bHLH Transcription Factor Math6 Antagonizes TGF-β Signalling in Reprogramming, Pluripotency and Early Cell Fate Decisions
Abstract
1. Introduction
2. Materials and Methods
2.1. Successful Generation of Math6Flag-Tag Mice using CRISPR-CAS9 Technology
2.2. Cell Culture
2.3. Generation of Mouse iPS Cells
2.4. Alkaline Phosphatase Staining
2.5. Calculation of Reprogramming Efficiency
2.6. Isolation of Mouse Blastocysts
2.7. Culturing iPSCs and ESCs
2.8. Chromosomal Analysis
2.9. Morphometric Analysis
2.10. RNA isolation, Reverse Transcription and Real-time PCR (RT-PCR)
2.11. Immunostaining
2.12. Western Blot
2.13. Statistical Analysis
3. Results
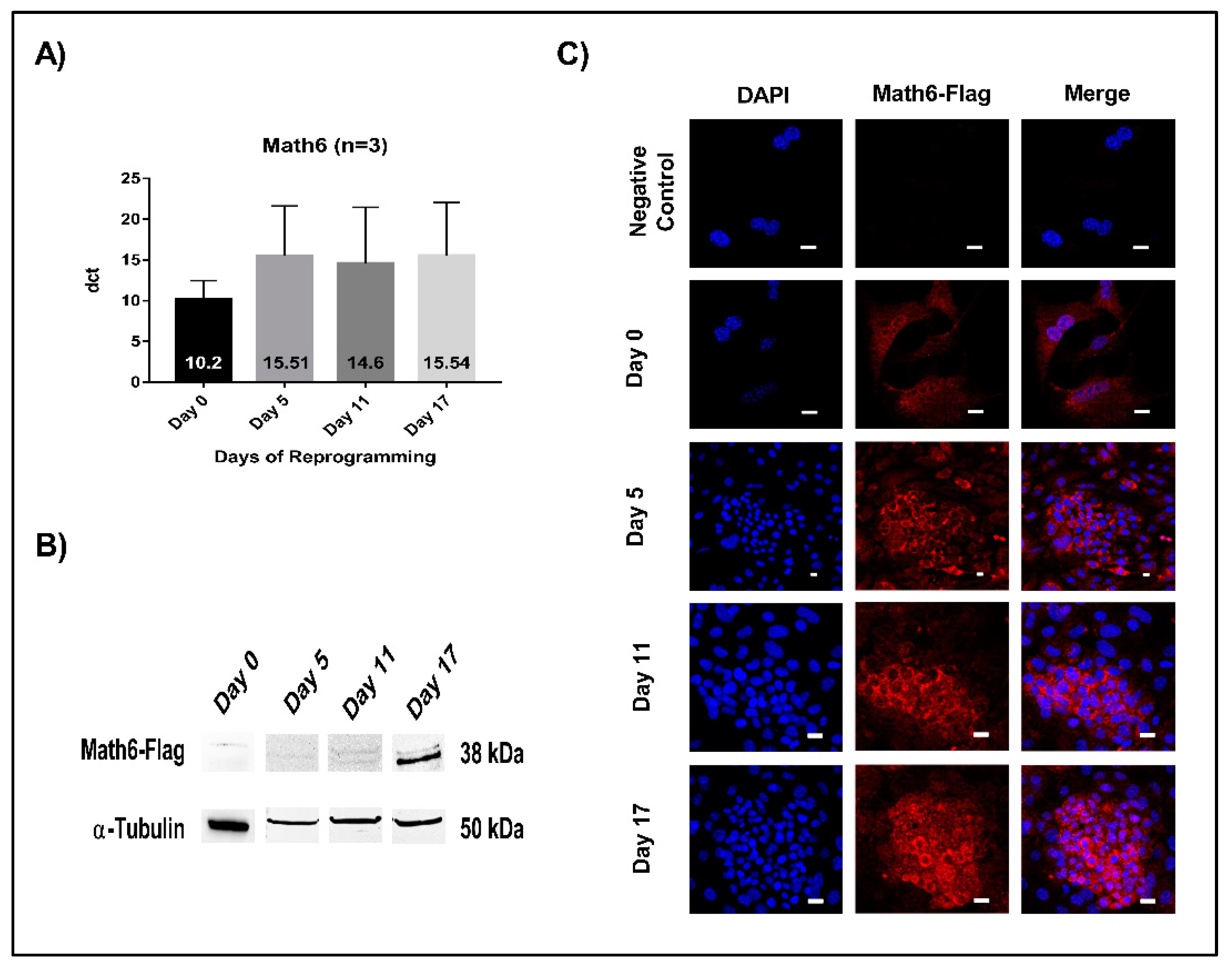
3.1. Math6 Expression during Somatic Cell Reprogramming
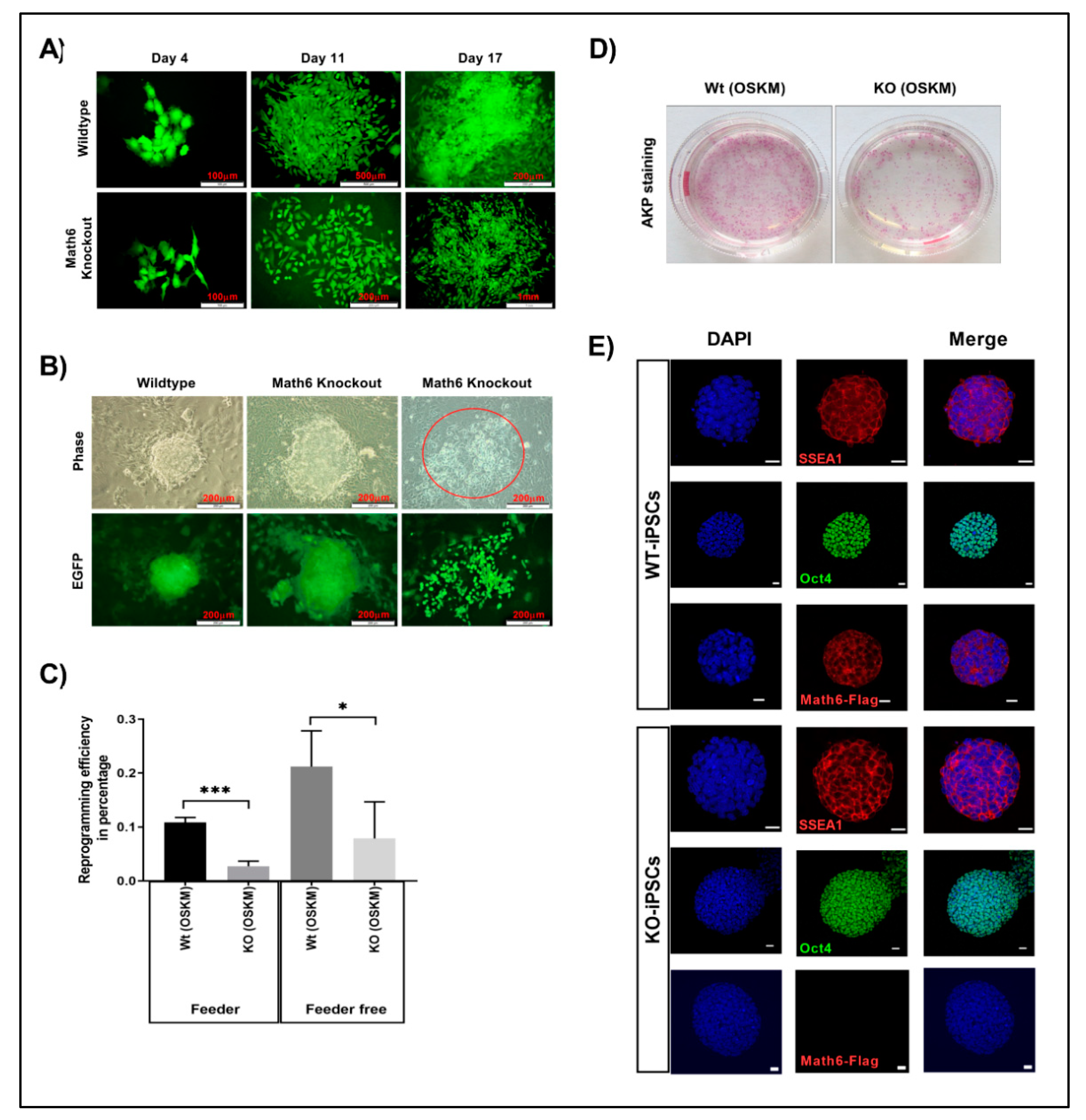
3.2. Math6 Promotes Somatic Cell Reprogramming
3.3. Lack of Math6 Disrupts Mesenchymal-to-Epithelial Transition (MET) and Somatic Cell Reprogramming
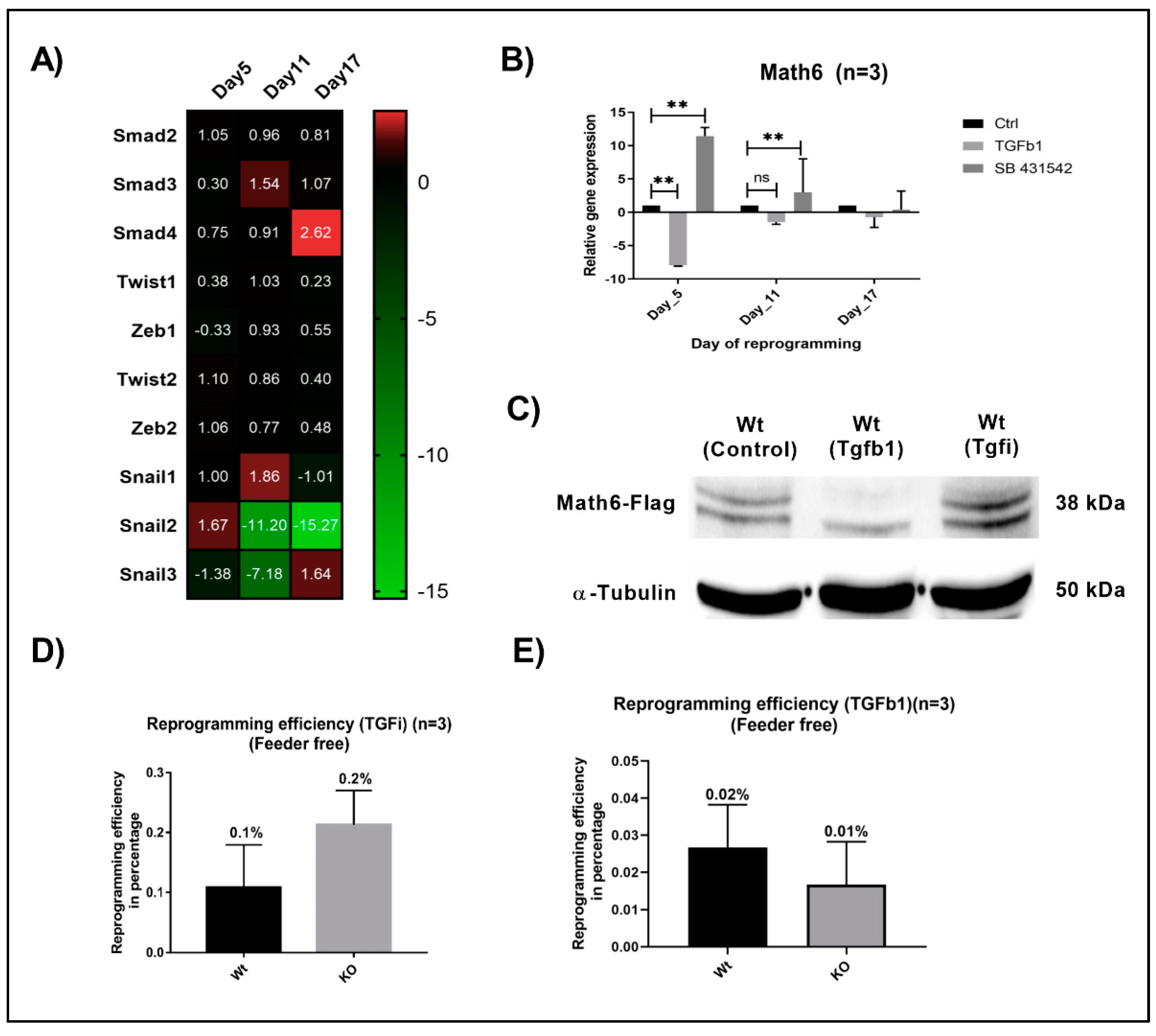
3.4. Math6 Enables MET by Antagonizing TGF-β Signalling
3.5. Math6 Sustains the Naive Pluripotent State of iPSCs
3.6. Isolation and Stabilization of WT and Math6 KO ESCs
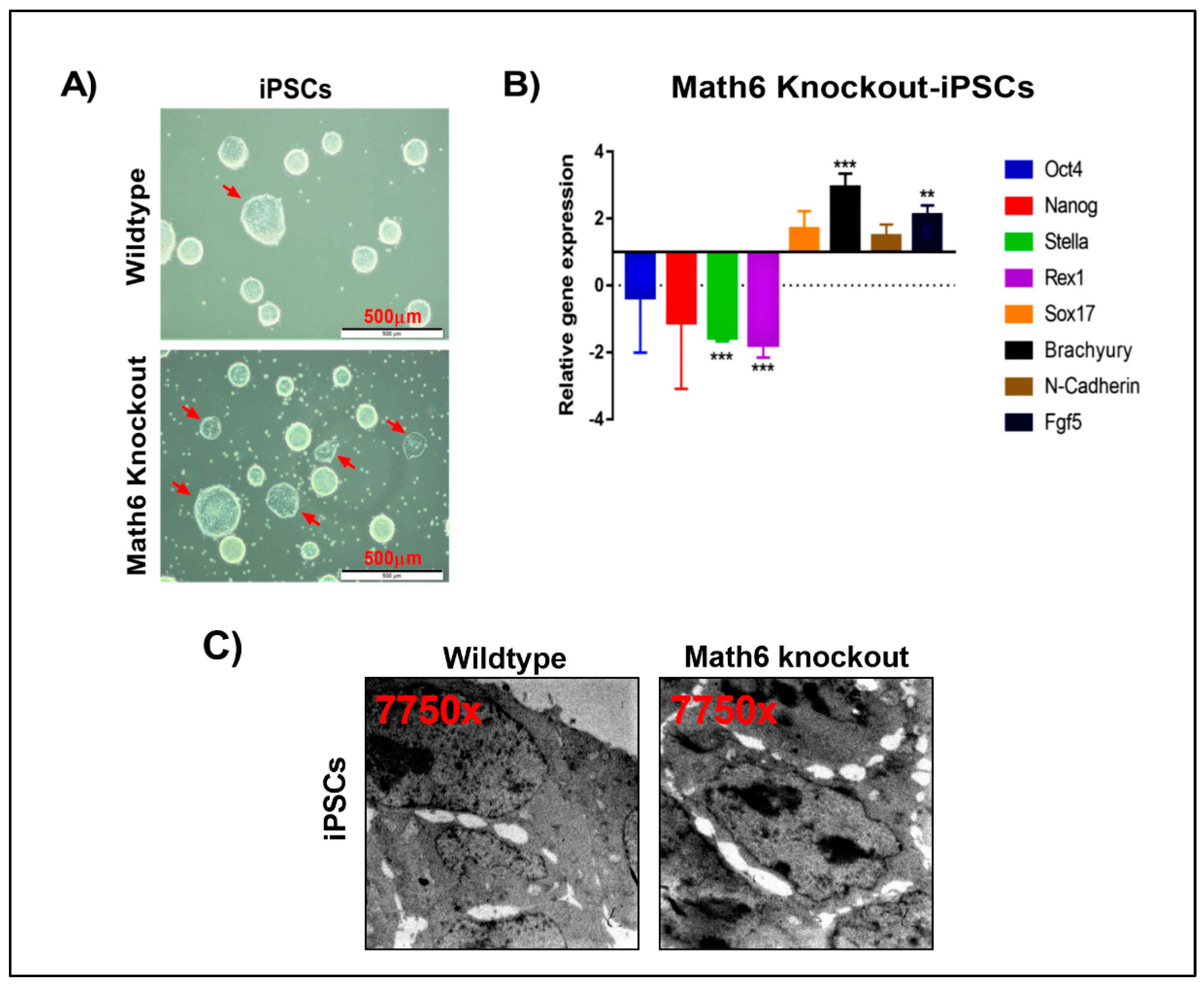
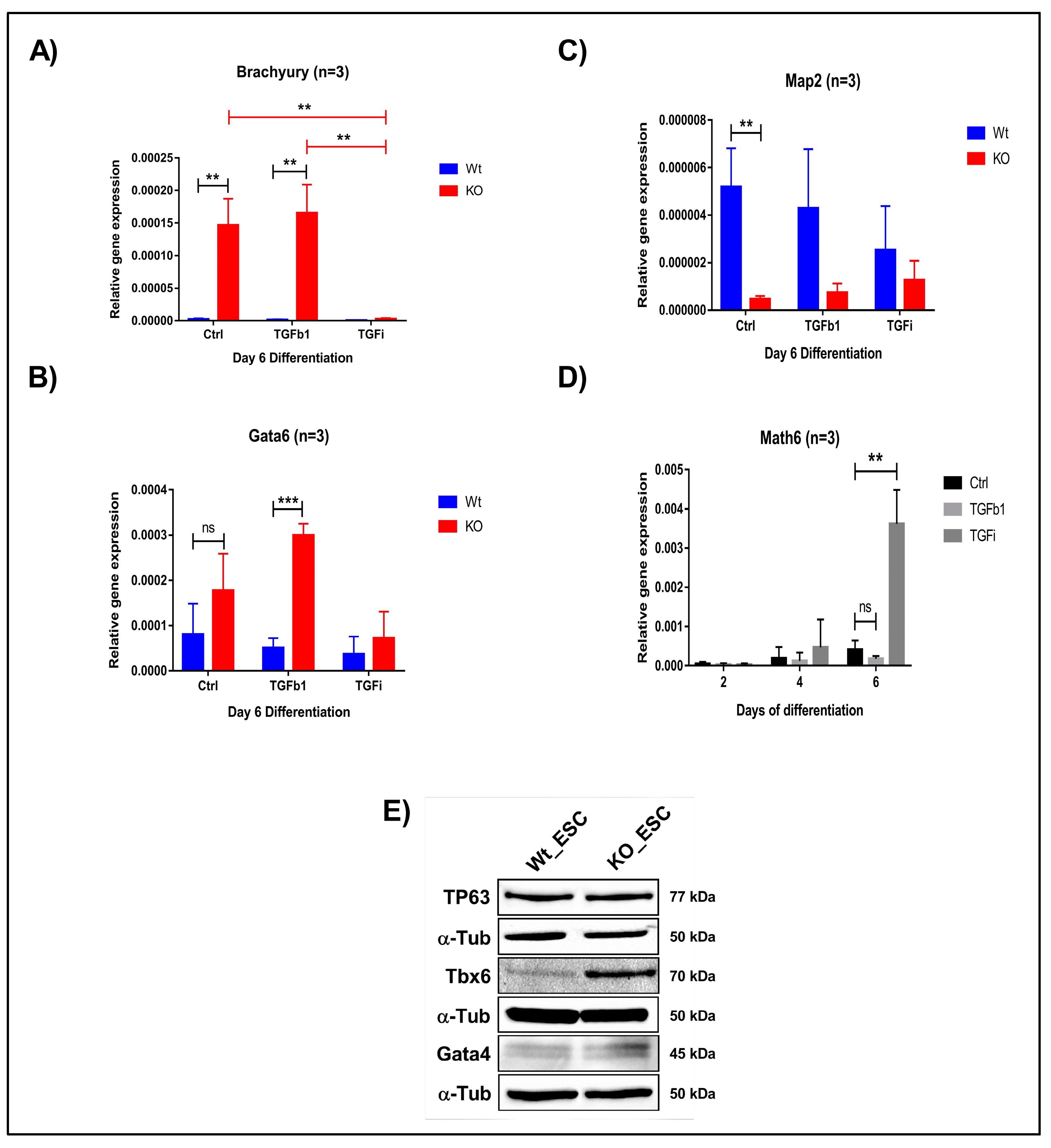
3.7. Lack of Math6 Primes Pluripotent Stem Cells into the Mesodermal Lineage
