Viroporins in the Influenza Virus
Abstract
1. Introduction
1.1. Influenza Viruses
1.2. The Proteins in Influenza Viruses
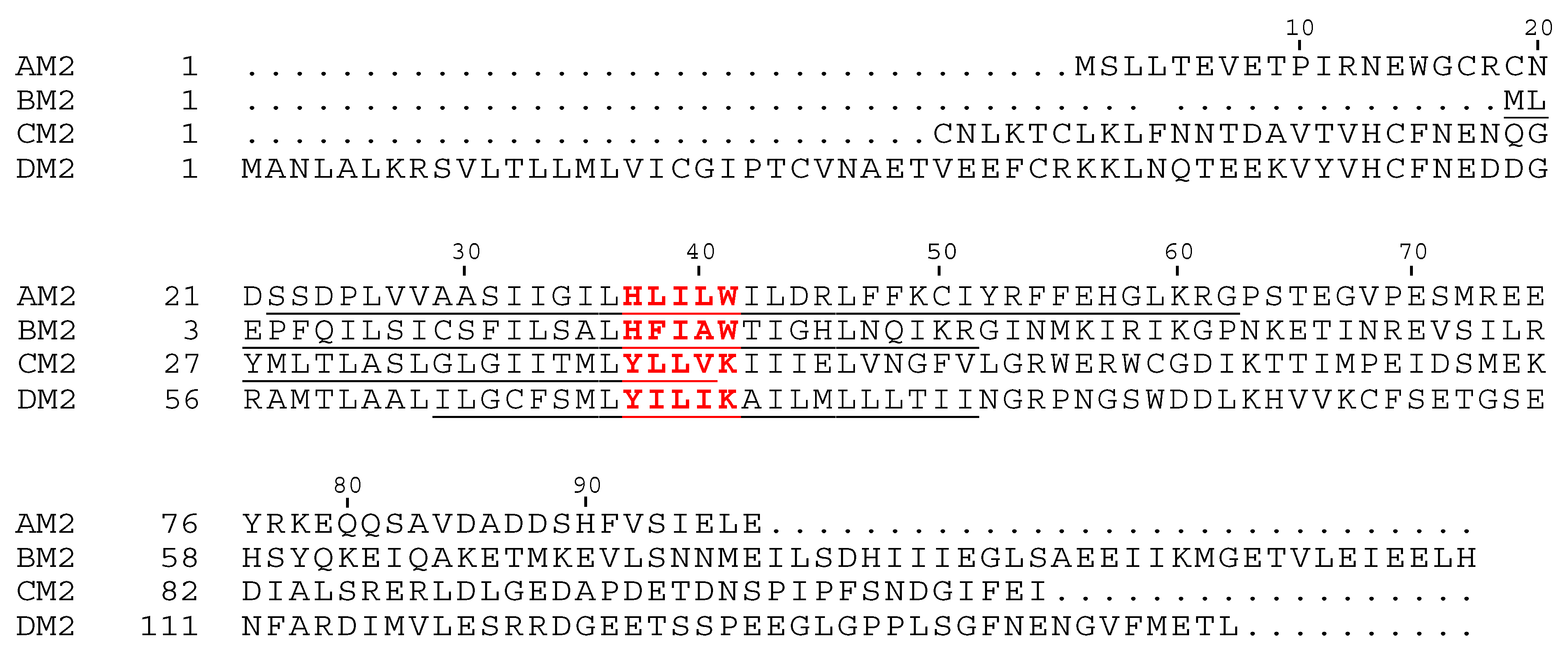
2. AM2 and BM2
2.1. AM2
2.2. BM2
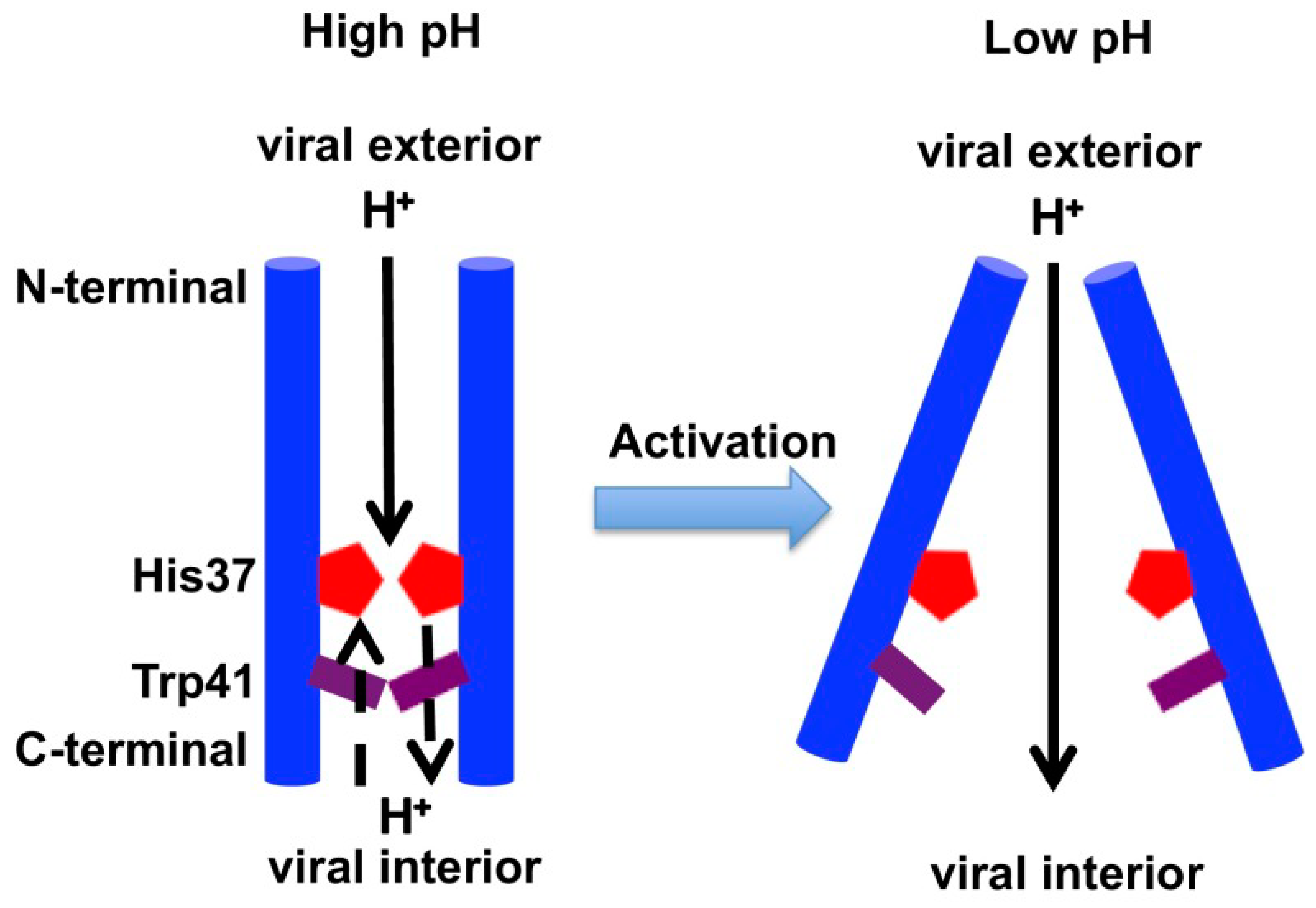
2.3. Acid Activation Mechanism of AM2
2.4. Rate of Proton Conductance in AM2 and BM2
2.5. Asymmetric Conductance in AM2 and BM2
3. CM2 and DM2
3.1. CM2
3.2. DM2
4. Other Influenza Viroporins
5. Influenza Viroporin Inhibition
6. M2-Mediated Disruption of Ion Homeostasis
7. Protein–Protein Interactions (PPIs)
7.1. Modulation of Host Autophagy
7.2. Interplay with Host Defense
7.3. Targeting by Host Restriction Factors
7.4. Modulation of Viral Replication
7.5. Modulation of Surface Expression
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ritchey, M.B.; Palese, P.; Kilbourne, E.D. RNAs of influenza A, B, and C viruses. J. Virol. 1976, 18, 738–744. [Google Scholar] [PubMed]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and swine: Proposal for a new genus in the Orthomyxoviridae family. MBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Ducatez, M.; Collin, E.A.; Ran, Z.; Liu, R.; Sheng, Z.; Armien, A.; Kaplan, B.; Chakravarty, S.; Hoppe, A.D.; et al. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013, 9, e1003176. [Google Scholar] [CrossRef] [PubMed]
- Parrish, C.R.; Murcia, P.R.; Holmes, E.C. Influenza virus reservoirs and intermediate hosts: Dogs, horses, and new possibilities for influenza virus exposure of humans. J. Virol. 2015, 89, 2990–2994. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [PubMed]
- Hay, A.J.; Gregory, V.; Douglas, A.R.; Yi, P.L. The evolution of human influenza viruses. Philos. Trans. R. Soc. B Biol. Sci. 2001, 356, 1861–1870. [Google Scholar] [CrossRef]
- Neumann, G.; Noda, T.; Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009, 459, 931–939. [Google Scholar] [CrossRef]
- Molinari, N.A.M.; Ortega-Sanchez, I.R.; Messonnier, M.L.; Thompson, W.W.; Wortley, P.M.; Weintraub, E.; Bridges, C.B. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine 2007, 25, 5086–5096. [Google Scholar] [CrossRef]
- Webster, R.G.; Monto, A.S.; Braciale, T.J.; Lamb, R.A. Textbook of Influenza; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Koutsakos, M.; Nguyen, T.H.; Barclay, W.S.; Kedzierska, K. Knowns and unknowns of influenza B viruses. Future Microbiol. 2016, 11, 119–135. [Google Scholar] [CrossRef]
- Ran, Z.; Shen, H.; Lang, Y.; Kolb, E.A.; Turan, N.; Zhu, L.; Ma, J.; Bawa, B.; Liu, Q.; Liu, H.; et al. Domestic pigs are susceptible to infection with influenza B viruses. J. Virol. 2015, 89, 4818–4826. [Google Scholar] [CrossRef]
- Osterhaus, A.D.M.E.; Rimmelzwaan, G.F.; Martina, B.E.E.; Bestebroer, T.M.; Fouchier, R.A.M. Influenza B virus in seals. Science 2000, 288, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.F. Pathogenesis of influenza D virus in cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef] [PubMed]
- Palese, P.; Shaw, M.L. Orthomyxoviridae: The viruses and their replication. Fields Virol. 2007, 1647–1689. [Google Scholar]
- Zebedee, S.L.; Lamb, R.A. Influenza A virus M2 protein: Monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 1988, 62, 2762–2772. [Google Scholar] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Avalos, R.T.; Sanderson, C.M.; Nayak, D.P. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 1996, 70, 6508–6515. [Google Scholar] [PubMed]
- Sakaguchi, T.; Tu, Q.A.; Pinto, L.H.; Lamb, R.A. The active oligomeric state of the minimalistic influenza virus M-2 ion channel is a tetramer. Proc. Natl. Acad. Sci. USA 1997, 94, 5000–5005. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Muraki, Y.; Hongo, S. The molecular virology and reverse genetics of influenza C virus. Jpn. J. Infect. Dis. 2010, 63, 157–165. [Google Scholar]
- Castano-Rodriguez, C.; Honrubia, J.M.; Gutierrez-Alvarez, J.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Verdia-Baguena, C.; Queralt-Martin, M.; et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. MBio 2018, 9. [Google Scholar] [CrossRef]
- Carrasco, L. Modification of membrane permeability by animal viruses. Adv. Virus Res. 1995, 45, 61–112. [Google Scholar]
- Nieva, J.L.; Carrasco, L. Viroporins: Structures and functions beyond cell membrane permeabilization. Viruses 2015, 7, 5169–5171. [Google Scholar] [CrossRef]
- Hyser, J.M.; Estes, M.K. Pathophysiological Consequences of Calcium-Conducting Viroporins. Annu. Rev. Virol. 2015, 2, 473–496. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Lamb, R.A.; Choppin, P.W. The gene structure and replication of influenza virus. Annu. Rev. Biochem. 1983, 52, 467–506. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, R.J.; Hay, A.J. Structural characteristics of the M2 protein of influenza a viruses: Evidence that it forms a tetrameric channel. Virology 1991, 180, 617–624. [Google Scholar] [CrossRef]
