Cytokinesis D is Mediated by Cortical Flow of Dividing Cells Instead of Chemotaxis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Microscopy
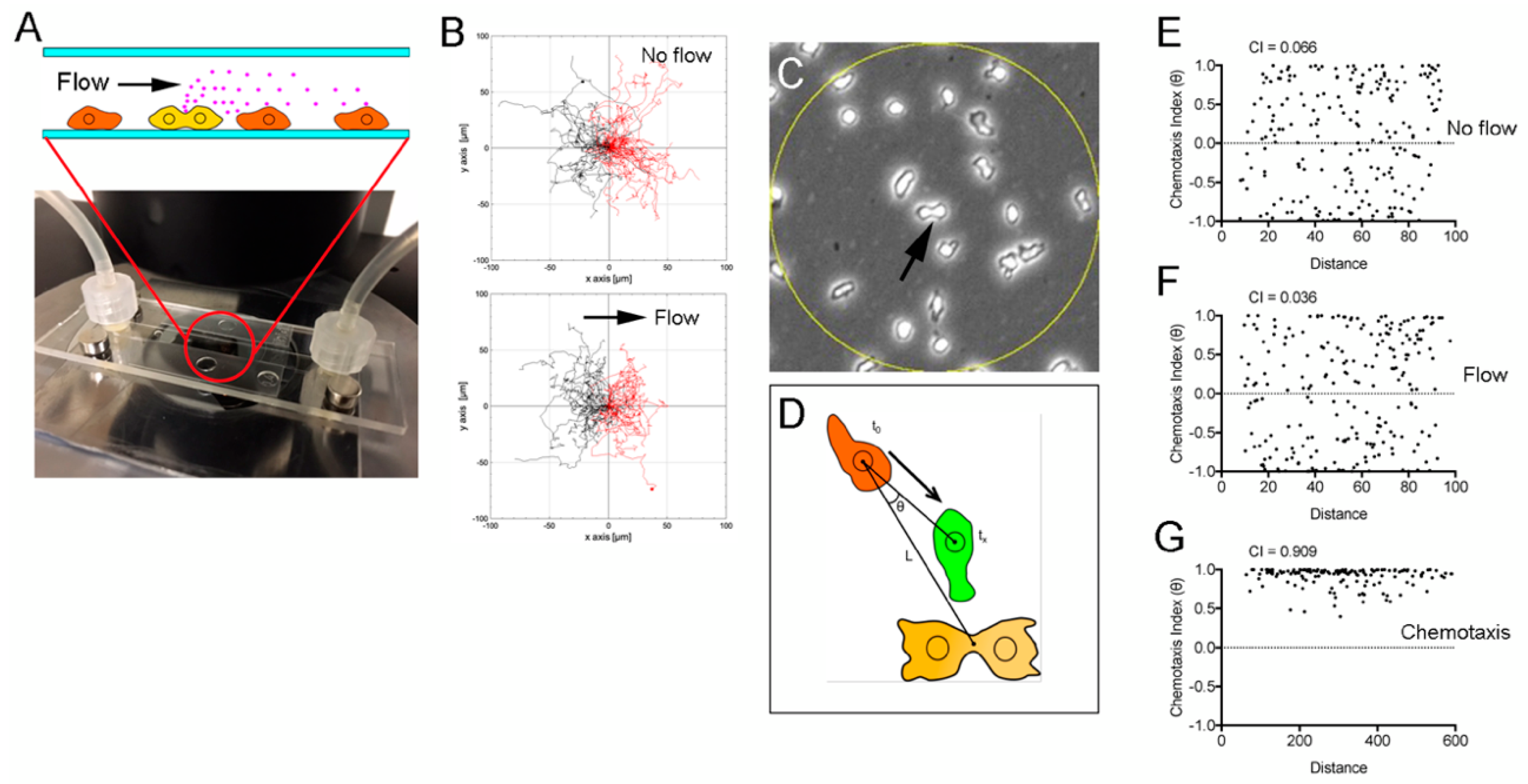
2.3. Microfluidic Experiments
2.4. Image Analysis
2.5. Statistical Analysis
3. Results and Discussion
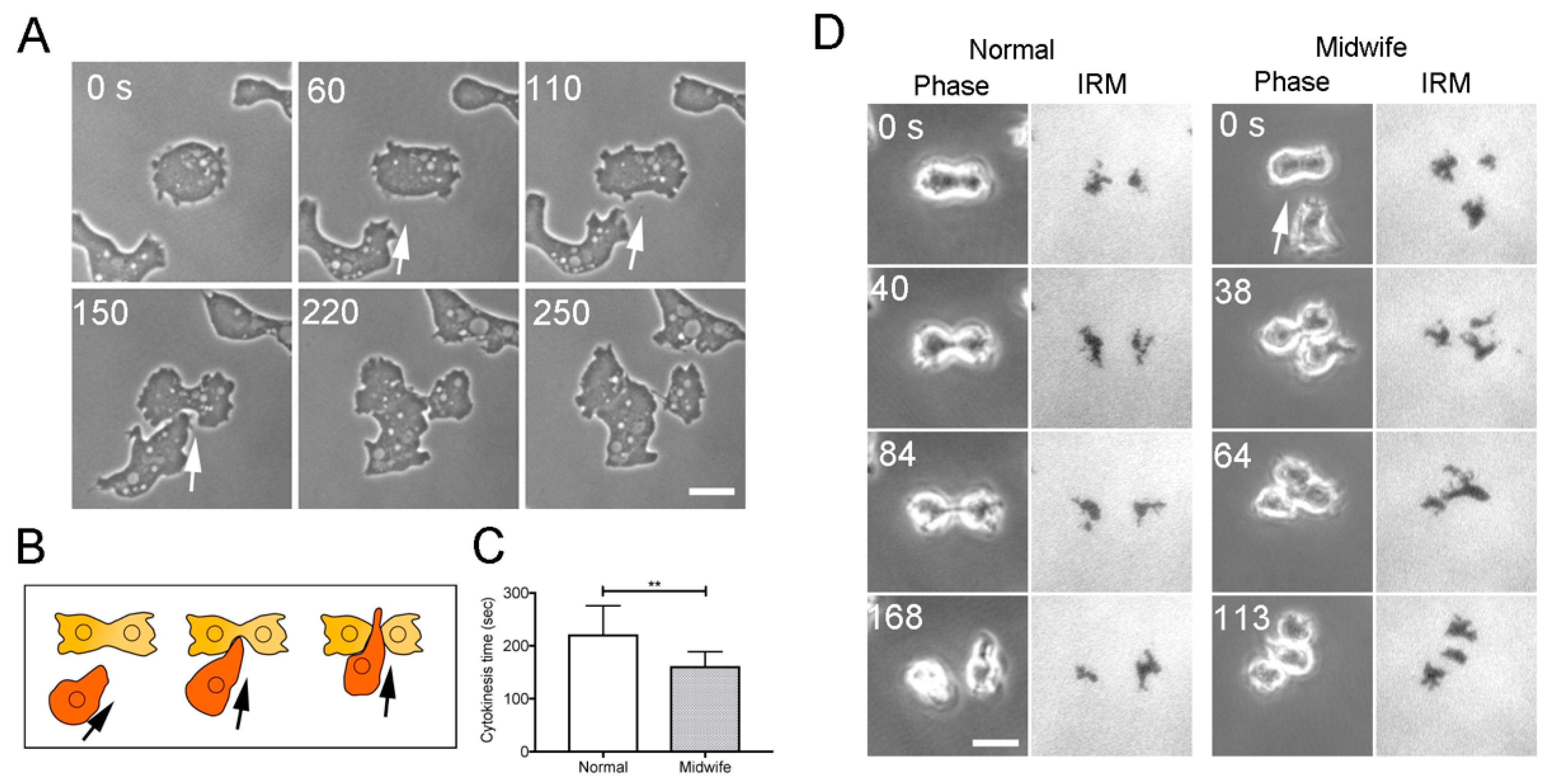
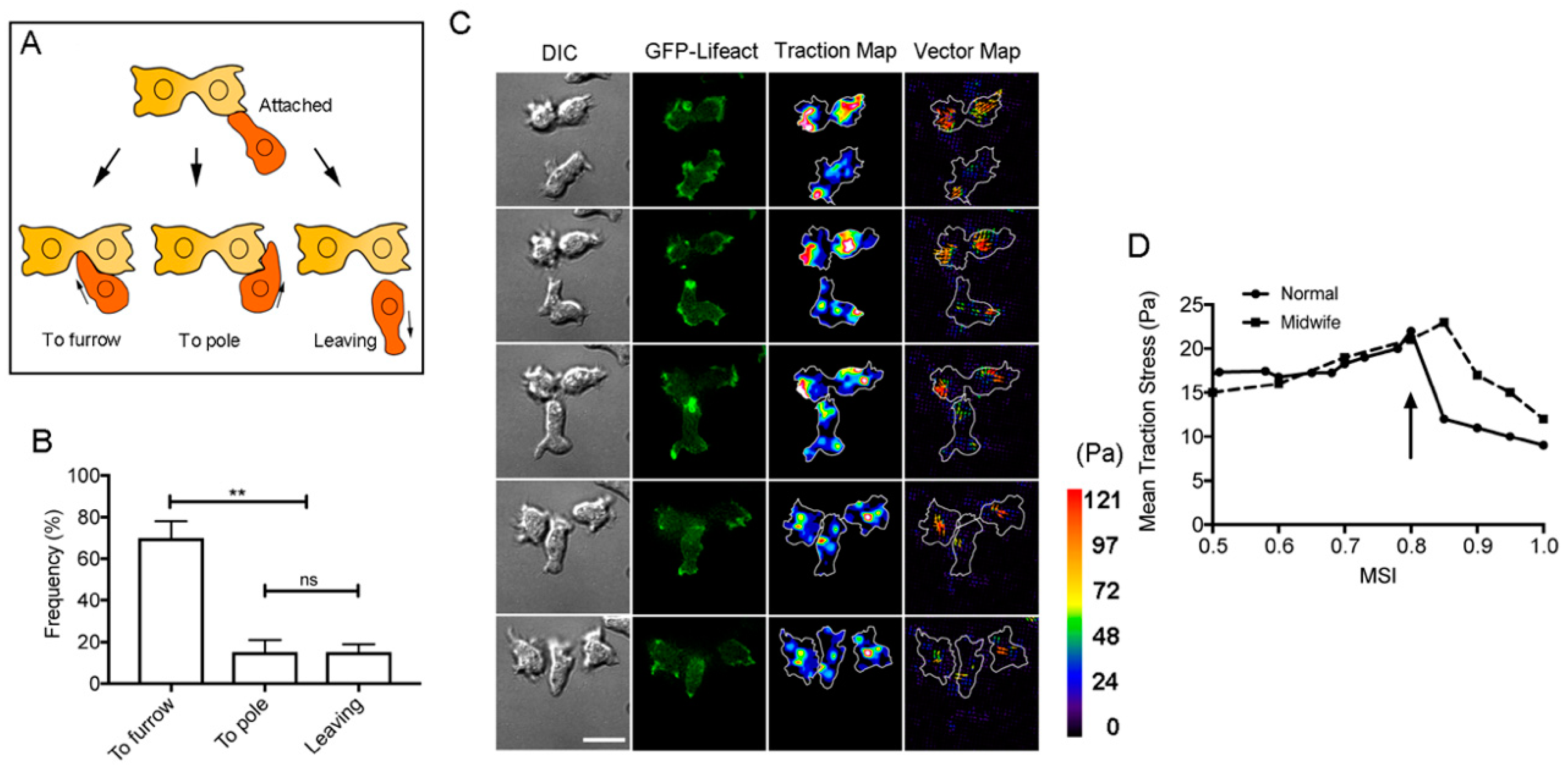
3.1. Neighboring Cells Facilitate Cell Division
3.2. Verification of the Chemotaxis Model (1)
3.3. Verification of the Chemotaxis Model (2)
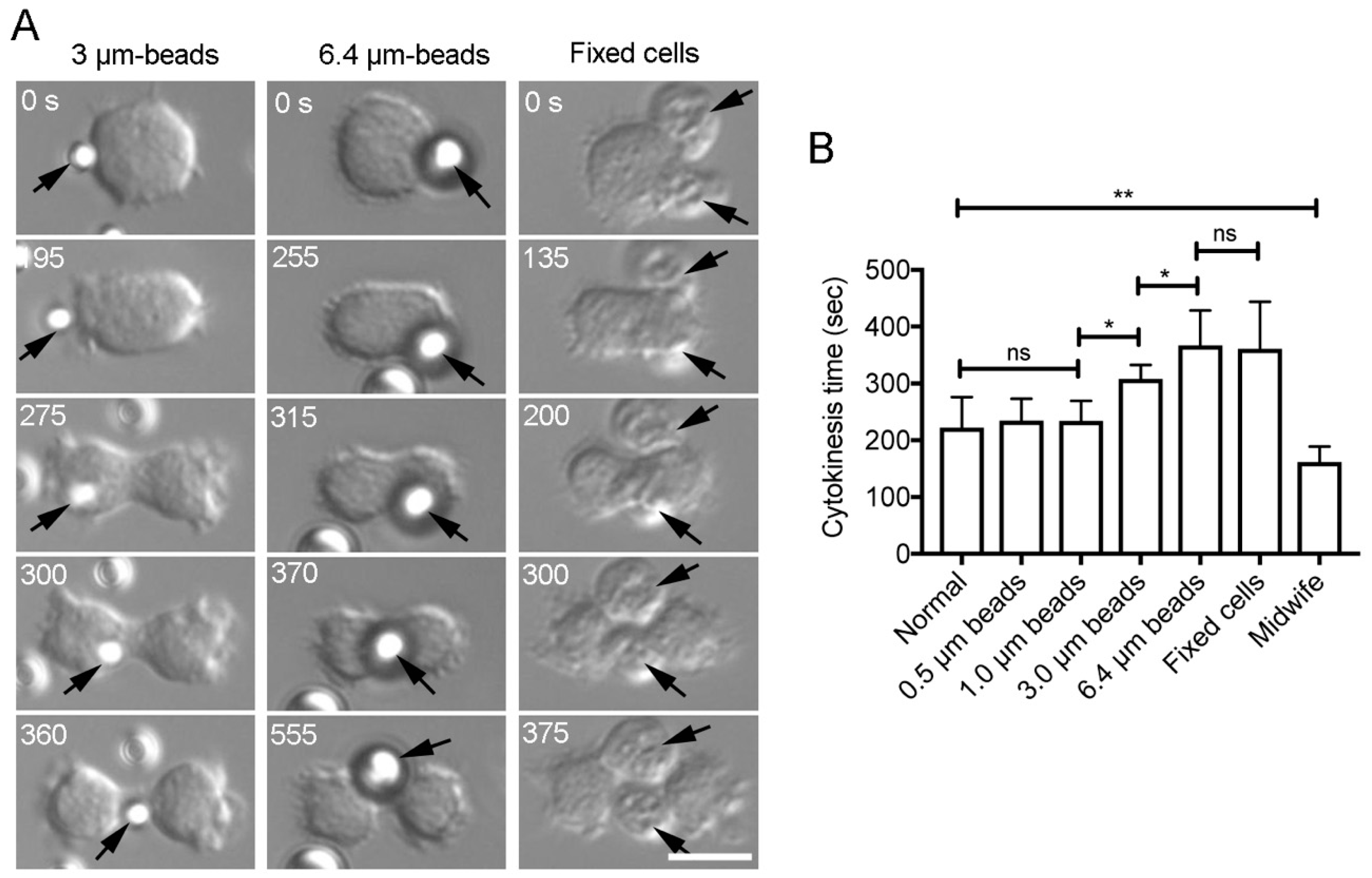
3.4. Verification of the Chemotaxis Model (3)
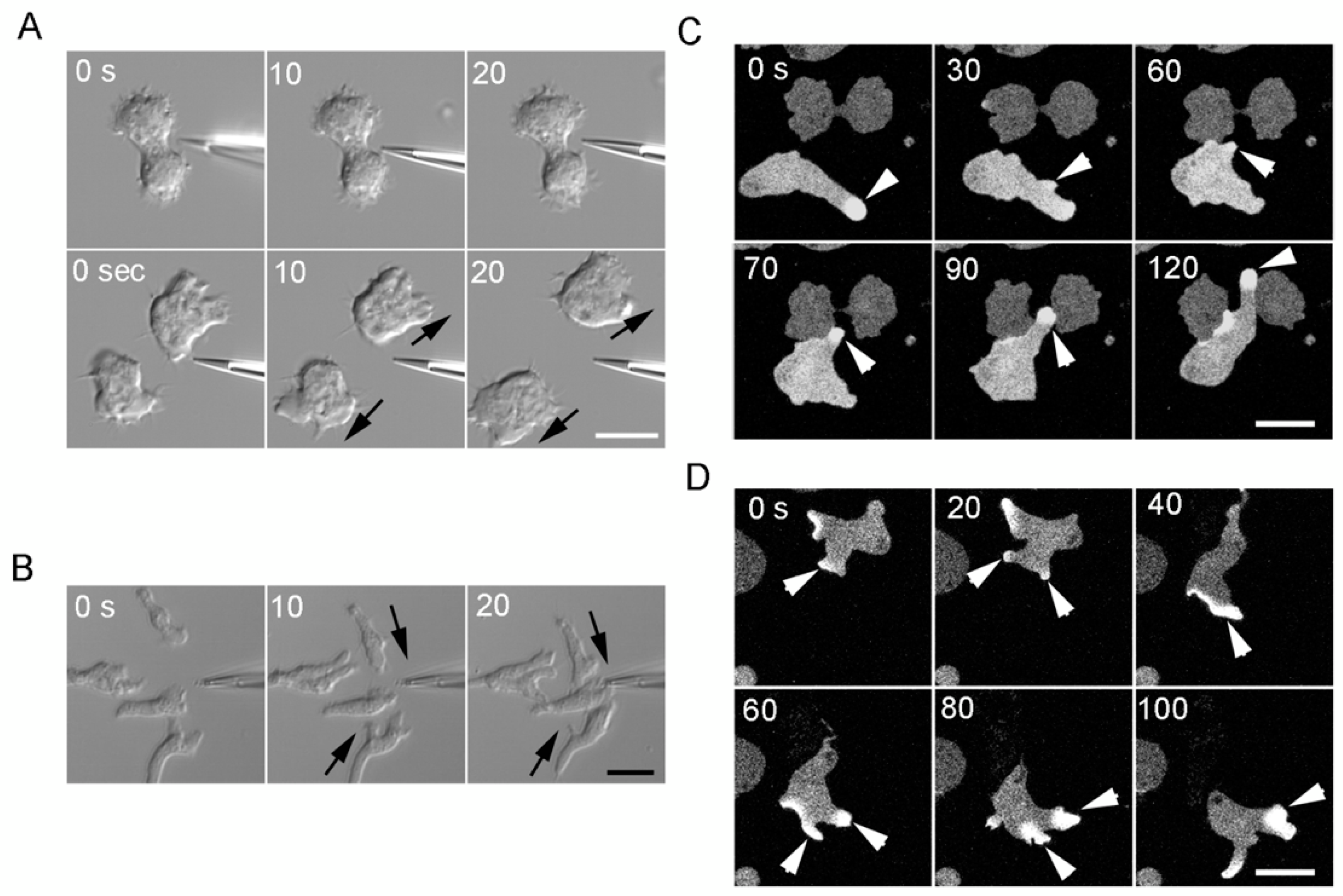
3.5. The Cortical-Flow Model
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uyeda, T.Q.; Nagasaki, A. Variations on a theme: The many modes of cytokinesis. Curr. Opin. Cell Biol. 2004, 16, 55–60. [Google Scholar] [CrossRef]
- Taira, R.; Yumura, S. A novel mode of cytokinesis without cell-substratum adhesion. Sci. Rep. 2017, 7, 17694. [Google Scholar] [CrossRef] [PubMed]