3.8. Inhibition of TGF-β Signalling Restored the Naive Pluripotent State in Math6 KO iPSCs and ESCs
3.9. Lack of Math6 Results in the Mesendodermal Specification of Pluripotent Stem Cells
4. Discussion
4.1. Math6 in Somatic Cell Reprogramming
4.2. Math6 in the Maintenance of Pluripotency and Early Differentiation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bergstrom, D.A.; Tapscott, S.J. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol. Cell. Biol. 2001, 21, 2404–2412. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Ohtsuka, T.; Hatakeyama, J.; Ohsawa, R. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 2005, 306, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, K.; Yao, Q.; Zheng, X.; Yang, Z. Phylogenetic analysis of zebrafish basic helix-loop-helix transcription factors. J. Mol. Evol. 2009, 68, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Simionato, E.; Ledent, V.; Richards, G.; Thomas-Chollier, M.; Kerner, P.; Coornaert, D.; Degnan, B.M.; Vervoort, M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: Insights from comparative genomics. BMC Evol. Biol. 2007, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Inoue, C.; Bae, S.K.; Takatsuka, K.; Inoue, T.; Bessho, Y.; Kageyama, R. Math6, a bHLH gene expressed in the developing nervous system, regulates neuronal versus glial differentiation. Genes Cells 2001, 6, 977–986. [Google Scholar] [CrossRef]
