Abstract
Inorganic polyphosphate (polyP) is crucial for adaptive reactions and stress response in microorganisms. A convenient model to study the role of polyP in yeast is the Saccharomyces cerevisiae strain CRN/PPN1 that overexpresses polyphosphatase Ppn1 with stably decreased polyphosphate level. In this study, we combined the whole-transcriptome sequencing, fluorescence microscopy, and polyP quantification to characterize the CRN/PPN1 response to manganese and oxidative stresses. CRN/PPN1 exhibits enhanced resistance to manganese and peroxide due to its pre-adaptive state observed in normal conditions. The pre-adaptive state is characterized by up-regulated genes involved in response to an external stimulus, plasma membrane organization, and oxidation/reduction. The transcriptome-wide data allowed the identification of particular genes crucial for overcoming the manganese excess. The key gene responsible for manganese resistance is PHO84 encoding a low-affinity manganese transporter: Strong PHO84 down-regulation in CRN/PPN1 increases manganese resistance by reduced manganese uptake. On the contrary, PHM7, the top up-regulated gene in CRN/PPN1, is also strongly up-regulated in the manganese-adapted parent strain. Phm7 is an unannotated protein, but manganese adaptation is significantly impaired in Δphm7, thus suggesting its essential function in manganese or phosphate transport.
Keywords:
polyphosphate; PPN1; PHM7; PHO84; manganese adaptation; oxidative stress; Saccharomyces cerevisiae 1. Introduction
Inorganic polyphosphate (polyP) is a linear anionic polymer containing from several to hundreds of orthophosphate residues linked by energy-rich phosphoanhydride bonds. PolyP participates in the regulation of many cellular events [1,2]. In mammals, polyP is involved in signal transduction cascades [3,4,5,6,7,8], serves as a phosphorus reserve for apatite formation in bone growth and development [9,10,11,12], and plays a role in blood coagulation [13,14] and inflammation [15,16,17]. In bacteria, polyP and its associated enzymes are involved in stress response and virulence, in addition to the function of phosphate and energy reserve [2,18,19,20,21]. Also, it is involved in cell motility, biofilm formation, and controls the level of the stringent response factor guanosine 5’-diphosphate 3’-diphosphate (ppGpp) [2,22], although it has been shown that ppGpp is not essential for polyP synthesis in Escherichia coli [23]. The role of polyP in oxidative stress response is associated with its protein-protective chaperone function [24,25], and the metal ion-complexing function [26]. In yeast, the polyP activity as a phosphate reserve is well studied [27], and the enzymes of polyP metabolism are well characterized [28,29,30]. However, the role of polyP in oxidative and toxic metal stresses in yeast remains notably less explored.
Knockout mutants lacking the genes encoding polyP-metabolizing enzymes serve as models for studying the role of polyP. For bacteria, the commonly used models are the ppk1 or ppk2-null strains lacking polyphosphate kinases and having a significantly decreased polyP level [2,31,32]. They provide the possibility of direct studying of the effect of a decreased polyP level on bacterial cells, including stress response.
In yeast, it is not trivial to limit polyP synthesis without non-intended disturbance in stress response due to the following. The synthesis of the major part of polyP pool is provided by the vacuolar transporter chaperone (VTC) complex of the vacuolar membrane [28,33]. The reduced polyP accumulation was observed in mutants without VTC4 [34] and VTC5 [33], and in knockout strains lacking subunits of vacuolar V-ATPase [35]. All such mutants either lack some components of the VTC complex, being deficient in V-ATPase assembly and stability [36], or lack V-ATPase per se. The V-ATPase itself plays a key role in the response to oxidative and heavy metal stress [37,38,39], thus its dysfunction hides possible polyP functions in stress response.
This is why we had to design strains with enhanced expression of polyP hydrolyzing enzymes. The S. cerevisiae strain overproducing the Ppn1 polyphosphatase is known to have a decreased polyP level [40], and can serve as a model for studying the role of polyP in stress response in yeast.
