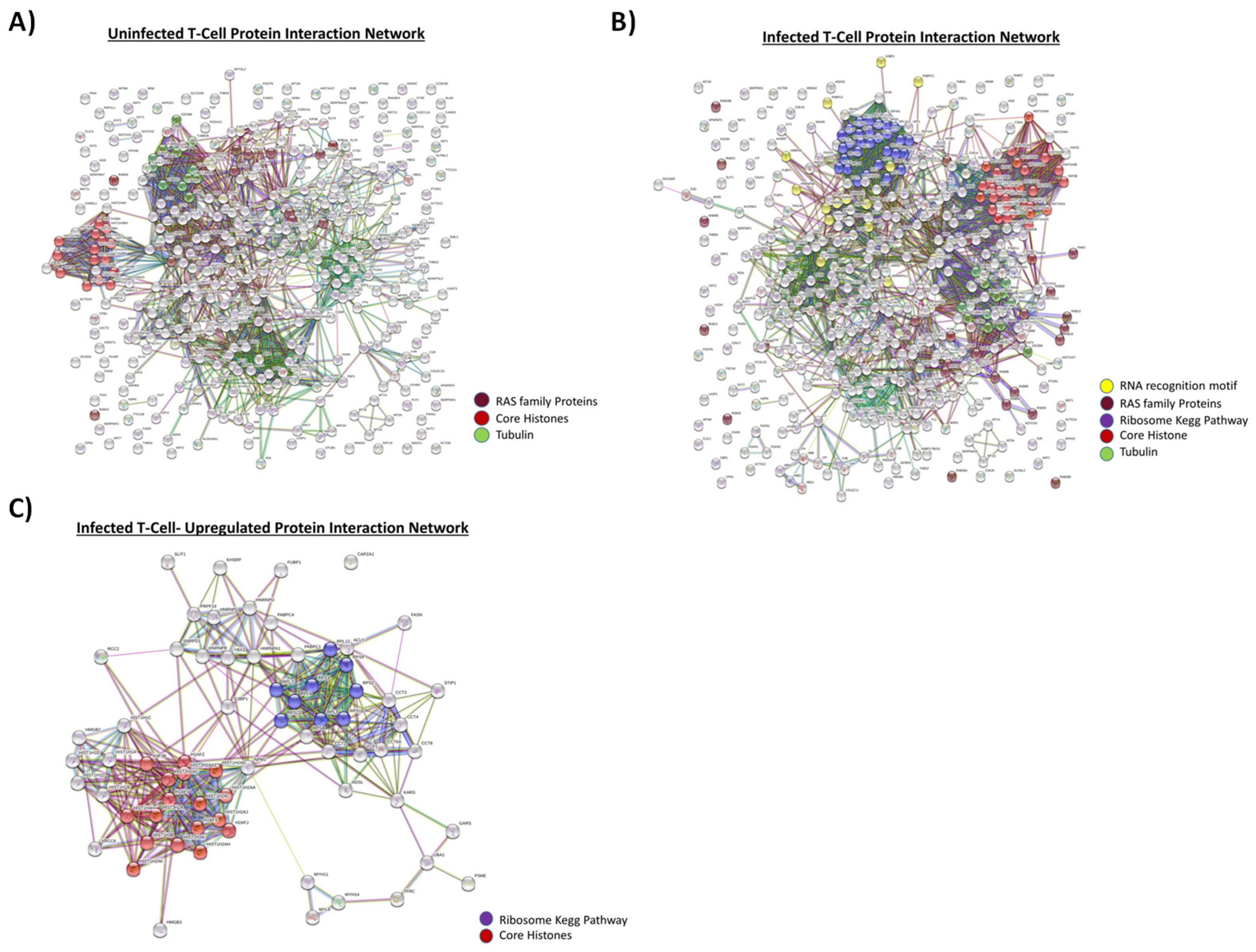
3.1. Proteomic Analysis of Exosomes from HIV-1-Infected and Uninfected T-cells
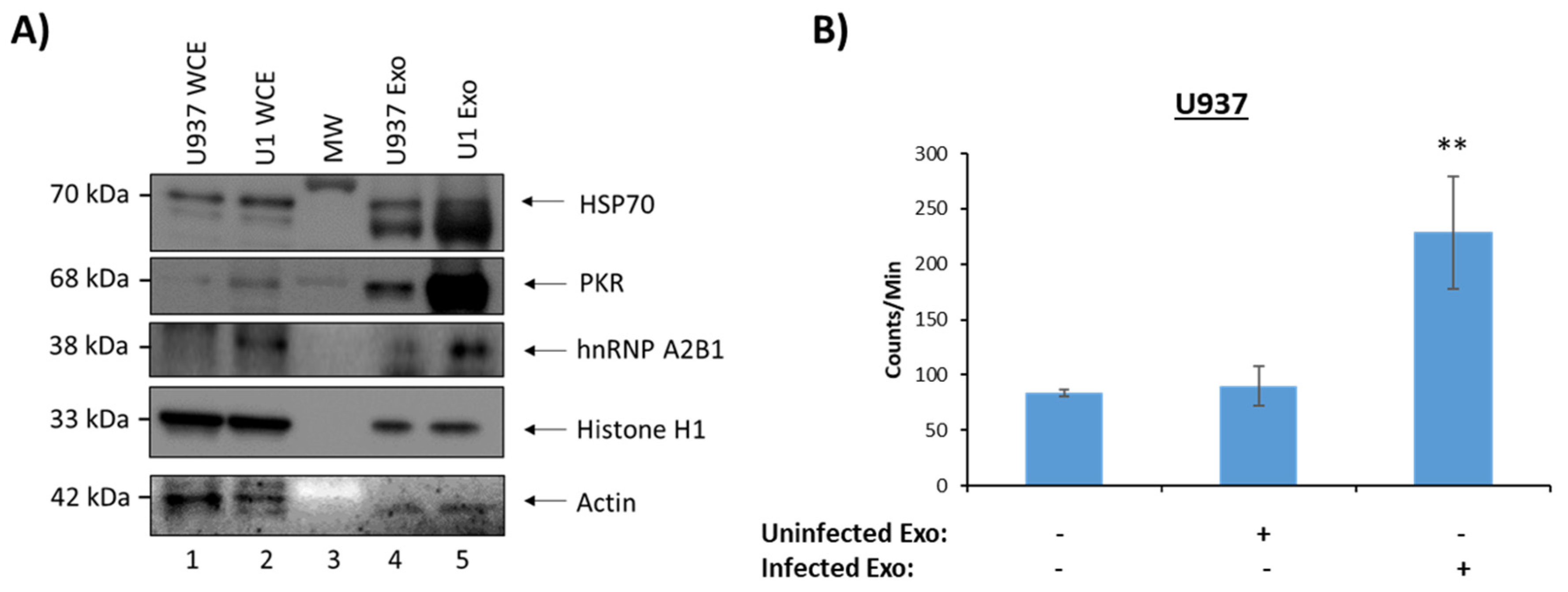
3.2. Functionality of EVs from HIV-1-Infected and Uninfected T-Cells
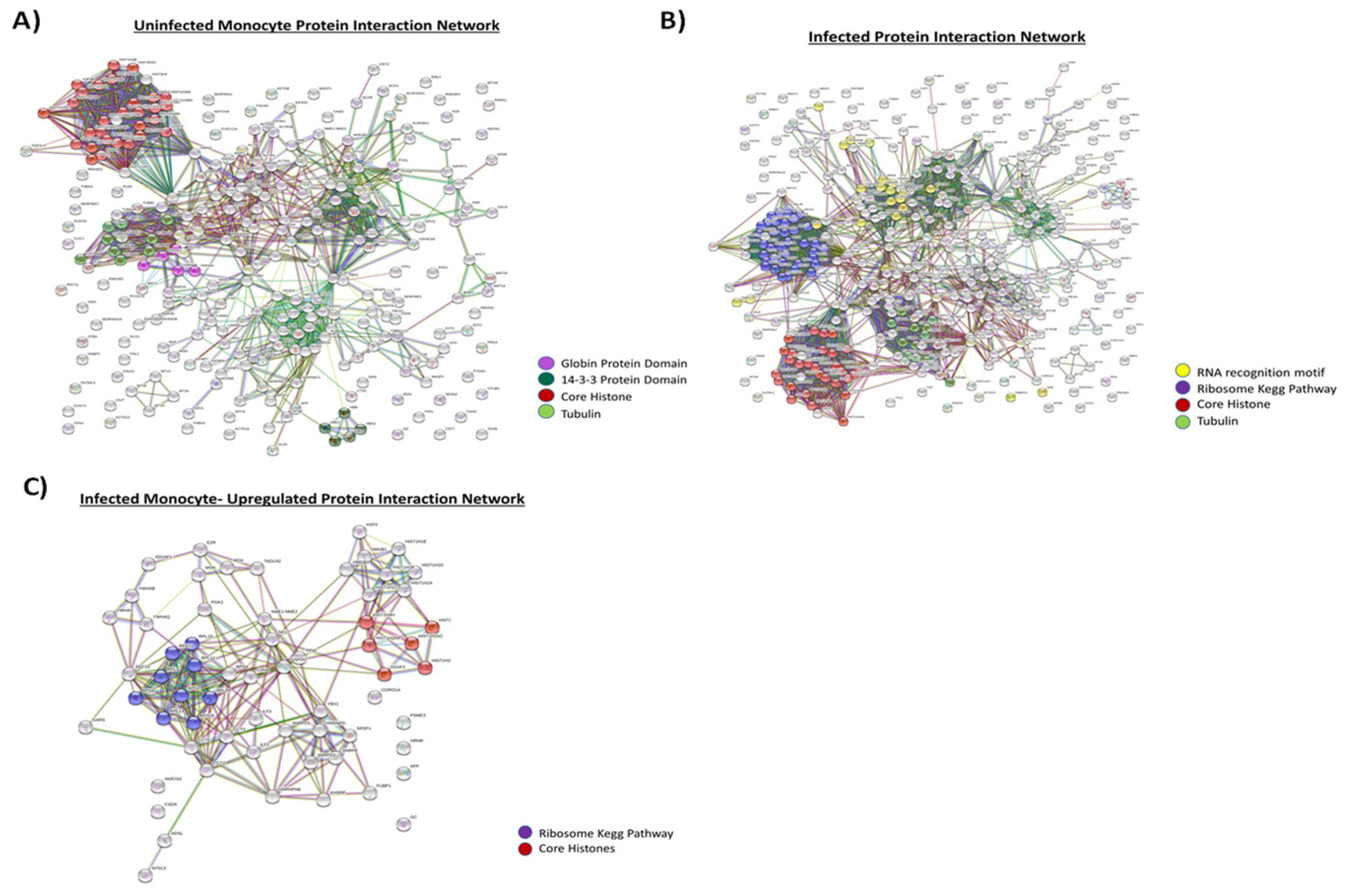
3.3. Proteomic Analysis of Exosomes from HIV-1-Infected and Uninfected Monocytes
3.4. Functionality of EVs from HIV-1-Infected and Uninfected Monocytes
3.5. Proteomic Analysis of EVs from PBMCs
3.6. EVs from HIV-1-Infected Cells Contain Similar Size RNAs
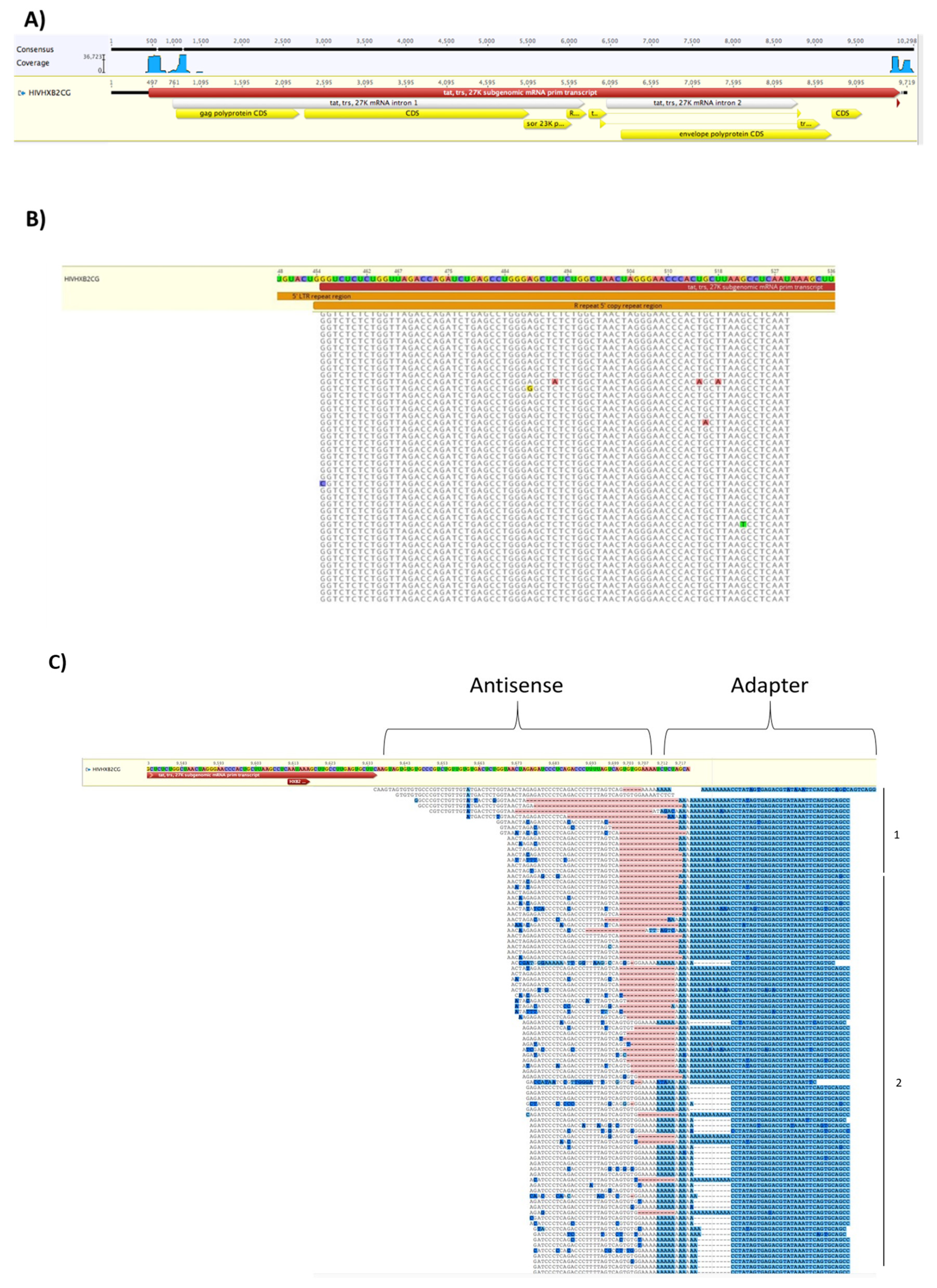
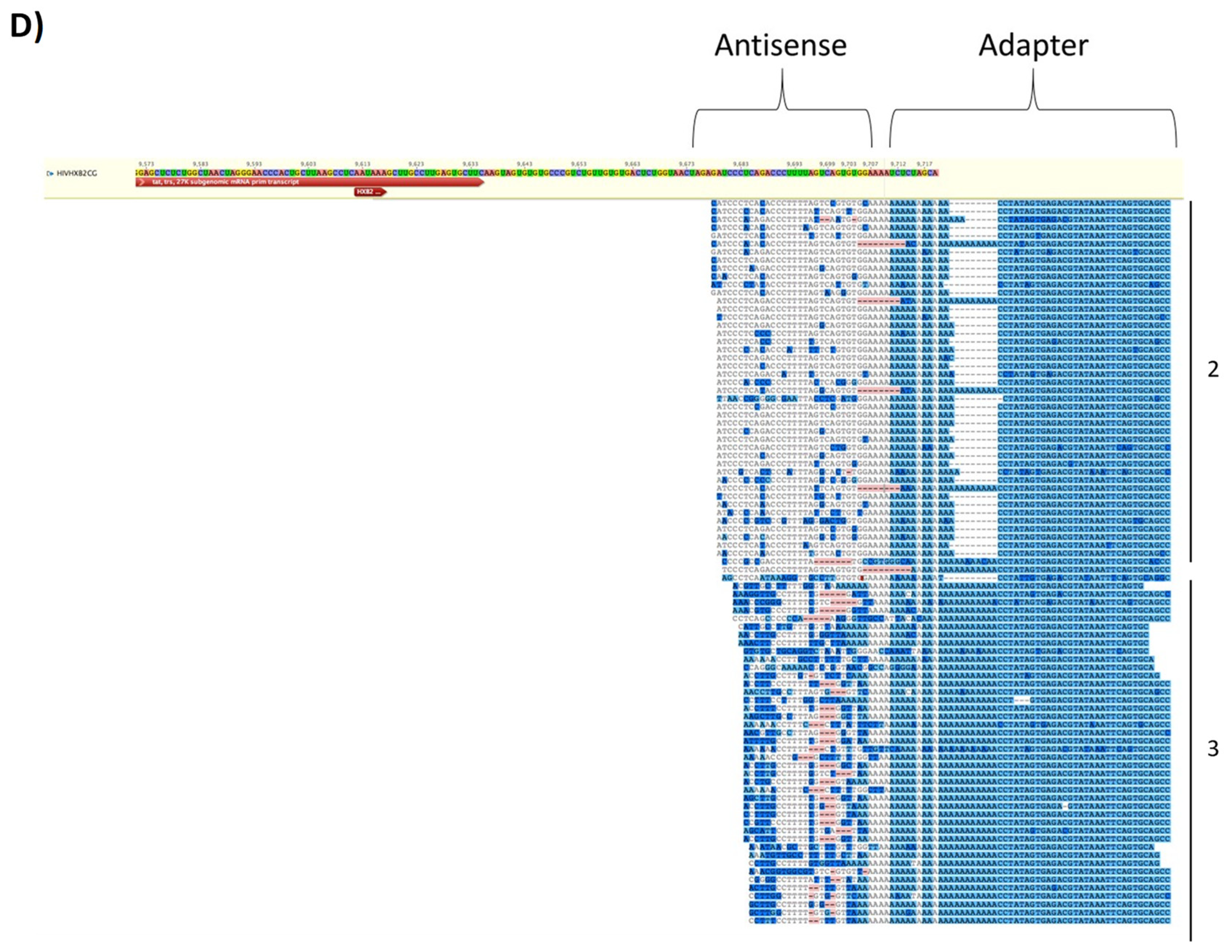
3.7. EVs from HIV-1 Infected T-Cells Contain Wild-Type and Modified Antisense RNA Transcripts
3.8. EVs from Infected Cells Contain Cellular Nucleic Acids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO|World Health Organization. Available online: http://www.who.int/hiv/en/ (accessed on 26 April 2017).
- HIV in the United States|Statistics Overview|Statistics Center|HIV/AIDS|CDC. Available online: https://www.cdc.gov/hiv/statistics/overview/ataglance.html (accessed on 26 April 2017).
- U.S. Statistics|HIV.gov. Available online: https://www.hiv.gov/hiv-basics/overview/data-and-trends/statistics (accessed on 4 January 2019).
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Abbas, W.; Herbein, G. HIV-1 Latency in Monocytes/Macrophages. Viruses 2014, 6, 1837–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahabieh, M.; Battivelli, E.; Verdin, E. Understanding HIV Latency: The Road to an HIV Cure. Annu. Rev. Med. 2015, 66, 407–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battistini, A.; Sgarbanti, M. HIV-1 Latency: An Update of Molecular Mechanisms and Therapeutic Strategies. Viruses 2014, 6, 1715–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davey, R.T.; Bhat, N.; Yoder, C.; Chun, T.W.; Metcalf, J.A.; Dewar, R.; Natarajan, V.; Lempicki, R.A.; Adelsberger, J.W.; Miller, K.D.; et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc. Natl. Acad. Sci. USA 1999, 96, 15109–15114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García, F.; Plana, M.; Vidal, C.; Cruceta, A.; O’Brien, W.A.; Pantaleo, G.; Pumarola, T.; Gallart, T.; Miró, J.M.; Gatell, J.M. Dynamics of viral load rebound and immunological changes after stopping effective antiretroviral therapy. AIDS 1999, 13, F79–F86. [Google Scholar] [CrossRef] [PubMed]
- Katlama, C.; Deeks, S.G.; Autran, B.; Martinez-Picado, J.; van Lunzen, J.; Rouzioux, C.; Miller, M.; Vella, S.; Schmitz, J.E.; Ahlers, J.; et al. Barriers to a cure for HIV: New ways to target and eradicate HIV-1 reservoirs. Lancet 2013, 381, 2109–2117. [Google Scholar] [CrossRef]
- Hatano, H.; Jain, V.; Hunt, P.W.; Lee, T.-H.; Sinclair, E.; Do, T.D.; Hoh, R.; Martin, J.N.; McCune, J.M.; Hecht, F.; et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J. Infect. Dis. 2013, 208, 50–56. [Google Scholar] [CrossRef]
