Sugar Transporter Proteins (STPs) in Gramineae Crops: Comparative Analysis, Phylogeny, Evolution, and Expression Profiling
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Plant Treatments
2.3. Identification and Phylogenetic Analysis of STP Genes
2.4. Gene Duplication Events, Orthologous Groups’ Analysis, Gene Structure, and Conserved Motifs
2.5. Quantitative Real-Time PCR (qRT-PCR)
3. Results
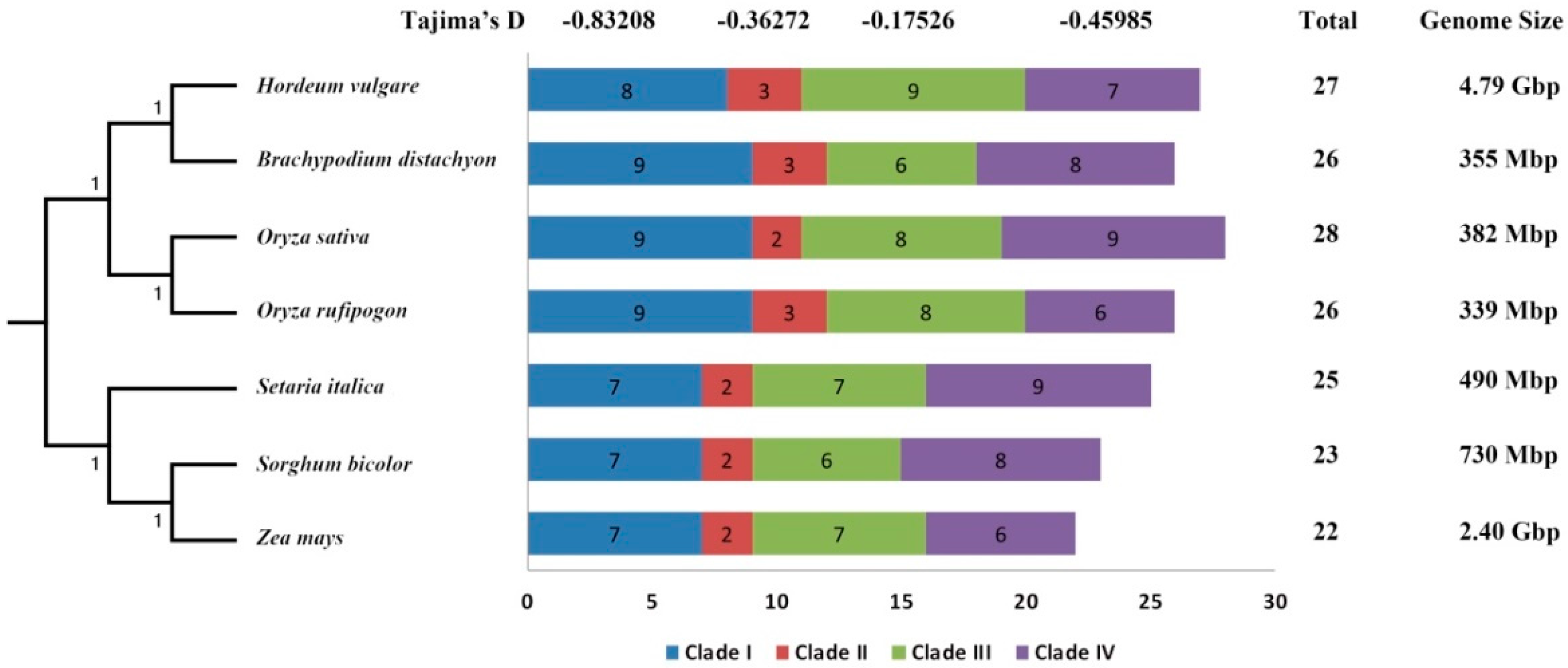
3.1. Identification and Comparative Phylogenetic of STP Genes in Seven Representative Gramineae Crops
3.2. Homologous Relationship, Gene Structure, and Selective Forces
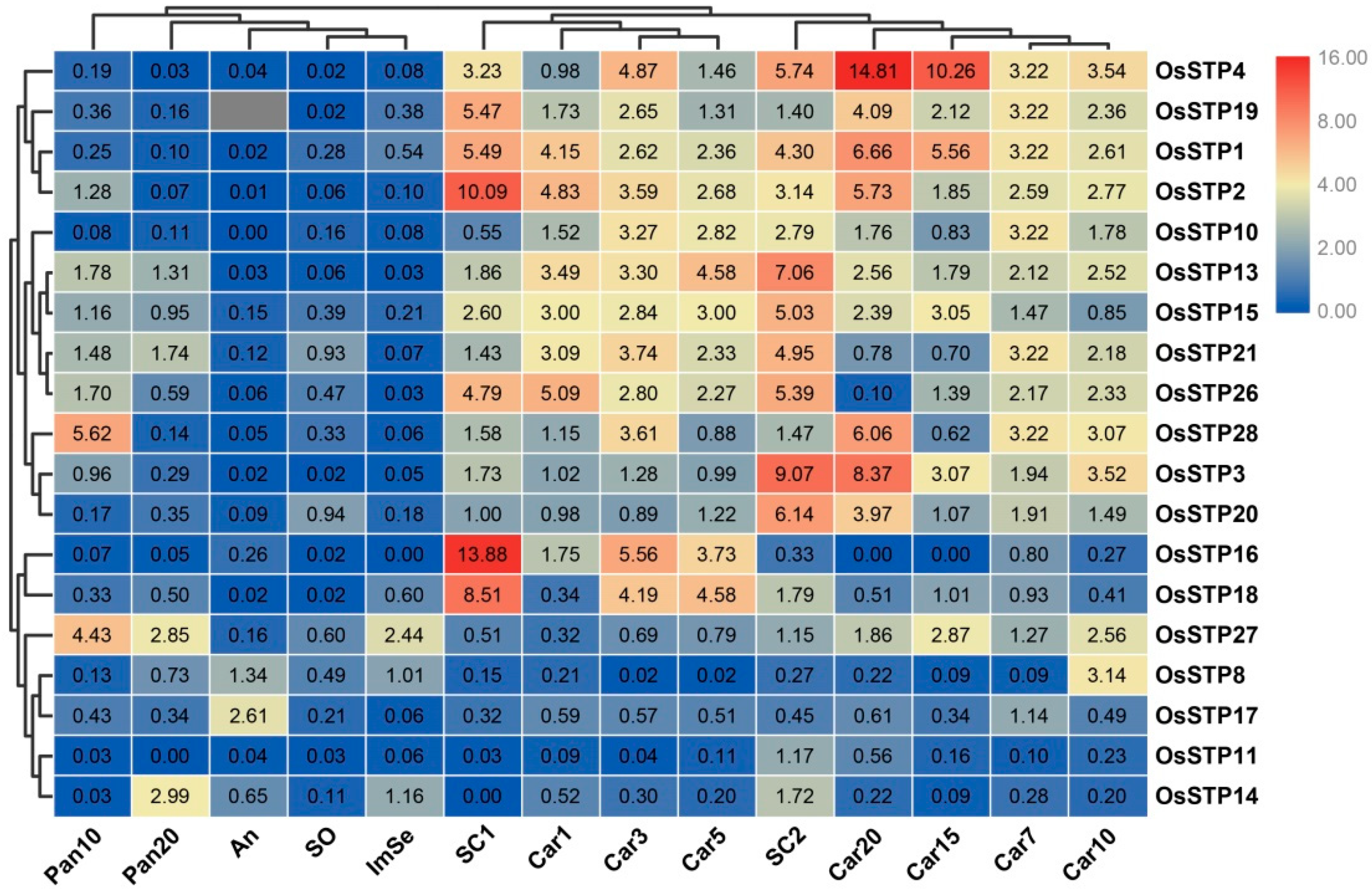
3.3. Expression Profiling of Rice STP Genes in 14 Tissues
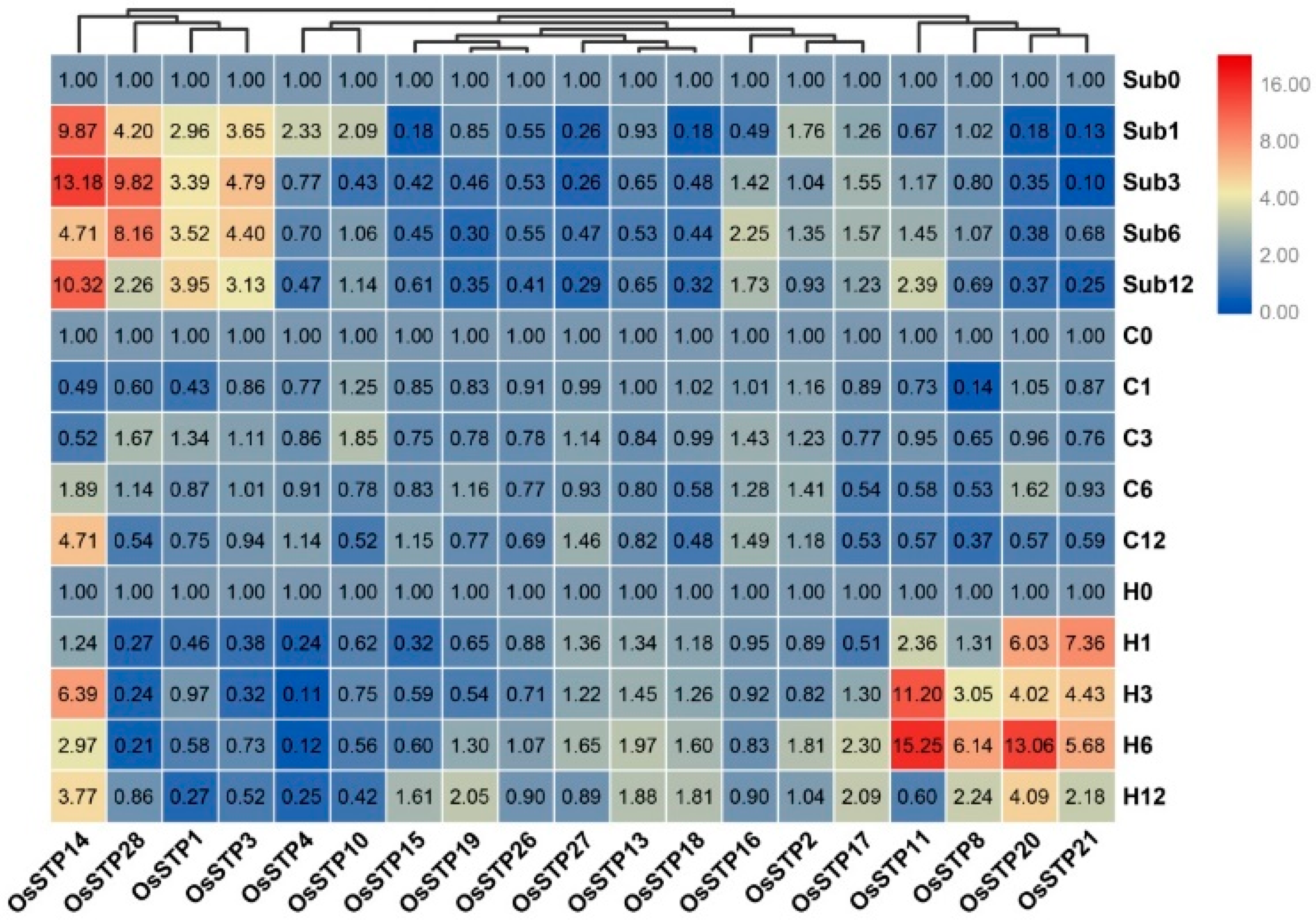
3.4. Expression Changes of STP Genes in Rice Seedling under Cold, High Temperature, and Submergence Stress
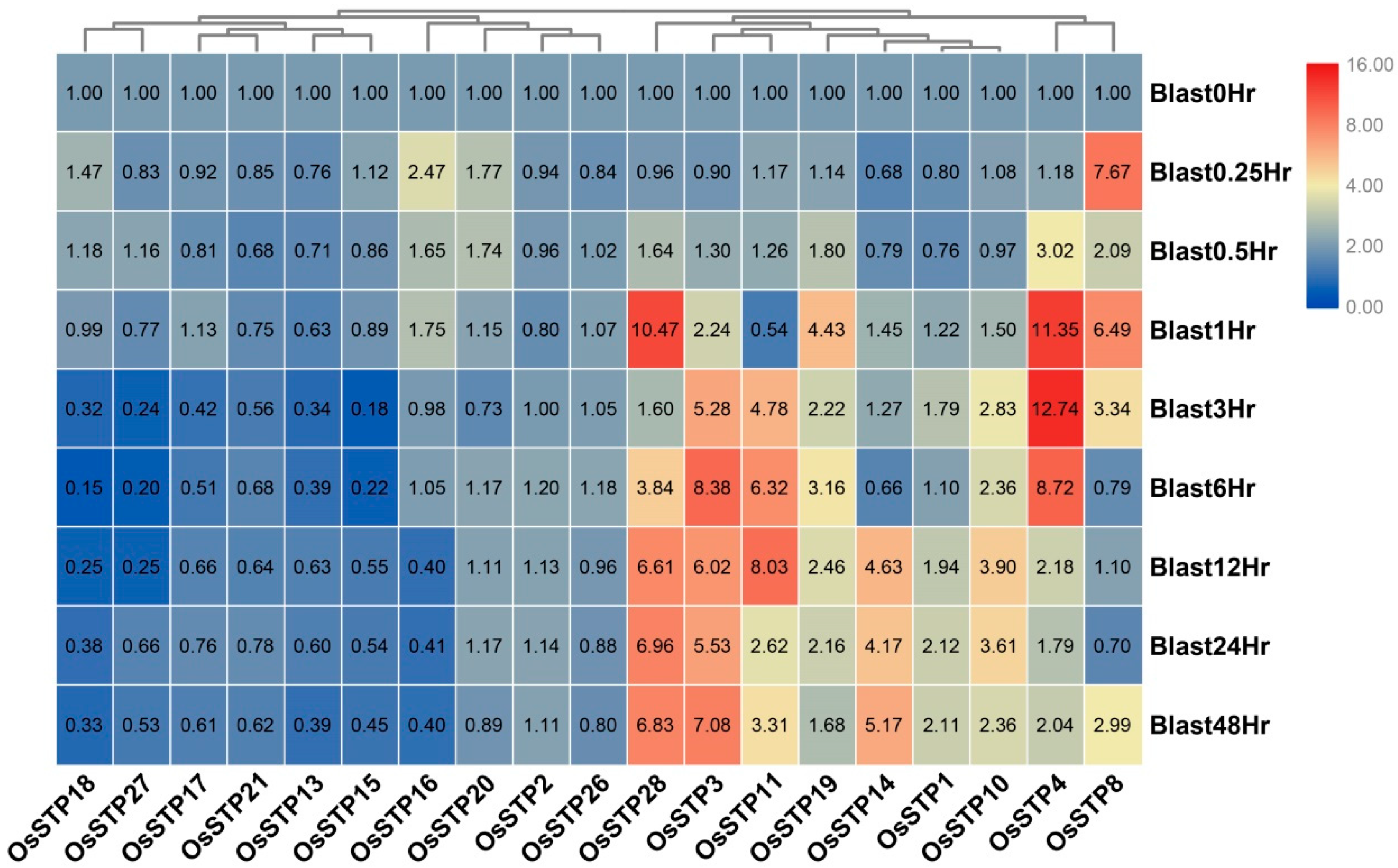
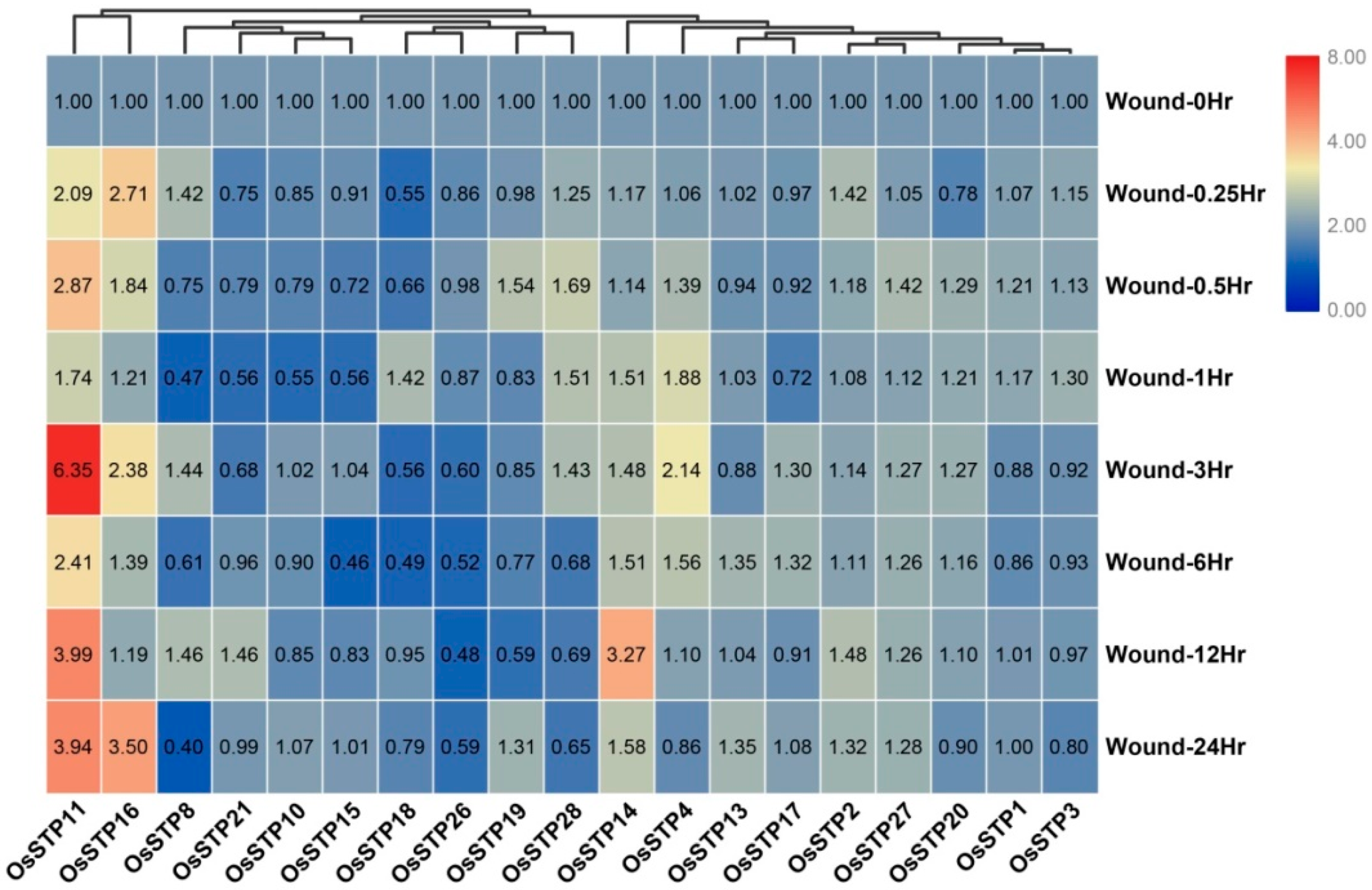
3.5. Expression Changes of STP Genes in Rice under Blast Infection and Wounding Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rolland, F.; Moore, B.; Sheen, J. Sugar sensing and signaling in plants. Plant Cell 2002, 14, S185–S205. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, H.A.; Smeekens, S. Sugar sensing and signaling in plants. Front. Plant Sci. 2014, 5, 113. [Google Scholar]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 38, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A novel insight into functional divergence of the MST gene family in rice based on comprehensive expression patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, S.; Yu, F.; Tang, J.; Yu, L.; Wang, H.; Li, J. Genome-wide