- Gasa, R.; Mrejen, C.; Leachman, N.; Otten, M.; Barnes, M.; Wang, J.; Chakrabarti, S.; Mirmira, R.; German, M. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 13245–13250. [Google Scholar] [CrossRef]
- Ross, M.D.; Martinka, S.; Mukherjee, A.; Sedor, J.R.; Vinson, C.; Bruggeman, L.A. Math6 expression during kidney development and altered expression in a mouse model of glomerulosclerosis. Dev. Dyn. 2006, 235, 3102–3109. [Google Scholar] [CrossRef]
- Balakrishnan-Renuka, A.; Morosan-Puopolo, G.; Yusuf, F.; Abduelmula, A.; Chen, J.; Zoidl, G.; Philippi, S.; Dai, F.; Brand-Saberi, B. ATOH8, a regulator of skeletal myogenesis in the hypaxial myotome of the trunk. Histochem. Cell Biol. 2014, 141, 289–300. [Google Scholar] [CrossRef]
- Rawnsley, D.R.; Xiao, J.; Lee, J.S.; Liu, X.; Mericko-Ishizuka, P.; Kumar, V.; He, J.; Basu, A.; Lu, M.; Lynn, F.C.; et al. The transcription factor Atonal homolog 8 regulates Gata4 and Friend of Gata-2 during vertebrate development. J. Biol. Chem. 2013, 288, 24429–24440. [Google Scholar] [CrossRef]
- Böing, M.; Brand-Saberi, B.; Napirei, M. Murine transcription factor Math6 is a regulator of placenta development. Sci. Rep. 2018, 8, 14997. [Google Scholar] [CrossRef]