- Biron, D.; Libros, P.; Sagi, D.; Mirelman, D.; Moses, E. Asexual reproduction: ‘Midwives’ assist dividing amoebae. Nature 2001, 410, 430. [Google Scholar] [CrossRef]
- Insall, R.; Müller-Taubenberger, A.; Machesky, L.; Köhler, J.; Simmeth, E.; Atkinson, S.J.; Weber, I.; Gerisch, G. Dynamics of the Dictyostelium Arp2/3 complex in endocytosis, cytokinesis, and chemotaxis. Cell Motil. Cytoskelet. 2001, 50, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Rohlfs, M.; Arasada, R.; Batsios, P.; Janzen, J.; Schleicher, M. The Ste20-like kinase SvkA of Dictyostelium discoideum is essential for late stages of cytokinesis. J. Cell Sci. 2007, 120, 4345–4354. [Google Scholar] [CrossRef]
- Nagasaki, A.; Uyeda, T.Q. Chemotaxis-mediated scission contributes to efficient cytokinesis in Dictyostelium. Cell Motil. Cytoskelet. 2008, 65, 896–903. [Google Scholar] [CrossRef]
- Yumura, S. Myosin II dynamics and cortical flow during contractile ring formation in Dictyostelium cells. J. Cell Biol. 2001, 154, 137–146. [Google Scholar] [CrossRef]