Given the role of polyP in metal ion resistance in microorganisms [41], we expected that the Ppn1 polyphosphatase overproducing strain (CRN/PPN1) would be more sensitive to manganese excess than the control strain (CRN). Instead, CRN/PPN1 had enhanced resistance to manganese. This is explained by the pre-adaptive state of CRN/PPN1 in the normal medium, revealed by the high-throughput transcriptome profiling. Furthermore, the pre-adaptive state was characterized by up-regulation of the oxidative stress response genes, which provided enhanced resistance to oxidative stress.
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
In this study we used the following S. cerevisiae strains: CRN (MATa ade2 his3 ura3 ppn1Δ::CgTRP1), CRN/PPN1 (CRN transformed with the pMB1/PPN1 Sc vector), and CRN/PPX1 (CRN transformed with the pMB1/PPX1 Sc vector). The source of the strains and the polyphosphatase activities in the cell-free extracts are given in Table S1.
The strains were maintained in the selective agarized medium [40]. The BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0), BY4743-derived Δphm7, and BY4743-derived Δvmr1 strains were obtained from the Dharmacon collection. For the experiments, the cells were cultivated in YPD containing 2% glucose, 2% peptone (Peptone from meat enzymatic digest, Fluka, Sigma-Aldrich, St. Louis, MO, USA), 1% yeast extract (CONDA/Pronadisa, Madrid, Spain), and 14 mM KH2PO4 at 29 °C and 145 rpm. For studying the resistance to manganese, the medium was supplemented with 5 mM MnSO4 [42], which provides notable but not complete growth inhibition of the CRN strain. In the control YPD medium, Mn2+ concentration was 0.004 mM. Growth curves were obtained by measuring the optical density (OD) at 530 nm in a 0.3 cm cuvette, 3 replicates per experiment. The cell concentration was measured in a hemocytometer, 10 samples for each experiment. For the analyses of polyphosphates and manganese content, the cells were harvested at 5000 g for 20 min, washed twice with sterile distilled water. The samples for RNA-Seq library construction were washed thrice.
2.2. RNA Extraction and Sequencing for Transcriptome Profiling
The CRN and CRN/PPN1 strains were grown in control YPD and YPD with 5 mM of MnSO4. After cultivation for 12 h (control, both strains), 40 h (manganese, CRN) and 16 h (manganese, CRN/PPN1), the cells were harvested by centrifugation at 5000× g for 10 min and washed three times with distilled water at 0 °C, centrifuged after each washing, and placed in RNALater. Two replicates were obtained from two simultaneous cultivations in control YPD and manganese-containing YPD. Total RNA extraction was performed using an RNeasy Plant Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol—with the addition of Plant RNA Isolation aid reagent (Ambion/ThermoFisher Scientific, Waltham, MA, USA) to lysis buffer RLT. Illumina cDNA libraries were constructed using the TruSeq RNA Sample Prep Kits v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. The libraries were sequenced on the Illumina NextSeq 500 (Illumina, San Diego, CA, USA). The total sequenced library size was no less than 3 million reads per replicate. The raw and processed RNA-Seq data are deposited in GEO under accession number GSE122987.
2.3. Processing and Analysis of RNA-Seq Data
Adapter sequences were trimmed with Trimmomatic v0.36 (ILLUMINACLIP:TruSeq3-SE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:30 MINLEN:36). Read mapping and counting was performed with STAR v2.5.3a (default parameters) using Ensembl S. cerevisiae R64-1-1.90 genome annotation (genome-build-accession GCA_000146045.2, UCSC sacCer3). For all samples, no less than 88% of the reads were mapped uniquely. The resulting gene lists were filtered to contain only genes reaching at least 2 counts per million in at least four of the eight libraries. Read counts were normalized using the edgeR [43] TMM approach. A principal component analysis of the resultant normalized and log-transformed matrices was done in the R environment. Differential expression was assessed with edgeR using a generalized linear model, accounting for the interaction between the strain (CRN, CRN/PPN1) and growth conditions (control or manganese excess medium). The genes with the FDR-adjusted p-value ≤ 0.05 and |log2 (Fold Change)| ≥ 1 were considered differentially expressed. The gene ontology (GO) enrichment analysis was performed with the YeastMine software [44]. The read counts and differential gene expression data are presented in Tables S3 and S4.