- Hull, J.D.; Gilmore, R.; Lamb, R.A. Integration of a small integral membrane protein, M2, of influenza virus into the endoplasmic reticulum: Analysis of the internal signal-anchor domain of a protein with an ectoplasmic NH2 terminus. J. Cell Biol. 1988, 106, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A. Unpacking the incoming influenza virus. Cell 1992, 69, 577–578. [Google Scholar] [CrossRef]
- Martin, K.; Helenius, A. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 1991, 65, 232–244. [Google Scholar] [CrossRef]
- Takeuchi, K.; Lamb, R.A. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 1994, 68, 911–919. [Google Scholar]
- Ciampor, F.; Bayley, P.M.; Nermut, M.V.; Hirst, E.M.A.; Sugrue, R.J.; Hay, A.J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans golgi compartment. Virology 1992, 188, 14–24. [Google Scholar] [CrossRef]
- Grambas, S.; Hay, A.J. Maturation of influenza a virus hemagglutinin-Estimates of the pH encountered during transport and its regulation by the M2 protein. Virology 1992, 190, 11–18. [Google Scholar] [CrossRef]
- Sugrue, R.J.; Bahadur, G.; Zambon, M.C.; Hall-Smith, M.; Douglas, A.R.; Hay, A.J. Specific structural alteration of the influenza haemagglutinin by amantadine. EMBO J. 1990, 9, 3469–3476. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Polishchuk, A.L.; Ohigashi, Y.; Stouffer, A.L.; Schön, A.; Magavern, E.; Jing, X.; Lear, J.D.; Freire, E.; Lamb, R.A.; et al. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl. Acad. Sci. USA 2009, 106, 12283–12288. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.H.; Dieckmann, G.R.; Gandhi, C.S.; Papworth, C.G.; Braman, J.; Shaughnessy, M.A.; Lear, J.D.; Lamb, R.A.; Degrado, W.F. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc. Natl. Acad. Sci. USA 1997, 94, 11301–11306. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zaitseva, F.; Lamb, R.A.; Pinto, L.H. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J. Biol. Chem. 2002, 277, 39880–39886. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.H.; Lamb, R.A. The M2 proton channels of influenza A and B viruses. J. Biol. Chem. 2006, 281, 8997–9000. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.; Goto, H.; Kawaoka, Y. Influenza B virus requires BM2 protein for replication. J. Virol. 2004, 78, 5576–5583. [Google Scholar] [CrossRef] [PubMed]
- Mould, J.A.; Paterson, R.G.; Takeda, M.; Ohigashi, Y.; Venkataraman, P.; Lamb, R.A.; Pinto, L.H. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell 2003, 5, 175–184. [Google Scholar] [CrossRef]
- Paterson, R.G.; Takeda, M.; Ohigashi, Y.; Pinto, L.H.; Lamb, R.A. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology 2003, 306, 7–17. [Google Scholar] [CrossRef]
- Wang, J.; Pielak, R.M.; McClintock, M.A.; Chou, J.J. Solution structure and functional analysis of the influenza B proton channel. Nat. Struct. Mol. Biol. 2009, 16, 1267–1271. [Google Scholar] [CrossRef]
- Ma, C.; Wang, J. Functional studies reveal the similarities and differences between AM2 and BM2 proton channels from influenza viruses. Biochim. Biophys. Acta Biomembr. 2018, 1860, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Odagiri, T.; Hong, J.; Ohara, Y. The BM2 protein of influenza B virus is synthesized in the late phase of infection and incorporated into virions as a subviral component. J. Gen. Virol. 1999, 80, 2573–2581. [Google Scholar] [CrossRef] [PubMed]
- Sugrue, R.J.; Belshe, R.B.; Hay, A.J. Palmitoylation of the influenza a virus M2 protein. Virology 1990, 179, 51–56. [Google Scholar] [CrossRef]
- Holsinger, L.J.; Alams, R. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology 1991, 183, 32–43. [Google Scholar] [CrossRef]
- Thomas, J.M.; Stevens, M.P.; Percy, N.; Barclay, W.S. Phosphorylation of the M2 protein of influenza A virus is not essential for virus viability. Virology 1998, 252, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.C.; Denham, E.M.; Thomas, B.; Trudgian, D.C.; Hester, S.S.; Ridlova, G.; York, A.; Turrell, L.; Fodor, E. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 2012, 8, e1002993. [Google Scholar] [CrossRef]
- Park, E.K.; Castrucci, M.R.; Portner, A.; Kawaoka, Y. The M2 ectodomain is important for its incorporation into influenza A virions. J. Virol. 1998, 72, 2449–2455. [Google Scholar]
- Acharya, R.; Carnevale, V.; Fiorin, G.; Levine, B.G.; Polishchuk, A.L.; Balannik, V.; Samish, I.; Lamb, R.A.; Pinto, L.H.; DeGrado, W.F.; et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. USA 2010, 107, 15075–15080. [Google Scholar] [CrossRef]
- Thomaston, J.L.; Polizzi, N.F.; Konstantinidi, A.; Wang, J.; Kolocouris, A.; Degrado, W.F. Inhibitors of the M2 Proton Channel Engage and Disrupt Transmembrane Networks of Hydrogen-Bonded Waters. J. Am. Chem. Soc. 2018, 140, 15219–15226. [Google Scholar] [CrossRef]
- Schnell, J.R.; Chou, J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451, 591–595. [Google Scholar] [CrossRef]