- Wang, B.; Balakrishnan-Renuka, A.; Napirei, M.; Theiss, C.; Brand-Saberi, B. Spatiotemporal expression of Math6 during mouse embryonic development. Histochem. Cell Biol. 2015, 143, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Freire, P.; Vilela, M.; Deus, H.; Kim, Y.-W.; Koul, D.; Colman, H.; Aldape, K.D.; Bogler, O.; Yung, W.K.A.; Coombes, K.; et al. Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS ONE 2008, 3, e4076. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Zhu, Y.; Han, C.; Xu, Q.; Wu, J.; Dai, B.; Zhang, H.; Shi, G.; Gu, W.; Ye, D. Identification and validation of an eight-gene expression signature for predicting high Fuhrman grade renal cell carcinoma. Int. J. Cancer 2017, 140, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.; Wang, S.; Sang, Y.; Sun, P.; Lal, B.; Goodwin, C.R.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Laterra, J.; Xia, S. Regulation of glioblastoma stem cells by retinoic acid: Role for Notch pathway inhibition. Oncogene 2011, 30, 3454–3467. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Pan, G.; Chen, L.; Ma, S.; Zeng, T.; Man Chan, T.H.; Li, L.; Lian, Q.; Chow, R.; Cai, X.; et al. Loss of ATOH8 Increases Stem Cell Features of Hepatocellular Carcinoma Cells. Gastroenterology 2015, 149, 1068-81.