- Kumar, A.M.; Borodowsky, I.; Fernandez, B.; Gonzalez, L.; Kumar, M. Human immunodeficiency virus type 1 RNA Levels in different regions of human brain: Quantification using real-time reverse transcriptase-polymerase chain reaction. J. Neurovirol. 2007, 13, 210–224. [Google Scholar] [CrossRef]
- Imamichi, H.; Dewar, R.L.; Adelsberger, J.W.; Rehm, C.A.; O’Doherty, U.; Paxinos, E.E.; Fauci, A.S.; Lane, H.C. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2016, 113, 8783–8788. [Google Scholar] [CrossRef] [Green Version]
- Maldarelli, F.; Kearney, M.; Palmer, S.; Stephens, R.; Mican, J.; Polis, M.A.; Davey, R.T.; Kovacs, J.; Shao, W.; Rock-Kress, D.; et al. HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. J. Virol. 2013, 87, 10313–10323. [Google Scholar] [CrossRef] [PubMed]
- Boltz, V.F.; Zheng, Y.; Lockman, S.; Hong, F.; Halvas, E.K.; McIntyre, J.; Currier, J.S.; Chibowa, M.C.; Kanyama, C.; Nair, A.; et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc. Natl. Acad. Sci. USA 2011, 108, 9202–9207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayanan, A.; Iordanskiy, S.; Das, R.; Van Duyne, R.; Santos, S.; Jaworski, E.; Guendel, I.; Sampey, G.; Dalby, E.; Iglesias-Ussel, M.; et al. Exosomes Derived from HIV-1-infected Cells Contain Trans-activation Response Element RNA. J. Biol. Chem. 2013, 288, 20014–20033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampey, G.C.; Saifuddin, M.; Schwab, A.; Barclay, R.; Punya, S.; Chung, M.-C.; Hakami, R.M.; Zadeh, M.A.; Lepene, B.; Klase, Z.A.; et al. Exosomes from HIV-1-infected Cells Stimulate Production of Pro-inflammatory Cytokines through Trans-activating Response (TAR) RNA. J. Biol. Chem. 2016, 291, 1251–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Feng, Z.; Yue, H.; Bazdar, D.; Mbonye, U.; Zender, C.; Harding, C.V.; Bruggeman, L.; Karn, J.; Sieg, S.F.; et al. Exosomes derived from HIV-1-infected cells promote growth and progression of cancer via HIV TAR RNA. Nat. Commun. 2018, 9, 4585. [Google Scholar] [CrossRef]
- Hu, G.; Yang, L.; Cai, Y.; Niu, F.; Mezzacappa, F.; Callen, S.; Fox, H.S.; Buch, S. Emerging roles of extracellular vesicles in neurodegenerative disorders: Focus on HIV-associated neurological complications. Cell Death Dis. 2016, 7, e2481. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta BBA-Gen. Subj. 2012, 1820, 940–948. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, R.M.; Mathew, A.; Mason, A.B.; Teng, K. Exosome formation during maturation of mammalian and avian reticulocytes: Evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 1991, 147, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Sampey, G.C.; Meyering, S.S.; Zadeh, M.A.; Saifuddin, M.; Hakami, R.M.; Kashanchi, F. Exosomes and their role in CNS viral infections. J. Neurovirol. 