identification and expression profiling of sugar transporter protein (STP) family genes in cabbage (Brassica oleracea var. capitata L.) reveals their involvement in clubroot disease responses. Genes 2019, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yuan, B.; Hou, S.; Li, X. Sugar transport played a more important role than sugar biosynthesis in fruit sugar accumulation during Chinese jujube domestication. Planta 2018, 248, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Durand, M.; Mainson, D.; Porcheron, B.; Maurousset, L.; Lemoine, R.; Pourtau, N. Carbon source–sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 2018, 247, 587–611. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527. [Google Scholar] [CrossRef]
- Doidy, J.; Grace, E.; Kühn, C.; Simon-Plas, F.; Casieri, L.; Wipf, D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012, 17, 413–422. [Google Scholar] [CrossRef]
- Ludewig, F.; Flügge, U.-I. Role of metabolite transporters in source-sink carbon allocation. Front. Plant Sci. 2013, 4, 231. [Google Scholar] [CrossRef]
- Liu, Q.; Dang, H.; Chen, Z.; Wu, J.; Chen, Y.; Chen, S.; Luo, L. Genome-wide identification, expression, and functional analysis of the sugar transporter gene family in Cassava (Manihot esculenta). Int. J. Mol. Sci. 2018, 19, 987. [Google Scholar] [CrossRef] [PubMed]
- Alexander, S.; Joachim, S.S.; Norbert, S.; Michael, B. AtSTP11, a pollen tube-specific monosaccharide transporter in Arabidopsis. Planta 2005, 221, 48–55. [Google Scholar]