- Wang, J.; Wu, Y.; Ma, C.; Fiorin, G.; Wang, J.; Pinto, L.H.; Lamb, R.A.; Klein, M.L.; DeGrado, W.F. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. USA 2013, 110, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Schmidt-Rohr, K.; Hong, M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J. Am. Chem. Soc. 2012, 134, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Yi, M.; Dong, H.; Qin, H.; Peterson, E.; Busath, D.D.; Zhou, H.X.; Cross, T.A. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 2010, 330, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Cady, S.D.; Schmidt-Rohr, K.; Wang, J.; Soto, C.S.; Degrado, W.F.; Hong, M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Liang, R.; Li, H.; Swanson, J.M.; Voth, G.A. Multiscale simulation reveals a multifaceted mechanism of proton permeation through the influenza A M2 proton channel. Proc. Natl. Acad. Sci. USA 2014, 111, 9396–9401. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fu, R.; Nishimura, K.; Zhang, L.; Zhou, H.X.; Busath, D.D.; Vijayvergiya, V.; Cross, T.A. Histidines, heart of the hydrogen ion channel from influenza A virus: Toward an understanding of conductance and proton selectivity. Proc. Natl. Acad. Sci. USA 2006, 103, 6865–6870. [Google Scholar] [CrossRef] [PubMed]
- Mandala, V.S.; Liao, S.Y.; Kwon, B.; Hong, M. Structural Basis for Asymmetric Conductance of the Influenza M2 Proton Channel Investigated by Solid-State NMR Spectroscopy. J. Mol. Biol. 2017, 429, 2192–2210. [Google Scholar] [CrossRef]
- Ma, C.; Fiorin, G.; Carnevale, V.; Wang, J.; Lamb, R.A.; Klein, M.L.; Wu, Y.; Pinto, L.H.; Degrado, W.F. Asp44 stabilizes the Trp41 gate of the M2 proton channel of influenza a virus. Structure 2013, 21, 2033–2041. [Google Scholar] [CrossRef]
- Pielak, R.M.; Chou, J.J. Kinetic analysis of the M2 proton conduction of the influenza virus. J. Am. Chem. Soc. 2010, 132, 17695–17697. [Google Scholar] [CrossRef]
- Okada, A.; Miura, T.; Takeuchi, H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza A virus. Biochemistry 2001, 40, 6053–6060. [Google Scholar] [CrossRef]
- Colvin, M.T.; Andreas, L.B.; Chou, J.J.; Griffin, R.G. Proton association constants of his 37 in the influenza-A M218-60dimer-of-dimers. Biochemistry 2014, 53, 5987–5994. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.Y.; Yang, Y.; Tietze, D.; Hong, M. The Influenza M2 Cytoplasmic Tail Changes the Proton-Exchange Equilibria and the Backbone Conformation of the Transmembrane Histidine Residue to Facilitate Proton Conduction. J. Am. Chem. Soc. 2015, 137, 6067–6077. [Google Scholar] [CrossRef] [PubMed]
- Stouffer, A.L.; Acharya, R.; Salom, D.; Levine, A.S.; Di Costanzo, L.; Soto, C.S.; Tereshko, V.; Nanda, V.; Stayrook, S.; DeGrado, W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 2008, 451, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Thomaston, J.L.; Alfonso-Prieto, M.; Woldeyes, R.A.; Fraser, J.S.; Klein, M.L.; Fiorin, G.; DeGrado, W.F. High-resolution structures of the M2 channel from influenza A virus reveal dynamic pathways for proton stabilization and transduction. Proc. Natl. Acad. Sci. USA 2015, 112, 14260–14265. [Google Scholar] [CrossRef] [PubMed]
- Thomaston, J.L.; DeGrado, W.F. Crystal structure of the drug-resistant S31N influenza M2 proton channel. Protein Sci. 2016, 1551–1554. [Google Scholar] [CrossRef] [PubMed]
- Thomaston, J.L.; Woldeyes, R.A.; Nakane, T.; Yamashita, A.; Tanaka, T.; Koiwai, K.; Brewster, A.S.; Barad, B.A.; Chen, Y.; Lemmin, T.; et al. XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction. Proc. Natl. Acad. Sci. USA 2017, 114, 13357–13362. [Google Scholar] [CrossRef] [PubMed]
- Agmon, N. The Grotthuss mechanism. Chem. Phys. Lett. 1995, 244, 456–462. [Google Scholar] [CrossRef]
- Liang, R.; Swanson, J.M.J.; Madsen, J.J.; Hong, M.; DeGrado, W.F.; Voth, G.A. Acid activation mechanism of the influenza A M2 proton channel. Proc. Natl. Acad. Sci. USA 2016, 113, E6955–E6964. [Google Scholar] [CrossRef]
- Lin, T.; Schroeder, C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J. Virol. 2001, 75, 3647–3656. [Google Scholar] [CrossRef]
- Chizhmakov, I.V.; Ogden, D.C.; Geraghty, F.M.; Hayhurst, A.; Skinner, A.; Betakova, T.; Hay, A.J. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J. Physiol. 2003, 546, 427–438. [Google Scholar] [CrossRef]