2014, 20, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.A.; Schwab, A.; DeMarino, C.; Akpamagbo, Y.; Lepene, B.; Kassaye, S.; Iordanskiy, S.; Kashanchi, F. Exosomes from uninfected cells activate transcription of latent HIV-1. J. Biol. Chem. 2017, 292, 11682–11701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Wittki, S.; Bräu, T.; Dreyer, F.S.; Krätzel, K.; Dindorf, J.; Johnston, I.C.D.; Gross, S.; Kremmer, E.; Zeidler, R.; et al. HIV Nef, paxillin, and Pak1/2 regulate activation and secretion of TACE/ADAM10 proteases. Mol. Cell 2013, 49, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Lenassi, M.; Cagney, G.; Liao, M.; Vaupotic, T.; Bartholomeeusen, K.; Cheng, Y.; Krogan, N.J.; Plemenitas, A.; Peterlin, B.M. HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 2010, 11, 110–122. [Google Scholar] [CrossRef]
- Raymond, A.D.; Diaz, P.; Chevelon, S.; Agudelo, M.; Yndart-Arias, A.; Ding, H.; Kaushik, A.; Jayant, R.D.; Nikkhah-Moshaie, R.; Roy, U.; et al. Microglia-derived HIV Nef+ exosome impairment of the blood-brain barrier is treatable by nanomedicine-based delivery of Nef peptides. J. Neurovirol. 2016, 22, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Arenaccio, C.; Chiozzini, C.; Columba-Cabezas, S.; Manfredi, F.; Affabris, E.; Baur, A.; Federico, M. Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. J. Virol. 2014, 88, 11529–11539. [Google Scholar] [CrossRef]
- Kulkarni, R.; Prasad, A. Exosomes Derived from HIV-1 Infected DCs Mediate Viral trans-Infection via Fibronectin and Galectin-3. Sci. Rep. 2017, 7, 14787. [Google Scholar] [CrossRef]
- Arenaccio, C.; Anticoli, S.; Manfredi, F.; Chiozzini, C.; Olivetta, E.; Federico, M. Latent HIV-1 is activated by exosomes from cells infected with either replication-competent or defective HIV-1. Retrovirology 2015, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, E.; Saifuddin, M.; Sampey, G.; Shafagati, N.; Van Duyne, R.; Iordanskiy, S.; Kehn-Hall, K.; Liotta, L.; Petricoin, E.; Young, M.; et al. The use of Nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS ONE 2014, 9, e96778. [Google Scholar] [CrossRef] [PubMed]
- DeMarino, C.; Pleet, M.L.; Cowen, M.; Barclay, R.A.; Akpamagbo, Y.; Erickson, J.; Ndembe, N.; Charurat, M.; Jumare, J.; Bwala, S.; et al. Antiretroviral Drugs Alter the Content of Extracellular Vesicles from HIV-1-Infected Cells. Sci. Rep. 2018, 8, 7653. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Suyasunanont, U.; Nakkuntod, M.; Mirasena, S. Mitochondrial DNA control region analysis of three ethnic populations in lower Northern part of Thailand. Genet. Mol. Res. GMR 2017, 16. [Google Scholar] [CrossRef]