- Yumura, S.; Matsuzaki, R.; Kitanishi-Yumura, T. Introduction of macromolecules into living Dictyostelium cells by electroporation. Cell Struct. Funct. 1995, 20, 185–190. [Google Scholar] [CrossRef]
- Yumura, S. A novel low-power laser-mediated transfer of foreign molecules into cells. Sci. Rep. 2016, 6, 22055. [Google Scholar] [CrossRef]
- Matsuda, S.; Harada, K.; Ito, M.; Takizawa, M.; Wongso, D.; Tsuboi, T.; Kitaguchi, T. Generation of a cGMP indicator with an expanded dynamic range by optimization of amino acid linkers between a fluorescent protein and PDE5α. ACS Sens. 2017, 2, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.G.S.; Yumura, S. Traction force and its regulation during cytokinesis in Dictyostelium cells. Eur. J. Cell Biol. 2017, 96, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.S.; Yumura, S. Dynamics of novel feet of Dictyostelium cells during migration. J. Cell Sci. 2004, 117, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Yumura, S.; Itoh, G.; Kikuta, Y.; Kikuchi, T.; Kitanishi-Yumura, T.; Tsujioka, M. Cell-scale dynamic recycling and cortical flow of the actin-myosin cytoskeleton for rapid cell migration. Biol. Open 2013, 2, 200–209. [Google Scholar] [CrossRef]
- Veltman, D.M.; Lemieux, M.G.; Knecht, D.A.; Insall, R.H. PIP₃-dependent macropinocytosis is incompatible with chemotaxis. J. Cell Biol. 2014, 204, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Parent, C.A.; Devreotes, P.N. A cell’s sense of direction. Science 1999, 284, 765–770. [Google Scholar] [CrossRef]
- Bagorda, A.; Das, S.; Rericha, E.C.; Chen, D.; Davidson, J.; Parent, C.A. Real-time measurements of cAMP production in live Dictyostelium cells. J. Cell Sci. 2009, 122, 3907–3914. [Google Scholar] [CrossRef] [PubMed]
- Hashimura, H.; Morimoto, Y.V.; Yasui, M.; Ueda, M. Collective cell migration of Dictyostelium without cAMP oscillations at multicellular stages. Commun. Biol. 