- Mould, J.A.; Li, H.C.; Dudlak, C.S.; Lear, J.D.; Pekosz, A.; Lamb, R.A.; Pinto, L.H. Mechanism for proton conduction of the M2 ion channel of influenza A virus. J. Biol. Chem. 2000, 275, 8592–8599. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.C.; Vijayvergiya, V.; Gao, P.F.; Cross, T.A.; Woodbury, D.J.; Busath, D.D. Proton transport through influenza A virus M2 protein reconstituted in vesicles. Biophys. J. 2008, 94, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Decoursey, T.E. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 2003, 83, 475–579. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Luo, W.; Hong, M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science 2010, 330, 505–508. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.K.; Zhang, Y.; Schmidt-Rohr, K.; Hong, M. PH-dependent conformation, dynamics, and aromatic interaction of the gating tryptophan residue of the influenza M2 proton channel from solid-state NMR. Biophys. J. 2013, 104, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- DiFrancesco, M.L.; Hansen, U.P.; Thiel, G.; Moroni, A.; Schroeder, I. Effect of cytosolic pH on inward currents reveals structural characteristics of the proton transport cycle in the influenza A protein M2 in cell-free membrane patches of Xenopus oocytes. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Zhou, H.X. A theory for the proton transport of the influenza virus M2 protein: Extensive test against conductance data. Biophys. J. 2011, 100, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Markiewicz, B.N.; Lemmin, T.; Zhang, W.; Ahmed, I.A.; Jo, H.; Fiorin, G.; Troxler, T.; DeGrado, W.F.; Gai, F. Infrared and fluorescence assessment of the hydration status of the tryptophan gate in the influenza A M2 proton channel. PCCP 2016, 18, 28939–28950. [Google Scholar] [CrossRef]
- Lin, C.W.; Mensa, B.; Barniol-Xicota, M.; DeGrado, W.F.; Gai, F. Activation pH and Gating Dynamics of Influenza A M2 Proton Channel Revealed by Single-Molecule Spectroscopy. Angew. Chem. Int. Ed. Engl. 2017, 56, 5283–5287. [Google Scholar] [CrossRef]
- Miao, Y.; Fu, R.; Zhou, H.X.; Cross, T.A. Dynamic Short Hydrogen Bonds in Histidine Tetrad of Full-Length M2 Proton Channel Reveal Tetrameric Structural Heterogeneity and Functional Mechanism. Structure 2015, 23, 2300–2308. [Google Scholar] [CrossRef]
- Li, C.; Qin, H.; Gao, F.P.; Cross, T.A. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim. Biophys. Acta 2007, 1768, 3162–3170. [Google Scholar] [CrossRef] [PubMed]
- Yi, M.; Cross, T.A.; Zhou, H.X. Conformational heterogeneity of the M2 proton channel and a structural model for channel activation. Proc. Natl. Acad. Sci. USA 2009, 106, 13311–13316. [Google Scholar] [CrossRef] [PubMed]
- Khurana, E.; Dal Peraro, M.; DeVane, R.; Vemparala, S.; DeGrado, W.F.; Klein, M.L. Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc. Natl. Acad. Sci. USA 2009, 106, 1069–1074. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Pohorille, A. M2 Proton Channel: Toward a Model of a Primitive Proton Pump. Orig. Life Evol. Biosph. 2015, 45, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Qiu, J.; DeGrado, W.F.; Hochstrasser, R.M. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 6115–6120. [Google Scholar] [CrossRef]
- Ma, C.; Soto, C.S.; Ohigashi, Y.; Taylor, A.; Bournas, V.; Glawe, B.; Udo, M.K.; DeGrado, W.F.; Lamb, R.A.; Pinto, L.H. Identification of the pore-lining residues of the BM2 ion channel protein of influenza B virus. J. Biol. Chem. 2008, 283, 15921–15931. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.K.; Tietze, D.; Lee, M.; Wang, J.; Hong, M. Solid-State NMR Investigation of the Conformation, Proton Conduction, and Hydration of the Influenza B Virus M2 Transmembrane Proton Channel. J. Am. Chem. Soc. 2016, 138, 8143–8155. [Google Scholar] [CrossRef]
- Williams, J.K.; Shcherbakov, A.A.; Wang, J.; Hong, M. Protonation equilibria and pore-opening structure of the dual-histidine influenza B virus M2 transmembrane proton channel from solid-state NMR. J. Biol. Chem. 2017, 292, 17876–17884. [Google Scholar] [CrossRef]
- Hongo, S.; Sugawara, K.; Nishimura, H.; Muraki, Y.; Kitame, F.; Nakamura, K. Identification of a second protein encoded by influenza C virus RNA segment 6. J. Gen. Virol. 1994, 75, 3503–3510. [Google Scholar] [CrossRef]
- Hongo, S.; Ishii, K.; Mori, K.; Takashita, E.; Muraki, Y.; Matsuzaki, Y.; Sugawara, K. Detection of ion channel activity in Xenopus laevis oocytes expressing Influenza C virus CM2 protein. Arch. Virol. 2004, 149, 35–50. [Google Scholar] [CrossRef]
- Pekosz, A.; Lamb, R.A. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc. Natl. Acad. Sci. USA 1998, 95, 13233–13238. [Google Scholar] [CrossRef] [PubMed]
- Hongo, S.; Sugawara, K.; Muraki, Y.; Matsuzaki, Y.; Takashita, E.; Kitame, F.; Nakamura, K. Influenza C virus CM2 protein is produced from a 374-amino-acid protein (P42) by signal peptidase cleavage. J. Virol. 1999, 73, 46–50. [Google Scholar] [PubMed]
- Pekosz, A.; Lamb, R.A. The CM2 protein of influenza C virus is an oligomeric integral membrane glycoprotein structurally analogous to influenza A virus M2 and influenza B virus NB proteins. Virology 1997, 237, 439–451. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hongo, S.; Sugawara, K.; Muraki, Y.; Kitame, F.; Nakamura, K. Characterization of a second protein (CM2) encoded by RNA segment 6 of influenza C virus. J. Virol. 1997, 71, 2786–2792. [Google Scholar] [PubMed]