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xie, J.; Yan, M.; Wang, J.; Wang, X.; Zhang, J.; Zhang, Y.; Li, P.; Lei, X.; Huang, Q.; et al. Downregulation of ATOH8 induced by EBV-encoded LMP1 contributes to the malignant phenotype of nasopharyngeal carcinoma. Oncotarget 2016, 7, 26765–26779. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Wasserman, S.M.; Torres-Vazquez, J.; Weinstein, B.; Cao, F.; Li, Z.; Wilson, K.D.; Yue, W.; Wu, J.C.; Xie, X.; et al. The role of Hath6, a newly identified shear-stress-responsive transcription factor, in endothelial cell differentiation and function. J. Cell Sci. 2014, 127, 1428–1440. [Google Scholar] [CrossRef] [PubMed]
- Van der Bogt, K.E.A.; Sheikh, A.Y.; Schrepfer, S.; Hoyt, G.; Cao, F.; Ransohoff, K.J.; Swijnenburg, R.-J.; Pearl, J.; Lee, A.; Fischbein, M.; et al. Comparison of different adult stem cell types for treatment of myocardial ischemia. Circulation 2008, 118, S121–S129. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, J.; Drews, K.; Adjaye, J. Preparation of mouse embryonic fibroblast cells suitable for culturing human embryonic and induced pluripotent stem cells. J. Vis. Exp. 2012. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.-L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Behringer, R. Manipulating the mouse embryo. A laboratory manual/Richard Behringer, University of Texas, M.D. Anderson Cancer Center [and three others], 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2014; ISBN 9781936113019. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Buganim, Y.; Faddah, D.A.; Jaenisch, R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013, 14, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.-K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Inman, G.J.; Nicolás, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002, 62, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Hjalt, T. Basic Helix–Loop–Helix Proteins Expressed During Early Embryonic Organogenesis; Elsevier: Amsterdam, The Netherlands, 2004; pp. 251–280. ISBN 9780123646408. [Google Scholar]
- Eiselleova, L.; Peterkova, I.; Neradil, J.; Slaninova, I.; Hampl, A.; Dvorak, P. Comparative study of mouse and human feeder cells for human embryonic stem cells. Int. J. Dev. Biol. 2008, 52, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Maherali, N.; Hochedlinger, K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009, 19, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lamouille, S.; Derynck, R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009, 19, 156–172. [Google Scholar] [CrossRef]
- Niwa, H.; Miyazaki, J.; Smith, A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000, 24, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Brennan, J.; Lu, C.C.; Norris, D.P.; Rodriguez, T.A.; Beddington, R.S.; Robertson, E.J. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 2001, 411, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Waldrip, W.R.; Bikoff, E.K.; Hoodless, P.A.; Wrana, J.L.; Robertson, E.J. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell 1998, 92, 797–808. [Google Scholar] [CrossRef]
- Dunn, N.R.; Vincent, S.D.; Oxburgh, L.; Robertson, E.J.; Bikoff, E.K. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development 2004, 131, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Sirard, C.; de La Pompa, J.L.; Elia, A.; Itie, A.; Mirtsos, C.; Cheung, A.; Hahn, S.; Wakeham, A.; Schwartz, L.; Kern, S.E.; et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998, 12, 107–119. [Google Scholar]
- Zeineddine, D.; Papadimou, E.; Chebli, K.; Gineste, M.; Liu, J.; Grey, C.; Thurig, S.; Behfar, A.; Wallace, V.A.; Skerjanc, I.S.; et al. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev. Cell 2006, 11, 535–546. [Google Scholar] [CrossRef]
- Kopp, J.L.; Ormsbee, B.D.; Desler, M.; Rizzino, A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 2008, 26, 903–911. [Google Scholar] [CrossRef]