- Eduardoff, M.; Xavier, C.; Strobl, C.; Casas-Vargas, A.; Parson, W. Optimized mtDNA Control Region Primer Extension Capture Analysis for Forensically Relevant Samples and Highly Compromised mtDNA of Different Age and Origin. Genes 2017, 8, 237. [Google Scholar] [CrossRef]
- Wang, J.; Yao, Y.; Wu, J.; Li, G. Identification and analysis of exosomes secreted from macrophages extracted by different methods. Int. J. Clin. Exp. Pathol. 2015, 8, 6135–6142. [Google Scholar]
- Schorey, J.S.; Bhatnagar, S. Exosome function: From tumor immunology to pathogen biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef]
- Li, W.; Mu, D.; Tian, F.; Hu, Y.; Jiang, T.; Han, Y.; Chen, J.; Han, G.; Li, X. Exosomes derived from Rab27a-overexpressing tumor cells elicit efficient induction of antitumor immunity. Mol. Med. Rep. 2013, 8, 1876–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malumbres, M.; Barbacid, M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005, 30, 630–641. [Google Scholar] [CrossRef] [PubMed]
- Boffo, S.; Damato, A.; Alfano, L.; Giordano, A. CDK9 inhibitors in acute myeloid leukemia. J. Exp. Clin. Cancer Res. CR 2018, 37, 36. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.P. The CDK Network: Linking Cycles of Cell Division and Gene Expression. Genes Cancer 2012, 3, 731–738. [Google Scholar] [CrossRef] [Green Version]
- Palacios, E.H.; Weiss, A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene 2004, 23, 7990–8000. [Google Scholar] [CrossRef] [Green Version]
- Kitase, Y.; Shuler, C.F. Palatal adhesion is dependent on Src family kinases and p38MAPK. Int. J. Dev. Biol. 2014, 58, 335–341. [Google Scholar] [CrossRef] [Green Version]
- Baruzzi, A.; Iacobucci, I.; Soverini, S.; Lowell, C.A.; Martinelli, G.; Berton, G. c-Abl and Src-family kinases cross-talk in regulation of myeloid cell migration. FEBS Lett. 2010, 584, 15–21. [Google Scholar] [CrossRef]
- Liu, H.; Herrmann, C.H. Differential localization and expression of the Cdk9 42k and 55k isoforms. J. Cell. Physiol. 2005, 203, 251–260. [Google Scholar] [CrossRef]
- Shore, S.M.; Byers, S.A.; Maury, W.; Price, D.H. Identification of a novel isoform of Cdk9. Gene 2003, 307, 175–182. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, L.; Leng, Y.; Dai, Y.; Orlowski, R.Z.; Grant, S. Positive transcription elongation factor b (P-TEFb) is a therapeutic target in human multiple myeloma. Oncotarget 2017, 8, 59476–59491. [Google Scholar] [CrossRef] [Green Version]
- Heras-Sandoval, D.; Pérez-Rojas, J.M.; Hernández-Damián, J.; Pedraza-Chaverri, J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell. Signal. 2014, 26, 2694–2701. [Google Scholar] [CrossRef]
- Rang, H.P. Pharmacology, 5th ed.; Churchill Livingstone: Edinburgh, UK; New York, NY, USA, 2003; ISBN 978-0-443-07145-4. [Google Scholar]
- García, M.A.; Meurs, E.F.; Esteban, M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie 2007, 89, 799–811. [Google Scholar] [CrossRef]