- Muraki, Y.; Okuwa, T.; Himeda, T.; Hongo, S.; Ohara, Y. Effect of Cysteine Mutations in the Extracellular Domain of CM2 on the Influenza C Virus Replication. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; Hongo, S.; Muraki, Y.; Matsuzaki, Y.; Sugawara, K.; Kitame, F.; Nakamura, K. Phosphorylation of influenza C virus CM2 protein. Virus Res. 1998, 58, 65–72. [Google Scholar] [CrossRef]
- Kukol, A.; Arkin, I.T. Structure of the Influenza C virus CM2 protein transmembrane domain obtained by site-specific infrared dichroism and global molecular dynamics searching. J. Biol. Chem. 2000, 275, 4225–4229. [Google Scholar] [CrossRef] [PubMed]
- Muraki, Y.; Washioka, H.; Sugawara, K.; Matsuzaki, Y.; Takashita, E.; Hongo, S. Identification of an amino acid residue on influenza C virus M1 protein responsible for formation of the cord-like structures of the virus. J. Gen. Virol. 2004, 85, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Kukol, A.; Arkin, I.T. Mapping the energy surface of transmembrane helix-helix interactions. Biophys. J. 2001, 81, 2681–2692. [Google Scholar] [CrossRef]
- Betakova, T.; Hay, A.J. Evidence that the CM2 protein of influenza C virus can modify the pH of the exocytic pathway of transfected cells. J. Gen. Virol. 2007, 88, 2291–2296. [Google Scholar] [CrossRef]
- Stewart, S.M.; Pekosz, A. The influenza C virus CM2 protein can alter intracellular pH, and: Its transmembrane domain can substitute for that of the influenza a virus M2 protein and support infectious virus production. J. Virol. 2012, 86, 1277–1281. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohuchi, M.; Ohuchi, R.; Mifune, K. Demonstration of hemolytic and fusion activities of influenza C virus. J. Virol. 1982, 42, 1076–1079. [Google Scholar] [PubMed]
- Chizhmakov, I.V.; Geraghty, F.M.; Ogden, D.C.; Hayhurst, A.; Antoniou, M.; Hay, A.J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996, 494, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Mould, J.A.; Drury, J.E.; Fring, S.M.; Kaupp, U.B.; Pekosz, A.; Lamb, R.A.; Pinto, L.H. Permeation and activation of the M2 ion channel of influenza A virus. J. Biol. Chem. 2000, 275, 31038–31050. [Google Scholar] [CrossRef] [PubMed]
- Betáková, T.; Kollerová, E. pH modulating activity of ion channels of influenza A, B, and C viruses. Acta Virol. 2006, 50, 187–193. [Google Scholar] [PubMed]
- Zhirnov, O.P.; Grigoriev, V.B. Disassembly of influenza C viruses, distinct from that of influenza A and B viruses requires neutral-alkaline pH. Virology 1994, 200, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Muraki, Y.; Noda, T.; Takashita, E.; Sho, R.; Sugawara, K.; Matsuzaki, Y.; Shimotai, Y.; Hongo, S. Role of the CM2 protein in the influenza C virus replication cycle. J. Virol. 2011, 85, 1322–1329. [Google Scholar] [CrossRef]
- Kesinger, E.; Liu, J.; Jensen, A.; Chia, C.P.; Demers, A.; Moriyama, H. Influenza D virus M2 protein exhibits ion channel activity in Xenopus laevis oocytes. PLoS ONE 2018, 13, e0199227. [Google Scholar] [CrossRef]
- Henkel, M.; Mitzner, D.; Henklein, P.; Meyer-Almes, F.J.; Moroni, A.; DiFrancesco, M.L.; Henkes, L.M.; Kreim, M.; Kast, S.M.; Schubert, U.; et al. Proapoptotic influenza A virus protein PB1-F2 forms a nonselective ion channel. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Chen, W.; Calvo, P.A.; Malide, D.; Gibbs, J.; Schubert, U.; Bacik, I.; Basta, S.; O’Neill, R.; Schickli, J.; Palese, P.; et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001, 7, 1306–1312. [Google Scholar] [CrossRef]
- Zamarin, D.; García-Sastre, A.; Xiao, X.; Wang, R.; Palese, P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Path. 2005, 1, 0040–0054. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.T.; Grant, A.; Manicassamy, B.; Palese, P. Influenza virus protein pb1-f2 inhibits the induction of type I interferon by binding to mavs and decreasing mitochondrial membrane potential. J. Virol. 2012, 86, 8359–8366. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.L.; Tate, M.D.; MacKenzie-Kludas, C.J.; Pinar, A.; Zeng, W.; Stutz, A.; Latz, E.; Brown, L.E.; Mansell, A. Activation of the NLRP3 Inflammasome by IAV Virulence Protein PB1-F2 Contributes to Severe Pathophysiology and Disease. PLoS Path. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.A.; Lamb, R.A. Effects of mutations and deletions in a bicistronic mRNA on the synthesis of influenza B virus NB and NA glycoproteins. J. Virol. 1989, 63, 28–35. [Google Scholar] [PubMed]