- Büttner, M. The Arabidopsis sugar transporter (AtSTP) family: An update. Plant Biol. 2010, 12, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, K.; Kasahara, M.; Yamaguchi, J. Characterization and expression of monosaccharide transporters (OsMSTs) in rice. Plant Cell Physiol. 2000, 41, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Afoufa-Bastien, D.; Medici, A.; Jeauffre, J.; Coutos-Thévenot, P.; Lemoine, R.; Atanassova, R.; Laloi, M. The Vitis vinifera sugar transporter gene family: Phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 2010, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Reuscher, S.; Akiyama, M.; Yasuda, T.; Makino, H.; Aoki, K.; Shibata, D.; Shiratake, K. The sugar transporter inventory of tomato: Genome-wide identification and expression analysis. Plant Cell Physiol. 2014, 55, 1123–1141. [Google Scholar] [CrossRef] [PubMed]
- Büttner, M. The monosaccharide transporter (-like) gene family in Arabidopsis. FEBS Lett. 2007, 581, 2318–2324. [Google Scholar] [CrossRef] [PubMed]
- Julius, B.T.; Leach, K.A.; Tran, T.M.; Mertz, R.A.; Braun, D.M. Sugar transporters in plants: New insights and discoveries. Plant Cell Physiol. 2017, 58, 1442–1460. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.I.; Burla, B.; Lee, D.W.; Ryoo, N.; Hong, S.K.; Kim, H.B.; Eom, J.S.; Choi, S.B.; Cho, M.H.; Bhoo, S.H. Expression analysis and functional characterization of the monosaccharide transporters, OsTMTs, involving vacuolar sugar transport in rice (Oryza sativa). New Phytologist 2010, 186, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, H.; Wei, X.; Chai, C.; Xiao, Y.; Zhang, Y.; Chen, B.; Xiao, G.; Ouwerkerk, P.B.; Wang, M. Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, Y.; Zhang, Y.; Chai, C.; Wei, G.; Wei, X.; Xu, H.; Wang, M.; Ouwerkerk, P.B.; Zhu, Z. Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa L.). Planta 2008, 228, 525. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.; Beyhl, D.; Hedrich, R.; Sauer, N. Functional and physiological characterization of Arabidopsis INOSITOL TRANSPORTER1, a novel tonoplast-localized transporter for myo-inositol. Plant Cell 2008, 20, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Poschet, G.; Hannich, B.; Büttner, M. Identification and characterization of AtSTP14, a novel galactose transporter from Arabidopsis. Plant Cell Physiol. 2010, 51, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Sherson, S.M.; Hemmann, G.; Wallace, G.; Forbes, S.; Germain, V.; Stadler, R.; Bechtold, N.; Sauer, N.; Smith, S.M. Monosaccharide/proton symporter AtSTP1 plays a major role in uptake and response of Arabidopsis seeds and seedlings to sugars. Plant J. 2000, 24, 849–857. [Google Scholar] [CrossRef]
- Truernit, E.; Schmid, J.; Epple, P.; Illig, J.; Sauer, N. The sink-specific and stress-regulated Arabidopsis STP4 gene: Enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 1996, 8, 2169–2182. [Google Scholar] [CrossRef] [PubMed]
- Büttner, M.; Truernit, E.; Baier, K.; Scholz-Starke, J.; Sontheim, M.; Lauterbach, C.; Huss, V.; Sauer, N. AtSTP3, a green leaf-specific, low affinity monosaccharide-H+ symporter of Arabidopsis thaliana. Plant Cell Environ. 2000, 23, 175–184. [Google Scholar] [CrossRef]