2019, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Mato, J.M.; Krens, F.A.; van Haastert, P.J.; Konijn, T.M. 3’:5’-cyclic AMP-dependent 3’:5’-cyclic GMP accumulation in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 1977, 74, 2348–2351. [Google Scholar] [CrossRef] [PubMed]
- Yumura, S.; Furuya, K.; Takeuchi, I. Intracellular free calcium responses during chemotaxis of Dictyostelium cells. J. Cell Sci. 1996, 109, 2673–2678. [Google Scholar]
- Dalous, J.; Burghardt, E.; Müller-Taubenberger, A.; Bruckert, F.; Gerisch, G.; Bretschneider, T. Reversal of cell polarity and actin-myosin cytoskeleton reorganization under mechanical and chemical stimulation. Biophys. J. 2008, 94, 1063–1074. [Google Scholar] [CrossRef]
- Décave, E.; Rieu, D.; Dalous, J.; Fache, S.; Brechet, Y.; Fourcade, B.; Satre, M.; Bruckert, F. Shear flow-induced motility of Dictyostelium discoideum cells on solid substrate. J. Cell Sci. 2003, 116, 4331–4343. [Google Scholar] [CrossRef] [PubMed]
- Bray, D.; White, J.G. Cortical flow in animal cells. Science 1988, 239, 883–888. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, Y.; Jahan, M.G.S.; Kondo, T.; Nakano, M.; Yumura, S. Cytokinesis D is Mediated by Cortical Flow of Dividing Cells Instead of Chemotaxis. Cells 2019, 8, 473. https://doi.org/10.3390/cells8050473
Tanaka Y, Jahan MGS, Kondo T, Nakano M, Yumura S. Cytokinesis D is Mediated by Cortical Flow of Dividing Cells Instead of Chemotaxis. Cells. 2019; 8(5):473. https://doi.org/10.3390/cells8050473
Chicago/Turabian StyleTanaka, Yuki, Md. Golam Sarowar Jahan, Tomo Kondo, Masaki Nakano, and Shigehiko Yumura. 2019. "Cytokinesis D is Mediated by Cortical Flow of Dividing Cells Instead of Chemotaxis" Cells 8, no. 5: 473. https://doi.org/10.3390/cells8050473
APA StyleTanaka, Y., Jahan, M. G. S., Kondo, T., Nakano, M., & Yumura, S. (2019). Cytokinesis D is Mediated by Cortical Flow of Dividing Cells Instead of Chemotaxis. Cells, 8(5), 473. https://doi.org/10.3390/cells8050473