- Demers, A.; Ran, Z.; Deng, Q.; Wang, D.; Edman, B.; Lu, W.; Li, F. Palmitoylation is required for intracellular trafficking of influenza B virus NB protein and efficient influenza B virus growth in vitro. J. Gen. Virol. 2014, 95, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Sunstrom, N.A.; Premkumar, L.S.; Premkumar, A.; Ewart, G.; Cox, G.B.; Gage, P.W. Ion channels formed by NB, an influenza B virus protein. J. Membr. Biol. 1996, 150, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Premkumar, A.; Ewart, G.D.; Cox, G.B.; Gage, P.W. An amino-acid substitution in the influenza-B NB protein affects ion-channel gating. J. Membr. Biol. 2004, 197, 135–143. [Google Scholar] [CrossRef]
- Hatta, M.; Kawaoka, Y. The NB protein of influenza B virus is not necessary for virus replication in vitro. J. Virol. 2003, 77, 6050–6054. [Google Scholar] [CrossRef]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef]
- Davies, W.L.; Grunert, R.R.; Haff, R.F.; McGahen, J.W.; Neumayer, E.M.; Paulshock, M.; Watts, J.C.; Wood, T.R.; Hermann, E.C.; Hoffmann, C.E. Antiviral Activity of 1-Adamantanamine (Amantadine). Science 1964, 144, 862–863. [Google Scholar] [CrossRef]
- Pielak, R.M.; Oxenoid, K.; Chou, J.J. Structural Investigation of Rimantadine Inhibition of the AM2-BM2 Chimera Channel of Influenza Viruses. Structure 2011, 19, 1655–1663. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.J.; Wolstenholme, A.J.; Skehel, J.J.; Smith, M.H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985, 4, 3021–3024. [Google Scholar] [CrossRef] [PubMed]
- Vanderlinden, E.; Naesens, L. Emerging Antiviral Strategies to Interfere with Influenza Virus Entry. Med. Res. Rev. 2014, 34, 301–339. [Google Scholar] [CrossRef] [PubMed]
- Deyde, V.M.; Xu, X.; Bright, R.A.; Shaw, M.; Smith, C.B.; Zhang, Y.; Shu, Y.; Gubareva, L.V.; Cox, N.J.; Klimov, A.I. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 2007, 196, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Bright, R.A.; Shay, D.K.; Shu, B.; Cox, N.J.; Klimov, A.I. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA 2006, 295, 891–894. [Google Scholar] [CrossRef] [PubMed]
- Bright, R.A.; Medina, M.J.; Xu, X.; Perez-Oronoz, G.; Wallis, T.R.; Davis, X.M.; Povinelli, L.; Cox, N.J.; Klimov, A.I. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: A cause for concern. Lancet 2005, 366, 1175–1181. [Google Scholar] [CrossRef]
- Hayden, F.G.; De Jong, M.D. Emerging influenza antiviral resistance threats. J. Infect. Dis. 2011, 203, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Fiore, A.E.; Fry, A.; Shay, D.; Gubareva, L.; Bresee, J.S.; Uyeki, T.M.; Centers for Disease Control Prevention. Antiviral agents for the treatment and chemoprophylaxis of influenza—Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 2011, 60, 1–24. [Google Scholar]
- Li, F.; Ma, C.; DeGrado, W.F.; Wang, J. Discovery of Highly Potent Inhibitors Targeting the Predominant Drug-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel. J. Med. Chem. 2016, 59, 1207–1216. [Google Scholar] [CrossRef]
- Li, F.; Ma, C.; Hu, Y.; Wang, Y.; Wang, J. Discovery of Potent Antivirals against Amantadine-Resistant Influenza A Viruses by Targeting the M2-S31N Proton Channel. ACS Infect. Dis. 2016, 2, 726–733. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, J.; Wang, J. Pharmacological Characterization of the Spectrum of Antiviral Activity and Genetic Barrier to Drug Resistance of M2-S31N Channel Blockers. Mol. Pharmacol. 2016, 90, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Musharrafieh, R.; Ma, C.; Zhang, J.; Smee, D.F.; DeGrado, W.F.; Wang, J. An M2-V27A channel blocker demonstrates potent in vitro and in vivo antiviral activities against amantadine-sensitive and -resistant influenza A viruses. Antivir. Res. 2017, 140, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Peng, C.; Luo, J.; Wang, C.; Han, L.; Wu, B.; Ji, G.; He, H. Adamantane-resistant influenza a viruses in the world (1902-2013): Frequency and distribution of M2 gene mutations. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Cross, T.A.; Zhou, H.X. Recent progress in structure-based anti-influenza drug design. Drug Discov. Today 2012, 17, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Gu, R.X.; Liu, L.A.; Wei, D.Q. Structural and energetic analysis of drug inhibition of the influenza A M2 proton channel. Trends Pharmacol. Sci. 2013, 34, 571–580. [Google Scholar] [CrossRef]
- To, J.; Surya, W.; Torres, J. Targeting the Channel Activity of Viroporins. Adv. Prot. Chem. Struct. Biol. 2016, 104, 307–355. [Google Scholar]
- Gandhi, C.S.; Shuck, K.; Lear, J.D.; Dieckmann, G.R.; DeGrado, W.F.; Lamb, R.A.; Pinto, L.H. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M 2 protein. J. Biol. Chem. 1999, 274, 5474–5482. [Google Scholar] [CrossRef]
- Su, Y.; Hu, F.; Hong, M. Paramagnetic Cu(II) for probing membrane protein structure and function: Inhibition mechanism of the influenza M2 proton channel. J. Am. Chem. Soc. 2012, 134, 8693–8702. [Google Scholar] [CrossRef][Green Version]