- Yamada, K.; Saijo, Y.; Nakagami, H.; Takano, Y. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science 2016, 354, 1427–1430. [Google Scholar] [CrossRef]
- Mendgen, K.; Hahn, M. Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 2002, 7, 352–356. [Google Scholar] [CrossRef]
- Tetlow, I.; Emes, M. Starch biosynthesis in the developing endosperms of grasses and cereals. Agronomy 2017, 7, 81. [Google Scholar] [CrossRef]
- Watanabe, N.; Nakazono, M.; Kanno, A.; Tsutsumi, N.; Hirai, A. Evolutionary variations in DNA sequences transferred from chloroplast genomes to mitochondrial genomes in the Gramineae. Curr. Genetics 1994, 26, 512. [Google Scholar] [CrossRef]
- An, B.; Lan, J.; Deng, X.; Chen, S.; Ouyang, C.; Shi, H.; Yang, J.; Li, Y. Silencing of D-lactate dehydrogenase impedes glyoxalase system and leads to methylglyoxal accumulation and growth inhibition in rice. Front. Plant Sci. 2017, 8, 2071. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhong, H.; Deng, X.; Gautam, M.; Gong, Z.; Zhang, Y.; Zhao, G.; Liu, C.; Li, Y. Evolutionary analysis of GH3 genes in six Oryza species/subspecies and their expression under salinity stress in Oryza sativa ssp. japonica. Plants 2019, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar]
- Londo, J.P.; Chiang, Y.C.; Hung, K.H.; Chiang, T.Y.; Schaal, B.A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Nat. Acad. Sci. USA 2006, 103, 9578–9583. [Google Scholar] [CrossRef]
- Swigoňová, Z.; Lai, J.; Ma, J.; Ramakrishna, W.; Llaca, V.; Bennetzen, J.L.; Messing, J. Close split of sorghum and maize genome progenitors. Genome Res. 2004, 14, 1916–1923. [Google Scholar] [CrossRef] [PubMed]