- Shaytan, A.K.; Landsman, D.; Panchenko, A.R. Nucleosome adaptability conferred by sequence and structural variations in histone H2A-H2B dimers. Curr. Opin. Struct. Biol. 2015, 32, 48–57. [Google Scholar] [CrossRef]
- Thiriet, C.; Hayes, J.J. Histone dynamics during transcription: Exchange of H2A/H2B dimers and H3/H4 tetramers during pol II elongation. Results Probl. Cell Differ. 2006, 41, 77–90. [Google Scholar]
- Guo, J.; Wang, W.; Yu, D.; Wu, Y. Spinoculation triggers dynamic actin and cofilin activity that facilitates HIV-1 infection of transformed and resting CD4 T cells. J. Virol. 2011, 85, 9824–9833. [Google Scholar] [CrossRef]
- Zapata, J.C.; Campilongo, F.; Barclay, R.A.; DeMarino, C.; Iglesias-Ussel, M.D.; Kashanchi, F.; Romerio, F. The Human Immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology 2017, 506, 34–44. [Google Scholar] [CrossRef]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef]
- Tirumuru, N.; Zhao, B.S.; Lu, W.; Lu, Z.; He, C.; Wu, L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 2016, 5, e15528. [Google Scholar] [CrossRef]
- Cao, L.; Xie, B.; Yang, X.; Liang, H.; Jiang, X.; Zhang, D.; Xue, P.; Chen, D.; Shao, Z. MiR-324-5p Suppresses Hepatocellular Carcinoma Cell Invasion by Counteracting ECM Degradation through Post-Transcriptionally Downregulating ETS1 and SP1. PLoS ONE 2015, 10, e0133074. [Google Scholar] [CrossRef]
- Louis, D.N.; von Deimling, A.; Chung, R.Y.; Rubio, M.P.; Whaley, J.M.; Eibl, R.H.; Ohgaki, H.; Wiestler, O.D.; Thor, A.D.; Seizinger, B.R. Comparative study of p53 gene and protein alterations in human astrocytic tumors. J. Neuropathol. Exp. Neurol. 1993, 52, 31–38. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, X.; Cui, H.; Luo, X.; Tang, Y.; Chen, S.; Wu, L.; Shen, N. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood 2010, 116, 5885–5894. [Google Scholar] [CrossRef]
- Tsilioni, I.; Theoharides, T.C. Extracellular vesicles are increased in the serum of children with autism spectrum disorder, contain mitochondrial DNA, and stimulate human microglia to secrete IL-1β. J. Neuroinflamm. 2018, 15, 239. [Google Scholar] [CrossRef]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [Green Version]
- Schwab, A.; Meyering, S.S.; Lepene, B.; Iordanskiy, S.; van Hoek, M.L.; Hakami, R.M.; Kashanchi, F. Extracellular vesicles from infected cells: Potential for direct pathogenesis. Front. Microbiol. 2015, 6, 1132. [Google Scholar] [CrossRef]
- Feng, X.; Hao, J.; Liu, Q.; Yang, L.; Lv, X.; Zhang, Y.; Xing, L.; Xu, N.; Liu, S. HMGB1 mediates IFN-γ-induced cell proliferation in MMC cells through regulation of cyclin D1/CDK4/p16 pathway. J. Cell. Biochem. 2012, 113, 2009–2019. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Agresti, A. HMG proteins: Dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005, 15, 496–506. [Google Scholar] [CrossRef]