- Gordon, N.A.; McGuire, K.L.; Wallentine, S.K.; Mohl, G.A.; Lynch, J.D.; Harrison, R.G.; Busath, D.D. Divalent copper complexes as influenza A M2 inhibitors. Antivir. Res. 2017, 147, 100–106. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Iwasaki, A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 2010, 11, 404–410. [Google Scholar] [CrossRef]
- Lazrak, A.; Iles, K.E.; Liu, G.; Noah, D.L.; Noah, J.W.; Matalon, S. Influenza virus M2 protein inhibits epithelial sodium channels by increasing reactive oxygen species. FASEB J. 2009, 23, 3829–3842. [Google Scholar] [CrossRef] [PubMed]
- Londino, J.D.; Lazrak, A.; Jurkuvenaite, A.; Collawn, J.F.; Noah, J.W.; Matalon, S. Influenza matrix protein 2 alters CFTR expression and function through its ion channel activity. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L582–L592. [Google Scholar] [CrossRef] [PubMed]
- Matalon, S.; O’Brodovich, H. Sodium channels in alveolar epithelial cells: Molecular characterization, biophysical properties, and physiological significance. Annu. Rev. Physiol. 1999, 61, 627–661. [Google Scholar] [CrossRef] [PubMed]
- Rogan, M.P.; Stoltz, D.A.; Hornick, D.B. Cystic fibrosis transmembrane conductance regulator intracellular processing, trafficking, and opportunities for mutation-specific treatment. Chest 2011, 139, 1480–1490. [Google Scholar] [CrossRef] [PubMed]
- Londino, J.D.; Lazrak, A.; Collawn, J.F.; Bebok, Z.; Harrod, K.S.; Matalon, S. Influenza virus infection alters ion channel function of airway and alveolar cells: Mechanisms and physiological sequelae. J. Physiol. Lung Cell. Mol. Physiol. 2017, 313, L845–L858. [Google Scholar] [CrossRef]
- To, J.; Torres, J. Beyond Channel Activity: Protein-Protein Interactions Involving Viroporins. Sub-Cell. Biochem. 2018, 88, 329–377. [Google Scholar] [CrossRef]
- Gannagé, M.; Dormann, D.; Albrecht, R.; Dengjel, J.; Torossi, T.; Rämer, P.C.; Lee, M.; Strowig, T.; Arrey, F.; Conenello, G.; et al. Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion with Lysosomes. Cell Host Microbe 2009, 6, 367–380. [Google Scholar] [CrossRef]
- Bourmakina, S.V.; García-Sastre, A. Reverse genetics studies on the filamentous morphology of influenza a virus. J. Gen. Virol. 2003, 84, 517–527. [Google Scholar] [CrossRef]
- Beale, R.; Wise, H.; Stuart, A.; Ravenhill, B.J.; Digard, P.; Randow, F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 2014, 15, 239–247. [Google Scholar] [CrossRef]
- Meurs, E.; Chong, K.; Galabru, J.; Thomas, N.S.B.; Kerr, I.M.; Williams, B.R.G.; Hovanessian, A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 1990, 62, 379–390. [Google Scholar] [CrossRef]
- Lu, Y.; Wambach, M.; Katze, M.G.; Krug, R.M. Binding of the Influenza Virus NS1 Protein to Double-Stranded RNA Inhibits the Activation of the Protein Kinase That Phosphorylates the eIF-2 Translation Initiation Factor. Virology 1995, 214, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Tripathi, S.; Ranjan, P.; Kumar, P.; Garten, R.; Deyde, V.; Katz, J.M.; Cox, N.J.; Lal, R.B.; Sambhara, S.; et al. Influenza a virus nucleoprotein exploits Hsp40 to inhibit PKR activation. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Liu, D.; Mi, S.; Zhang, J.; Ye, Q.; Wang, M.; Gao, G.F.; Yan, J. Interaction of Hsp40 with influenza virus M2 protein: Implications for PKR signaling pathway. Protein Cell 2010, 1, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Le Tortorec, A.; Willey, S.; Neil, S.J.D. Antiviral inhibition of enveloped virus release by Tetherin/BST-2: Action and counteraction. Viruses 2011, 3, 520–540. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yin, L.; Mei, S.; Li, J.; Xu, F.; Sun, H.; Liu, X.; Cen, S.; Liang, C.; Li, A.; et al. BST-2 restricts IAV release and is countered by the viral M2 protein. Biochem. J. 2017, 474, 715–730. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Kien, F.; Manière, M.; Zhang, Y.; Lagarde, N.; Tse, K.S.; Poon, L.L.M.; Nal, B. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J. Virol. 2012, 86, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Mok, C.K.P.; Chan, M.C.W.; Zhang, Y.; Nal, B.; Kien, F.; Bruzzone, R.; Sanyal, S. Cell Cycle-independent Role of Cyclin D3 in Host Restriction of Influenza Virus Infection. J. Biol. Chem. 2017, 292, 5070–5088. [Google Scholar] [CrossRef]
- Rudick, M.; Anderson, R.G.W. Multiple functions of caveolin-1. J. Biol. Chem. 2002, 277, 41295–41298. [Google Scholar]
- Sun, L.; Hemgård, G.V.; Susanto, S.A.; Wirth, M. Caveolin-1 influences human influenza A virus (H1N1) multiplication in cell culture. Virol. J. 2010, 7. [Google Scholar] [CrossRef]
- Couet, J.; Li, S.; Okamoto, T.; Ikezu, T.; Lisanti, M.P. Identification of peptide and protein ligands for the caveolin- scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J. Biol. Chem. 1997, 272, 6525–6533. [Google Scholar] [CrossRef]