- Coghlan, A.; Eichler, E.E.; Oliver, S.G.; Paterson, A.H.; Stein, L. Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genetics 2005, 21, 673–682. [Google Scholar] [CrossRef]
- Kong, W.; Bendahmane, M.; Fu, X. Genome-wide identification and characterization of aquaporins and their role in the flower opening processes in carnation (Dianthus caryophyllus). Molecules 2018, 23, 1895. [Google Scholar] [CrossRef]

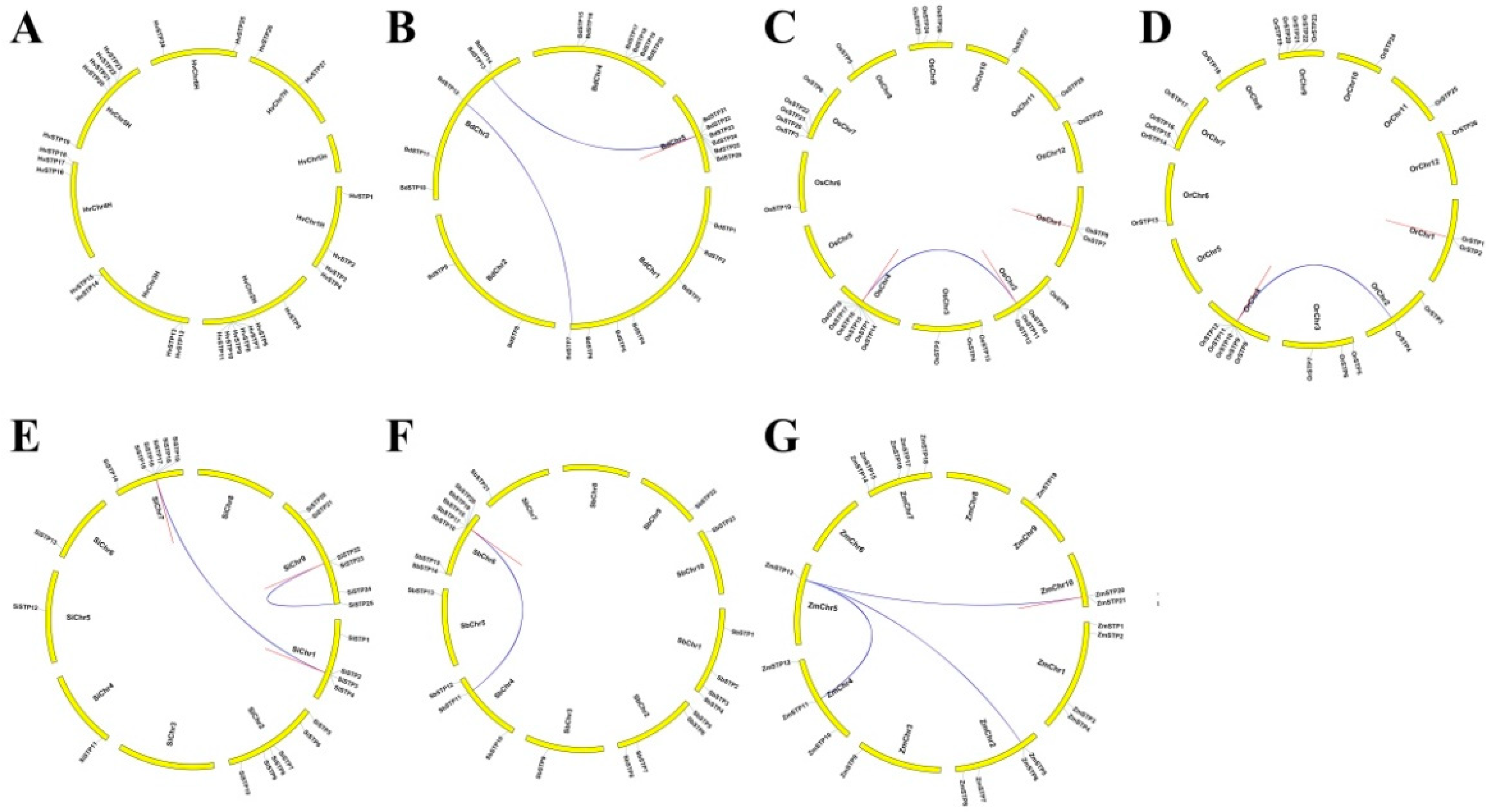
| Seq_1 | Seq_2 | Ka/Ks | Duplication Type | Date (MY) | Purifying Selection |
|---|---|---|---|---|---|
| BdSTP21 | BdSTP22 | 0.4413 | Tandem duplication | 25.29 | YES |
| BdSTP7 | BdSTP12 | 0.1415 | WGD or segmental duplication | 60.90 | YES |
| BdSTP13 | BdSTP21 | 0.6124 | WGD or segmental duplication | 30.94 | YES |
| OrSTP1 | OrSTP2 | 0.5062 | Tandem duplication | 6.58 | YES |
| OrSTP8 | OrSTP9 | 0.6519 | Tandem duplication | 28.34 | YES |
| OrSTP9 | OrSTP10 | 0.6026 | Tandem duplication | 20.73 | YES |
| OrSTP10 | OrSTP11 | 0.7493 | Tandem duplication | 25.73 | YES |
| OrSTP4 | OrSTP10 | 0.6994 | WGD or segmental duplication | 31.39 | YES |
| OsSTP8 | OsSTP7 | 0.5424 | Tandem duplication | 6.24 | YES |
| OsSTP11 | OsSTP12 | 0.3240 | Tandem duplication | 5.57 | YES |
| OsSTP14 | OsSTP1 | 0.5108 | Tandem duplication | 21.25 | YES |
| OsSTP1 | OsSTP15 | 0.7193 | Tandem duplication | 23.42 | YES |
| OsSTP10 | OsSTP18 | 0.5693 | WGD or segmental duplication | 31.75 | YES |
| SbSTP17 | SbSTP18 | 0.4488 | Tandem duplication | 23.42 | YES |
| SbSTP18 | SbSTP19 | 0.7516 | Tandem duplication | 25.57 | YES |
| SbSTP11 | SbSTP16 | 0.6396 | WGD or segmental duplication | 30.31 | YES |