- Storchova, Z.; Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004, 5, 45–54. [Google Scholar] [CrossRef]
- Fox, D.T.; Duronio, R.J. Endoreplication and polyploidy: Insights into development and disease. Development 2013, 140, 3–12. [Google Scholar] [CrossRef]
- Lee, H.O.; Davidson, J.M.; Duronio, R.J. Endoreplication: Polyploidy with purpose. Genes Dev. 2009, 23, 2461–2477. [Google Scholar] [CrossRef]
- Yi, F.; Guo, J.; Dabbagh, D.; Spear, M.; He, S.; Kehn-Hall, K.; Fontenot, J.; Yin, Y.; Bibian, M.; Park, C.M.; et al. Discovery of Novel Small-Molecule Inhibitors of LIM Domain Kinase for Inhibiting HIV-1. J. Virol. 2017, 91, e02418-16. [Google Scholar] [CrossRef] [Green Version]
- Collin, D.; van Heijenoort, C.; Boiziau, C.; Toulmé, J.J.; Guittet, E. NMR characterization of a kissing complex formed between the TAR RNA element of HIV-1 and a DNA aptamer. Nucleic Acids Res. 2000, 28, 3386–3391. [Google Scholar] [CrossRef]
- Akpamagbo, Y.A.; DeMarino, C.; Pleet, M.L.; Schwab, A.; Rodriguez, M.; Barclay, R.A.; Sampey, G.; Iordanskiy, S.; El-Hage, N.; Kashanchi, F. HIV-1 Transcription Inhibitors Increase the Synthesis of Viral Non-Coding RNA that Contribute to Latency. Curr. Pharm. Des. 2017, 23, 4133–4144. [Google Scholar] [CrossRef]
- Imashimizu, M.; Takahashi, H.; Oshima, T.; McIntosh, C.; Bubunenko, M.; Court, D.L.; Kashlev, M. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 2015, 16, 98. [Google Scholar] [CrossRef]
- Carey, L.B. RNA polymerase errors cause splicing defects and can be regulated by differential expression of RNA polymerase subunits. eLife 2015, 4, e09945. [Google Scholar] [CrossRef] [Green Version]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [Green Version]
- Walkley, C.R.; Li, J.B. Rewriting the transcriptome: Adenosine-to-inosine RNA editing by ADARs. Genome Biol. 2017, 18, 205. [Google Scholar] [CrossRef]
- Lal, A.; Kim, H.H.; Abdelmohsen, K.; Kuwano, Y.; Pullmann, R.; Srikantan, S.; Subrahmanyam, R.; Martindale, J.L.; Yang, X.; Ahmed, F.; et al. p16INK4a Translation Suppressed by miR-24. PLoS ONE 2008, 3, e1864. [Google Scholar] [CrossRef]
- Martin, S.J. Cell death and inflammation: The case for IL-1 family cytokines as the canonical DAMPs of the immune system. FEBS J. 2016, 283, 2599–2615. [Google Scholar] [CrossRef]
- Chahar, H.S.; Corsello, T.; Kudlicki, A.S.; Komaravelli, N.; Casola, A. Respiratory Syncytial Virus Infection Changes Cargo Composition of Exosome Released from Airway Epithelial Cells. Sci. Rep. 2018, 8, 387. [Google Scholar] [CrossRef]



| Protein | Uninfected T-cell Exosome (CEM) | Infected T-cell Exosome (ACH2) | |
|---|---|---|---|
| Controls | Nucleophosmin | - | +++ |
| 14-3-3 Protein | + | - | |
| HSP70 | + | + | |