- Amarelle, L.; Lecuona, E. The antiviral effects of na,K-ATPase inhibition: A minireview. Int. J. Mol. Sci. 2018, 19, 2154. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.W.; Kuok, D.I.T.; Leung, C.Y.H.; Hui, K.P.Y.; Valkenburg, S.A.; Lau, E.H.Y.; Nicholls, J.M.; Fang, X.; Guan, Y.; Lee, J.W.; et al. Human mesenchymal stromal cells reduce influenza A H5N1-associated acute lung injury in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 3621–3626. [Google Scholar] [CrossRef]
- Hoffmann, H.H.; Palese, P.; Shaw, M.L. Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antivir. Res. 2008, 80, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Dowall, S.D.; Bewley, K.; Watson, R.J.; Vasan, S.S.; Ghosh, C.; Konai, M.M.; Gausdal, G.; Lorens, J.B.; Long, J.; Barclay, W.; et al. Antiviral screening of multiple compounds against ebola virus. Viruses 2016, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.F.; Li, Y.; Yan, J.H.; Gao, G.F. Na+/K+-ATPase β1 subunit interacts with M2 proteins of influenza A and B viruses and affects the virus replication. Sci. China Life. Sci. 2010, 53, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Doms, R.W.; Lamb, R.A.; Rose, J.K.; Helenius, A. Folding and assembly of viral membrane proteins. Virology 1993, 193, 545–562. [Google Scholar] [CrossRef] [PubMed]
- Hughey, P.G.; Compans, R.W.; Zebedee, S.L.; Lamb, R.A. Expression of the influenza A virus M2 protein Is restricted to apical surfaces of polarized epithelial cells. J. Virol. 1992, 66, 5542–5552. [Google Scholar] [PubMed]
- Tasaki, T.; Mulder, L.C.F.; Iwamatsu, A.; Lee, M.J.; Davydov, I.V.; Varshavsky, A.; Muesing, M.; Kwon, Y.T. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005, 25, 7120–7136. [Google Scholar] [CrossRef]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; Dejesus, P.; Che, J.; Mulder, L.C.F.; et al. Meta- and Orthogonal Integration of Influenza “oMICs“ Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18, 723–735. [Google Scholar] [CrossRef]
- Bruce, E.A.; Digard, P.; Stuart, A.D. The Rab11 pathway is required for influenza A virus budding and filament formation. J. Virol. 2010, 84, 5848–5859. [Google Scholar] [CrossRef]
- Chen, W.; Feng, Y.; Chen, D.; Wandinger-Ness, A. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol. Biol. Cell 1998, 9, 3241–3257. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, O.; Reinsch, S.; Urbé, S.; Zerial, M.; Parton, R.G. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 1996, 135, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Urbé, S.; Huber, L.A.; Zerial, M.; Tooze, S.A.; Parton, R.G. Rab11, a small GTPase associated with both constitutive and regulated secretory pathways in PC12 cells. FEBS Lett. 1993, 334, 175–182. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Liang, L.; Shao, X.; Luo, W.; Jiang, S.; Zhao, Q.; Sun, N.; Zhao, Y.; Li, J.; Wang, J.; et al. Host cellular protein TRAPPC6AΔ interacts with influenza A virus M2 protein and regulates viral propagation by modulating M2 trafficking. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Sacher, M.; Jiang, Y.; Barrowman, J.; Scarpa, A.; Burston, J.; Zhang, L.; Schieltz, D.; Yates Iii, J.R.; Abeliovich, H.; Ferro-Novick, S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998, 17, 2494–2503. [Google Scholar] [CrossRef] [PubMed]
- Heaton, N.S.; Moshkina, N.; Fenouil, R.; Gardner, T.J.; Aguirre, S.; Shah, P.S.; Zhao, N.; Manganaro, L.; Hultquist, J.F.; Noel, J.; et al. Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity 2016, 44, 46–58. [Google Scholar] [CrossRef]
- Wang, L.; Fu, B.; Li, W.; Patil, G.; Liu, L.; Dorf, M.E.; Li, S. Comparative influenza protein interactomes identify the role of plakophilin 2 in virus restriction. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Watanabe, T.; Kawakami, E.; Shoemaker, J.E.; Lopes, T.J.S.; Matsuoka, Y.; Tomita, Y.; Kozuka-Hata, H.; Gorai, T.; Kuwahara, T.; Takeda, E.; et al. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 2014, 16, 795–805. [Google Scholar] [CrossRef]
| Influenza Type | Host Range | Viroporin | Coding RNA | Typical Length | Functional Motif | Ion Conductance |
|---|---|---|---|---|---|---|
| A | Humans, aquatic birds, porcine, bovine, canine | AM2 PB1-F2 | Segment 7 Segment 2 | 97 aa 90 aa | HxxxW -- | H+, K+ Ca2+, MVC |
| B | Humans, pigs, harbor seals | BM2 NB | Segment 7 Segment 6 | 109 aa 100 aa | HxxxW -- | H+, K+ ND |
| C | Humans, pigs | CM2 | Segment 6 | 115 aa | YxxxK | Cl− |
| D | Cattle, pigs | DM2 | Segment 6 | 152 aa | YxxxK | Cl− |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
To, J.; Torres, J. Viroporins in the Influenza Virus. Cells 2019, 8, 654. https://doi.org/10.3390/cells8070654
To J, Torres J. Viroporins in the Influenza Virus. Cells. 2019; 8(7):654. https://doi.org/10.3390/cells8070654
Chicago/Turabian StyleTo, Janet, and Jaume Torres. 2019. "Viroporins in the Influenza Virus" Cells 8, no. 7: 654. https://doi.org/10.3390/cells8070654
APA StyleTo, J., & Torres, J. (2019). Viroporins in the Influenza Virus. Cells, 8(7), 654. https://doi.org/10.3390/cells8070654