| SiSTP22 | SiSTP23 | 0.2625 | Tandem duplication | 26.71 | YES |
| SiSTP16 | SiSTP17 | 0.5320 | Tandem duplication | 20.80 | YES |
| SiSTP17 | SiSTP18 | 0.4016 | Tandem duplication | 23.18 | YES |
| SiSTP18 | SiSTP19 | 0.6358 | Tandem duplication | 28.01 | YES |
| SiSTP2 | SiSTP3 | 0.4998 | Tandem duplication | 20.83 | YES |
| SiSTP3 | SiSTP4 | 0.3743 | Tandem duplication | 15.09 | YES |
| SiSTP2 | SiSTP15 | 0.6225 | WGD or segmental duplication | 30.39 | YES |
| SiSTP4 | SiSTP18 | 0.5679 | WGD or segmental duplication | 33.62 | YES |
| SiSTP22 | SiSTP25 | 0.1653 | WGD or segmental duplication | 50.11 | YES |
| ZmSTP20 | ZmSTP21 | 0.5103 | Tandem duplication | 35.43 | YES |
| ZmSTP20 | ZmSTP13 | 0.5835 | WGD or segmental duplication | 34.12 | YES |
| ZmSTP5 | ZmSTP13 | 0.5345 | WGD or segmental duplication | 35.53 | YES |
| ZmSTP11 | ZmSTP13 | 0.3853 | WGD or segmental duplication | 29.68 | YES |
| Bd | Hv | Or | Os | Sb | Si | Zm | Total | Tajima’s D | |
|---|---|---|---|---|---|---|---|---|---|
| OG1 | 6 | 4 | 4 | 4 | 4 | 4 | 3 | 29 | −0.6892 |
| OG2 | 2 | 4 | 3 | 3 | 2 | 1 | 2 | 17 | −0.49247 |
| OG3 | 2 | 1 | 2 | 2 | 4 | 4 | 2 | 17 | −0.57229 |
| OG4 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 15 | −0.56846 |
| OG5 | 2 | 1 | 1 | 3 | 1 | 3 | 2 | 13 | −0.75888 |
| OG6 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 10 | −0.20414 |
| OG7 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 10 | −0.17804 |
| OG8 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 9 | −1.00743 |
| OG9 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 9 | −0.72613 |