2.4. Polyphosphate and Polyphosphatase Activity Assay
For polyphosphate assay, the cells were cultivated for 24 h (CRN, CRN/PPN1 and CRN/PPX1 strains, control growth; CRN/PPN1 strain, growth in the presence of 5 mM MnSO4) and for 50 h (CRN and CRN/PPX1 strains, growth in the presence of 5 mM MnSO4).
The acid-soluble polyphosphate was extracted from biomass samples by the twofold treatment, with 0.5 M HClO4 at 0 °C for 15 min with continuous stirring. The cells were precipitated at 4000 g. In supernatants, the Pi and polyP amounts were assayed as described earlier [45]. The amount of acid-insoluble polyphosphate in the remaining biomass was assayed by the Pi content after twofold treatment with 0.5 M HClO4 at 90 °C for 20 min. The polyphosphatase activities were measured in cell-free extracts with the polyphosphate of 65 phosphate residues as described earlier [40].
2.5. Manganese Assay
The CRN, CRN/PPN1, and CRN/PPX1 strains were cultivated in YPD medium supplemented with 5 mM MnSO4. The cells were cultivated to the logarithmic growth stage: For 40 h (CRN and CRN/PPX1) and for 16 h (CRN/PPN1). The manganese content was assayed by atomic absorption spectroscopy after biomass sample burning at 150 °C in 32% HClO4.
2.6. The Assay of H2O2 MICs
The minimum inhibitory concentrations (MICs) of H2O2 were determined by inoculation of cell samples (standardized by culture density) in sterile plates with covers in YPD medium supplemented with H2O2. After 24-h cultivation, culture densities were measured using Sapphir plate photometers (Moscow, Russia) at 600 nm.
2.7. Fluorescence and Light Microscopy
Live and dead cells were stained using a LIVE/DEAD Fungal Light Yeast Viability Kit (Molecular Probes Inc. / ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Yeast cultures were stained without washing and incubated with the staining reagent for 15 min at 37 °C. In case of growth inhibition in the presence of manganese, the cell culture was concentrated by centrifugation before staining. The samples were examined using a fluorescent microscope AXIO Imager A1 ZEISS (Oberkochen, Germany) with a filter kit 56 (ZEISS) at a wavelength of 450–500 nm (excitation) and 600–650 nm (emission). The number of stained cells was calculated on 3 to 5 micrographs.
1,2,3-dihydrorhodamine was used for ROS staining [46]. Cell culture samples (1 mL) were centrifuged at 5000× g for 5 min, washed twice with MilliQ water and suspended in 1 mL of MilliQ water. After the addition of 0.02 mL 1,2,3-dihydrorhodamine (Sigma-Aldrich, St. Louis, MO, USA) (stock solution 0.5 mg/mL in DMSO), the cells were incubated at 30 °C and 1000 rpm in a Thermomixer (Biosan, Riga, Latvia). Then the samples were centrifuged and washed twice with MilliQ water. The Nikon Eclipse FN 1 microscope with Intenselight C-HGFI HG Precentered Fiber Illuminator (Nikon, Tokyo, Japan) was used to make micrographs. The emission was registered with the bandpass filter 420–620 nm.
3. Results
3.1. CRN/PPN1 Strain Exhibits a Decreased PolyP Level and Enhanced Resistance to Manganese
The parent CRN strain (Δppn1 mutant), the CRN/PPN1 strain (overproducing Ppn1 polyphosphatase), and the CRN/PPX1 strain (transformed by the same vector but carrying the PPX1 gene encoding Ppx1 polyphosphatase) showed no difference in the control growth in YPD medium (Figure 1A). The polyphosphatase activity in the cell-free extracts was higher in the respective overproducing strains (Table S1). The polyP level in CRN/PPN1 cells was lower as compared to CRN and CRN/PPX1 (Figure 1B). A decrease in the polyP level was more pronounced in the acid-soluble polyP fraction (Table S2), as it was observed in selective YNB medium [40]. In the presence of excess manganese, the polyP level was increased in CRN cells, remained almost unchanged in CRN/PPX1 cells, and showed a further decrease in CRN/PPN1 cells (Figure 1B).