- Teo, A.K.K.; Arnold, S.J.; Trotter, M.W.B.; Brown, S.; Ang, L.T.; Chng, Z.; Robertson, E.J.; Dunn, N.R.; Vallier, L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011, 25, 238–250. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Divvela, S.S.K.; Nell, P.; Napirei, M.; Zaehres, H.; Chen, J.; Gerding, W.M.; Nguyen, H.P.; Gao, S.; Brand-Saberi, B. bHLH Transcription Factor Math6 Antagonizes TGF-β Signalling in Reprogramming, Pluripotency and Early Cell Fate Decisions. Cells 2019, 8, 529. https://doi.org/10.3390/cells8060529
Divvela SSK, Nell P, Napirei M, Zaehres H, Chen J, Gerding WM, Nguyen HP, Gao S, Brand-Saberi B. bHLH Transcription Factor Math6 Antagonizes TGF-β Signalling in Reprogramming, Pluripotency and Early Cell Fate Decisions. Cells. 2019; 8(6):529. https://doi.org/10.3390/cells8060529
Chicago/Turabian StyleDivvela, Satya Srirama Karthik, Patrick Nell, Markus Napirei, Holm Zaehres, Jiayu Chen, Wanda Maria Gerding, Huu Phuc Nguyen, Shaorong Gao, and Beate Brand-Saberi. 2019. "bHLH Transcription Factor Math6 Antagonizes TGF-β Signalling in Reprogramming, Pluripotency and Early Cell Fate Decisions" Cells 8, no. 6: 529. https://doi.org/10.3390/cells8060529
APA StyleDivvela, S. S. K., Nell, P., Napirei, M., Zaehres, H., Chen, J., Gerding, W. M., Nguyen, H. P., Gao, S., & Brand-Saberi, B. (2019). bHLH Transcription Factor Math6 Antagonizes TGF-β Signalling in Reprogramming, Pluripotency and Early Cell Fate Decisions. Cells, 8(6), 529. https://doi.org/10.3390/cells8060529