| HSP90 | + | + | |
| RNA Binding Proteins | hnRNP A1 | - | + |
| hnRNP L | - | ++ | |
| hnRNP R | + | ++ | |
| hnRNP D0 | - | ++ | |
| hnRNP Q | + | + | |
| hnRNP K | + | - | |
| Histones | Histone H1 | + | ++ |
| Histone H2A | - | ++ | |
| Histone H2B | + | + | |
| Histone H3 | + | ++ | |
| Histone H4 | - | +++ | |
| Kinases (Cdks) | Cdk1 | - | + |
| Cdk2 | - | + | |
| Cdk3 | - | + | |
| Cdk4 | - | + | |
| Cdk5 | - | + | |
| Cdk6 | - | + | |
| Cdk9 | - | + | |
| Cdk12 | - | + | |
| Cdk13 | - | + | |
| Cdk14 | - | + | |
| Cdk15 | - | + | |
| Cdk16 | - | + | |
| Cdk17 | - | + | |
| Cdk18 | - | + | |
| Cdk19 | - | + | |
| Cdk20 | - | + | |
| Kinases (RTKs) | Blk | - | + |
| Fgr | - | + | |
| Fyn | - | + | |
| Hck | - | + | |
| Lck | - | + | |
| Lyn | - | + | |
| Yes | - | + |
| Protein | Uninfected Monocyte Exosome (U937) | Infected Monocyte Exosome (U1) | |
|---|---|---|---|
| Controls | TGF-β binding protein 4 | +++ | - |
| Nucleolin | - | +++ | |
| HSP70 | + | + | |
| HSP90 | + | + | |
| Complement Protein C1 | + | + | |
| Complement Protein C3 | + | + | |
| Complement Protein C4A | + | + | |
| Complement Protein C5 | + | + | |
| Complement Protein C7 | + | - | |
| Complement Protein C8 | + | + | |
| Complement Protein C9 | + | + | |
| Complement Protein H | + | - | |
| RNA Binding Proteins | hnRNP A1 | - | ++ |
| hnRNP R | - | + | |
| hnRNP D0 | - | ++ | |
| hnRNP Q | - | + | |
| hnRNP K | - | + | |
| hnRNP U | - | + | |
| hnRNP DL | - | + | |
| hnRNP C1/2 | - | + | |
| hnRNP A1/B1 | - | + | |
| hnRNP A/B | - | + | |
| Histones | Histone H1 | - | + |
| Histone H2A | - | + | |
| Histone H2B | + | + | |
| Histone H3 | + | + | |
| Histone H4 | + | + | |
| Kinases | PKM | +++ | +++ |
| PGK1 | + | + | |
| NDK | + | + | |
| Int Kinase | + | + | |
| Adeno Kinase | + | - | |
| cAMP Kinase | + | - | |
| PI3K | + | - | |
| Xylulose Kinase | + | - | |
| BADDA | + | - | |
| Infected Cell Exosomes | PKR | - | + |
| HMGB1 | - | ++ | |
| Interleukin enhancer-BF3 | - | ++ | |
| FVEB1 | - | ++ | |
| T-Complex Protein | - | ++ | |
| Adenylosuccinate lyase | - | ++ | |
| Guanine nuc-binding protein | - | ++ | |
| Uninfected Cell Exosomes | Spondin-1 | ++ | - |
| ILGBP | ++ | - | |
| EMILIN-2 | ++ | - | |
| ATP-citrate synthase | ++ | - | |
| Proteoglycan 4 | + | - | |
| Exostosin-1 | + | - |
| Protein | Uninfected PBMC 1 | Infected PBMC 1 | Uninfected PBMC 2 | Infected PBMC 2 | Uninfected PBMC 3 | Infected PBMC 3 | |
|---|---|---|---|---|---|---|---|
| Controls | Integrin Alpha-II B Preprotein | +++ | - | +++ | - | +++ | - |
| Sodium/Calcium Exchanger 3 Preprotein | - | ++ | - | ++ | - | ++ | |
| Talin-1 | + | + | + | + | + | + | |
| HSP 90 | ++ | + | ++ | + | ++ | + | |
| RNA Binding Proteins | hnRNP K | + | + | ++ | + | ++ | + |
| hnRNP C1/C2 | + | + | + | + | +++ | - | |