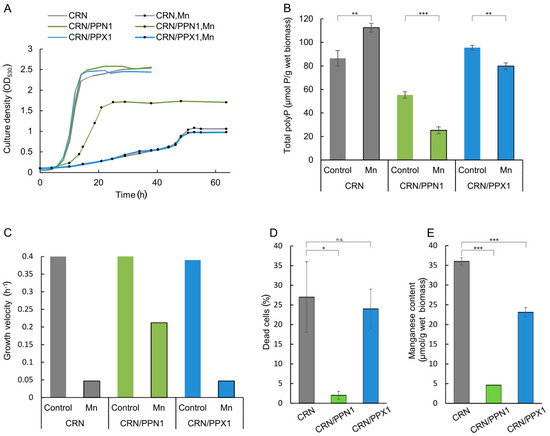
Figure 1.
Growth characteristics and polyphosphate content in the cells of CRN, CRN/PPN1, and CRN/PPX1 strains of S. cerevisiae cultivated in control YPD medium with 14 mM KH2PO4 and in the same medium supplemented with 5 mM MnSO4. (A) growth curves; control medium—lines without dots; manganese excess medium—lines with dots; CRN is shown in gray, CRN/PPN1 in green, and CRN/PPX1 in blue; (B) growth velocities, logarithmic growth stage; (C) manganese content in the cells grown in the presence of 5 mM MnSO4, logarithmic growth stage: CRN (40 h growth), CRN/PPN1 (16 h growth), and CRN/PPX1 (40 h growth); (D) the percentage of dead cells in the logarithmic growth stage in the presence of 5 mM MnSO4; (E) total polyphosphate content in control and manganese grown cells, stationary growth stage. Three replicates were performed for each experiment; mean ± s.d. is shown on panels (B,D,E). Two-sample t-test was used to assess the statistical significance: *** p-value < 0.001, ** p-value < 0.01, * p-value < 0.05, n.s.—not significant.
The manganese excess inhibited the growth of CRN and CRN/PPX1 strains (Figure 1A,C) similar to other S. cerevisiae strains [42]. Surprisingly, the growth inhibition in the medium with manganese excess was dramatically lower for polyP-deficient CRN/PPN1. Furthermore, manganese excess induced a notable cell death of the CRN and CRN/PPX1 strains (Figure 1D and Figure S1), while CRN/PPN1 was more resistant and accumulated a lower amount of manganese (Figure 1E).
3.2. Transcriptome Profiling Reveals the Pre-Adapted State of the CRN/PPN1 Strain
To explore the mechanism of CRN/PPN1 resistance to manganese, we performed transcriptome profiling (RNA-Seq) for the CRN and CRN/PPN1 strains in the control and in manganese excess media.
To investigate global gene expression changes in CRN and CRN/PPN1 strains, we used a principal component analysis (Figure 2A) of the quantitative gene expression data (Table S3). The first and the second principal components (PCs) account for 70% of variance and clearly segregate the samples: PC1 reflects the expression changes arising from the adaptation to manganese, while PC2 reflects the changes resulting from Ppn1 overexpression. Interestingly, CRN/PPN1 samples were shifted towards the manganese adapted state even in the control medium. This observation is also supported by a comparison of gene sets up- and down-regulated in CRN/PPN1 and upon manganese adaptation (Figure 2B), which are strikingly similar, with only a few genes uniquely differentially expressed in CRN/PPN1.