| OG10 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | −0.15393 |
| OG11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | −0.23779 |
| OG12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | −0.44667 |
| OG13 | 1 | 1 | 1 | 0 | 0 | 1 | 2 | 6 | −0.72003 |
| OG14 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 6 | −0.14611 |
| OG15 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 6 | −0.23085 |
| OG16 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 4 | −0.84257 |
| OG17 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, W.; An, B.; Zhang, Y.; Yang, J.; Li, S.; Sun, T.; Li, Y. Sugar Transporter Proteins (STPs) in Gramineae Crops: Comparative Analysis, Phylogeny, Evolution, and Expression Profiling. Cells 2019, 8, 560. https://doi.org/10.3390/cells8060560
Kong W, An B, Zhang Y, Yang J, Li S, Sun T, Li Y. Sugar Transporter Proteins (STPs) in Gramineae Crops: Comparative Analysis, Phylogeny, Evolution, and Expression Profiling. Cells. 2019; 8(6):560. https://doi.org/10.3390/cells8060560
Chicago/Turabian StyleKong, Weilong, Baoguang An, Yue Zhang, Jing Yang, Shuangmiao Li, Tong Sun, and Yangsheng Li. 2019. "Sugar Transporter Proteins (STPs) in Gramineae Crops: Comparative Analysis, Phylogeny, Evolution, and Expression Profiling" Cells 8, no. 6: 560. https://doi.org/10.3390/cells8060560
APA StyleKong, W., An, B., Zhang, Y., Yang, J., Li, S., Sun, T., & Li, Y. (2019). Sugar Transporter Proteins (STPs) in Gramineae Crops: Comparative Analysis, Phylogeny, Evolution, and Expression Profiling. Cells, 8(6), 560. https://doi.org/10.3390/cells8060560