| hnRNP R | + | - | ++ | - | +++ | - | |
| hnRNP Q | + | + | +++ | + | ++ | + | |
| hnRNP U | + | + | + | +++ | + | +++ | |
| hnRNP D0 | ++ | + | +++ | + | +++ | + | |
| Histones | Histone H1 | +++ | - | +++ | + | +++ | + |
| Histone H2A | ++ | + | ++ | + | ++ | + | |
| Histone H2B | + | + | ++ | + | + | + | |
| Histone H3 | ++ | + | +++ | + | +++ | + | |
| Histone H4 | + | + | + | + | +++ | + | |
| Annexins | Annexin A1 | +++ | + | ++ | + | +++ | + |
| Annexin A2 | ++ | + | +++ | + | ++ | - | |
| Annexin A5 | +++ | - | - | - | + | - | |
| Annexin A6 | ++ | + | +++ | + | +++ | + | |
| Misc. Proteins | Proflin | + | + | + | +++ | + | +++ |
| Cofilin | + | + | ++ | + | +++ | + | |
| Elongation Factor 1 | ++ | - | + | + | ++ | - | |
| Elongation Factor 2 | + | + | + | + | ++ | + | |
| Calponin 2 | ++ | - | ++ | - | ++ | - | |
| Cathepsin G | + | - | + | - | - | - | |
| Plectin | + | - | + | - | + | - | |
| 14-3-3 Protein Zeta/Delta | + | ++ | + | + | + | + | |
| Rap-1B | + | ++ | + | + | + | ++ | |
| HSP60 | + | +++ | + | + | + | +++ |
| (a) | |||
| RNA | Infected T-Cell Exosome | Infected Myeloid Exosome | P-Value |
| MS12 | + | + | 0.0097 |
| SON | + | + | 0.0054 |
| ADAM10 | + | + | 0.0006 |
| GAK | + | + | 0.0107 |
| (b) | (c) | ||
| Unique to T-Cell Exosome (3065 total RNAs) | Unique to Myeloid Exosome (136 total RNAs) | ||
| LINC00184 | IFI44L (Interferon-Stimulated) | ||
| LINC00152 | IFI16 (Interferon-Stimulated) | ||
| LINC01234 | IRF8 (Interferon-Stimulated) | ||
| LINC00969 | SLFN11 (Interferon-Stimulated) | ||
| miRNA | Source | Target |
|---|---|---|
| miR-635 | T-cell | ICAM-1 |
| miR-324 | T-cell | SP-1 |
| miR-621 | T-cell | FBXO11 |
| miR-31 | T-cell | p53 |
| miR-590 | T-cell | TGF-βR2 |
| miR-221 | T-cell | c-Kit |
| miR-25 | T-cell | SRC-3 |
| miR-29A | T-cell | CDK6 |
| miR-24A | T-cell | INK4A |
| miR-34A | T-cell | SIRT-1 |
| miR-155HG | Myeloid | IRAK3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barclay, R.A.; Khatkar, P.; Mensah, G.; DeMarino, C.; Chu, J.S.C.; Lepene, B.; Zhou, W.; Gillevet, P.; Torkzaban, B.; Khalili, K.; et al. An Omics Approach to Extracellular Vesicles from HIV-1 Infected Cells. Cells 2019, 8, 787. https://doi.org/10.3390/cells8080787
Barclay RA, Khatkar P, Mensah G, DeMarino C, Chu JSC, Lepene B, Zhou W, Gillevet P, Torkzaban B, Khalili K, et al. An Omics Approach to Extracellular Vesicles from HIV-1 Infected Cells. Cells. 2019; 8(8):787. https://doi.org/10.3390/cells8080787
Chicago/Turabian StyleBarclay, Robert A., Pooja Khatkar, Gifty Mensah, Catherine DeMarino, Jeffery S. C. Chu, Benjamin Lepene, Weidong Zhou, Patrick Gillevet, Bahareh Torkzaban, Kamel Khalili, and et al. 2019. "An Omics Approach to Extracellular Vesicles from HIV-1 Infected Cells" Cells 8, no. 8: 787. https://doi.org/10.3390/cells8080787