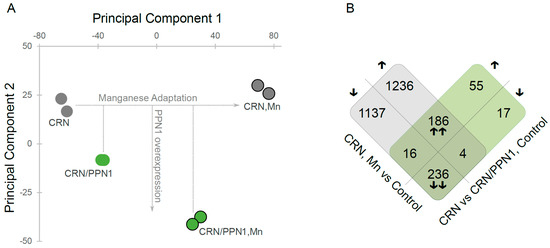
Figure 2.
Gene expression profile of the CRN/PPN1 strain is similar to that of the manganese-adapted CRN strain. CRN and CRN/PPN1 were grown in control YPD with 14 mM KH2PO4 and in the same medium supplemented with 5 mM MnSO4. (A) Principal component analysis of gene expression data: (green circles) CRN/PPN1 samples; (gray circles) CRN samples. Circles denoting the samples grown in manganese excess conditions (Mn) are outlined. (B) Venn-like diagram for genes up- and down-regulated in (gray) CRN in control YPD versus CRN in manganese-supplemented YPD; (green) CRN/PPN1 as compared to CRN, both in control YPD. Each cell shows the number of genes; the direction of expression change (up- or down-regulation) is shown with arrows. Up- or down-regulated genes in both comparisons are indicated by double arrows.
To investigate this phenomenon, we assessed differential gene expression (Table S4) for the samples grown in manganese versus control medium, and used the gene ontology enrichment analysis.
The adaptation of the parent CRN strain to manganese follows the same pattern as we have observed earlier for the CRY strain [47]. The up-regulated genes were enriched with multiple GO terms reflecting stress response, cell wall, and membrane organization (multiple GO terms, p < 10−7). The down-regulated genes were enriched with all types of protein biosynthesis and ribosome-related GO terms, which clearly agrees with growth suppression under manganese excess (Figure 1A,C).
The CRN/PPN1 adaptation follows the same pattern as that of the CRN strain, with a few additions of GO-terms related to phosphate metabolic processes (GO:0006796 and GO:0006793, p < 10−4), which is associated with changes in the polyP content (Figure 1B).
Additionally, we estimated differential gene expression in the control medium for CRN/PPN1 versus CRN samples. This analysis confirmed the pre-adapted state of the CRN/PPN1 strain, with up-regulation of the genes responsible for cellular response to external stimulus (GO:0071496 and GO:0009991, p < 10−5), plasma membrane components (GO:0005886, p < 10−4), and, importantly, oxidation-reduction process (GO:0055114, p < 10−5). Of note, in manganese-adapted CRN/PPN1 samples, slight enrichment of the genes responsible for oxidoreductase activity (GO:0016491, p ~0.01) was also detected.
Thus, Ppn1 overexpression leads to general adaptive processes and, specifically, oxidation-reduction pathways. This particular feature facilitates CRN/PPN1 adaptation to manganese excess.
3.3. CRN/PPN1 Strain Has Increased ROS Level and H2O2 Resistance
The global transcriptome analysis suggested that the CRN/PPN1 strain has a pre-adapted state realized through up-regulation of the oxidation-reduction pathways. We estimated MICs of H2O2 for CRN, CRN/PPX1, and CRN/PPN1 growth. Indeed, the CRN/PPN1 strain showed enhanced H2O2 resistance as compared to CRN or CRN/PPX1. The average MICs of H2O2 were 2.5, 1.5, and 4.0 mM for CRN, CRN/PPX1, and CRN/PPN1, respectively (Figure 3). Additionally, we compared the reactive oxygen species (ROS) levels in CRN and CRN/PPN1—both in the control and manganese excess media. The staining with 1,2,3-dihydrorhodamine showed a higher ROS level in CRN/PPN1 cells in both cases (Figure 4, Supplementary Figure S2).
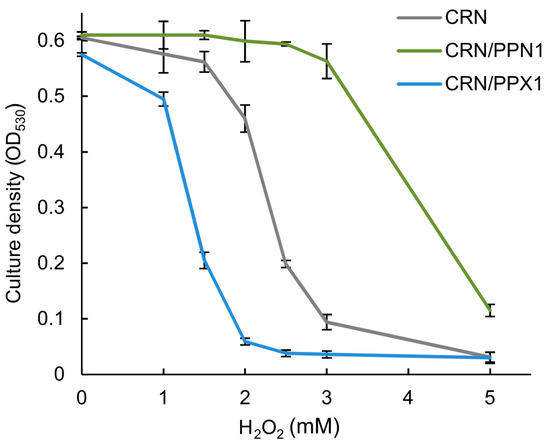
Figure 3.
The effect of H2O2 on the growth of the CRN, CRN/PPN1, and CRN/PPX1 strains of S. cerevisiae in YPD medium. CRN is shown in gray, CRN/PPN1 in green, and CRN/PPX1 in blue. Three replicates were performed for each experiment; mean ± s.d. is given.
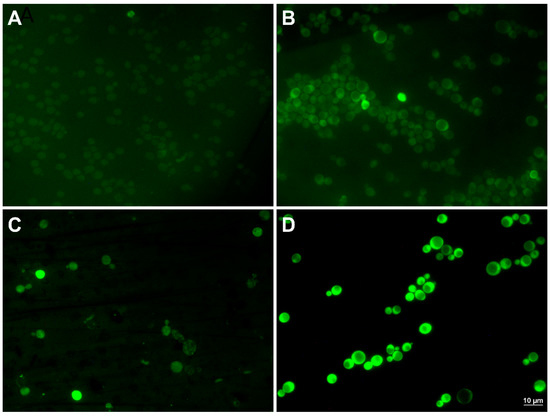
Figure 4.
The micrographs of the cells of CRN and CRN/PPN1 strains of S. cerevisiae stained with 1,2,3-dihydrorhodamine. The strains were cultivated in control YPD medium with 14 mM KH2PO4, and in the same medium supplemented with 5 mM MnSO4 to the logarithmic stage (control—12 h growth, both strains; 5 mM manganese—40 h growth, CRN; 16 h growth, CRN/PPN1). (A) CRN, control YPD; (B) CRN/PPN1, control YPD; (C) CRN, manganese excess; (D) CRN/PPN1, manganese excess.
3.4. An Increased Manganese Resistance of CRN/PPN1 Is Realized through PHO-Pathway Genes
To understand the enhanced manganese resistance of the CRN/PPN1 strain, we paid special attention to particular groups of genes (Table 1).

Table 1.
Differential expression of the selected genes responsible for manganese transport and belonging to the PHO pathway. The cases of differential expression with FDR-corrected p-value ≤ 0.05 are marked in bold.
First of all, with manganese excess, the genes encoding manganese transport of the plasma membrane, ER and Golgi (SMF1, SMF2, PMR1), are up-regulated, while the genes of vacuolar transporters are either stable (CCC1) or down-regulated (GDT1P). In the control medium, there are no major changes in expression of the above genes in CRN/PPN1 versus CRN. The expression of the putative manganese chelator encoded by MNC1 [47] is specific to manganese excess.
The plasma membrane ATPase Pma2 is essential for manganese adaptation [47]. It is up-regulated in manganese-grown cells of both strains and in CRN/PPN1 grown in the control medium, although with a weak significance (FDR ~0.08).
Among the genes of polyP hydrolysis enzymes, we should mention DDP1, which is down-regulated in the presence of manganese excess, which accounts for the increase in polyP content in CRN cells. All subunits of the polyP synthesis VTC complex are down-regulated in all cases.
Regarding the known phosphate transporters, the genes encoding Pho89 and Pho84 are known to be regulated by the transcription factor Pho4. However, in our case, the changes in Pho89 and Pho84 expression correlate neither with Pho4 differential expression, nor with each other. It is noteworthy that PHO84 is down-regulated both under manganese excess and in CRN/PPN1 in the control medium, which is in agreement with decreased manganese uptake in CRN/PPN1 cells.
Additionally, we have found two unannotated genes, PHM6 and PHM7, that were supposed to be regulated by phosphate levels [48]. Upon manganese excess, they are strongly differentially expressed in the opposite directions: PHM6 is down-regulated, while PHM7 is up-regulated, displaying the highest reachable fold change among the genes—up to 5% FDR-corrected p-value in the comparisons CRN/PPN1 versus CRN (control) and CRN (control versus manganese excess). Strong up-regulation of PHM7 suggests its essential role in manganese adaptation, and suggests studying the survival of the Δphm7-null mutant under manganese excess.
3.5. Δphm7 Has Reduced Resistance to Manganese
We have compared the growth characteristics of the Δphm7-null mutant under the control and manganese excess conditions to confirm the role of Phm7 in resistance to manganese. The Δvmr1 strain was used as a positive control. VMR1 encodes the vacuolar membrane protein that is an ABC transporter of the MRP subfamily. The deletion of this gene enhanced the sensitivity to cadmium [49] and lead [50], and, according to our data, VMR1 was significantly up-regulated in CRN/PPN1 (as compared to CRN) and in manganese excess conditions.
The tested strains grew similarly in the YPD medium; in the presence of 5 mM manganese, the growth of Δphm7 and Δvmr1 was inhibited more than that of the parent BY4743 strain, the cell concentrations of the mutants were lower, and the number of dead cells was significantly higher than in the parent strain (Figure 5). The light microscopy of the mutant cultures revealed large-scale cell lysis in the stationary phase (Figure 6).
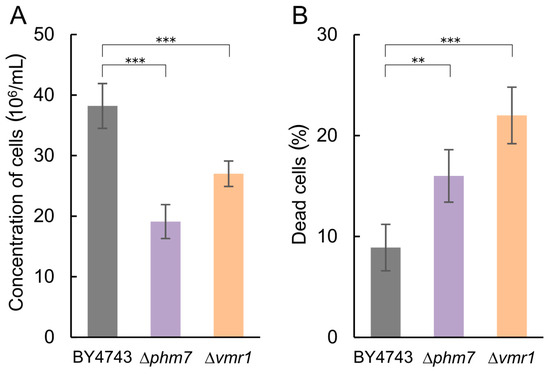
Figure 5.
Growth characteristics of the BY4743 parent strain and Δphm7 and Δvmr1 knockout strains of S. cerevisiae cultivated in YPD medium with 14 mM KH2PO4 and 5 mM MnSO4 to the stationary growth stage (72 h). (A) cell concentration, (B) percentage of dead cells. Three replicates were performed for each experiment; mean ± s.d. is shown. Two-sample t-test was used to assess the statistical significance: *** p-value < 0.001, ** p-value < 0.01.
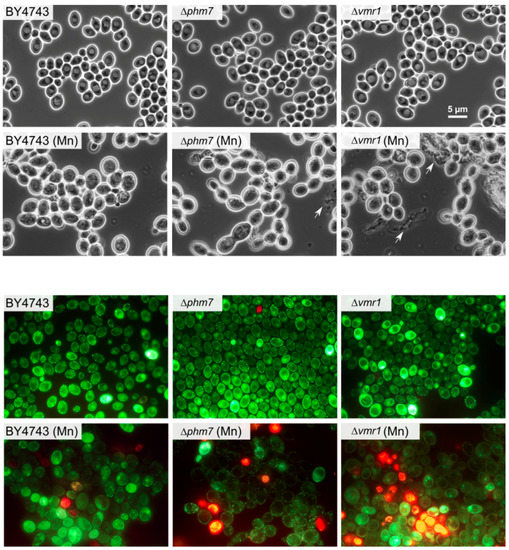
Figure 6.
The micrographs of the cells of the parent BY4743 strain and Δphm7 and Δvmr1 knockout strains of S. cerevisiae cultivated in YPD medium with (Mn) and without 5 mM MnSO4 to the stationary growth stage (72 h). Top—phase contrast, bottom—LIVE/DEAD Fungal Light Yeast Viability Kit staining.
4. Discussion
PolyP is widely recognized as an anionic polymer that binds and de-toxifies heavy metal ions [41]. However, in yeast, particular mechanisms of polyP-involving detoxification are understudied. Manganese-adapted yeast cells contain longer polyP chains [42], but this could be a side effect of VTC4 activation by manganese ions [28]. PolyP content increases during the elongated lag phase in manganese excess conditions and decreases at the active growth stage [42]. It is known that the excess of manganese ions results in oxidative stress, while manganese-phosphate complexes provide antioxidant activity [26]. Thus, phosphate released through polyP degradation in yeast cells could allow for manganese adaptation.
In bacteria, polyP is known to be necessary for oxidative stress adaptation [2] due to its ability to act as a chaperone [24,25]. In our study, we found evidence for polyP involvement in stress response in yeast: The reduced polyP content through Ppn1 polyphosphatase overexpression leads to a significant alteration in expression of genes responsible for manganese and oxidative stress adaptation. Manganese homeostasis in S. cerevisiae involves numerous proteins [26,51,52,53]. The adaptation of yeast cells to toxic concentrations of manganese ions involves—in particular, the transport systems responsible for manganese compartmentalization, the systems of phosphorus transport and polyphosphate accumulation, as well as other membrane proteins. In this study, we identified new proteins contributing to manganese adaptation in S. cerevisiae. The protein encoded by PHM7 is essential for this process. The Phm7 protein sequence contains several transmembrane domains [54], and the protein is localized in the plasma membrane [55]. Its ortholog CaPhm7 in Candida albicans is also localized in the plasma membrane, and participates in drug resistance and filamentous growth [56,57]. The cells lacking CaPHM7 are sensitive to SDS and ketoconazole, but resistant to rapamycin and zinc [56]. This evidence suggests that Phm7 is a membrane transporter of divalent metal cations.
Another important observation is the role of Pho84 in oxidative stress response, in addition to its known role in phosphate and manganese transport. Pho84 is the major high-affinity plasma membrane phosphate importer of S. cerevisiae and an important element of phosphate homeostasis [58]. Pho84 is a phosphate transceptor: It transports phosphate and mediates rapid phosphate activation of the protein kinase A (PKA) pathway [59]. This protein also acts as a divalent cation transporter [52]. The knockout strain shows multiple phenotypic changes: Poor growth on a low-phosphate medium, constitutive up-regulation of the PHO pathway, and negligible levels of polyP, enhanced arsenate [60], and manganese resistance [61]. The overexpression of PHO84 induces enhanced heavy metal accumulation [62].
In our case, PHO84 is down-regulated in the CRN/PPN1 strain that exhibits manganese and resistance to peroxide. In C. albicans, Pho84 participates in oxidative stress response: The Δpho84 mutant accumulates intracellular ROS in the absence of extrinsic oxidative stress and hyperactivates oxidative stress signaling [63]. Thus, PHO84 down-regulation enhances resistance to manganese by decreasing metal ion uptake and mobilizing oxidative stress signaling. This down-regulation can also cause resistance to peroxide.
The mechanisms of PHO84 down-regulation in CRN/PPN1 are still in question. The intracellular phosphate level may be increased as a result of polyP hydrolysis in the PPN1 overproducing strain CRN/PPN1, which leads both to PHO84 down-regulation and an increase of the ROS level. A decrease of the polyP level reduces the antioxidant potential of the cell, because polyP plays an antioxidant role [24], additionally increasing the ROS level. As a result, the cells mobilize their stress response pathways, thereby providing, together with PHO84 down-regulation, the manganese and peroxide resistance of the CRN/PPN1 strain.
The reduced polyP of CRN/PPN1 was expected to lead to decreased resistance to manganese. Instead, we found the opposite—i.e., the increased resistance achieved through the pre-adaptive state. PolyP deficiency does not affect the cell phenotype and growth in normal conditions, but acts as a cellular signal leading to major changes in the gene expression pattern that facilitates stress adaptation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/8/5/461/s1, Figure S1: The micrographs of S. cerevisiae cells cultivated in YPD medium with 5 mM MnSO4 for logarithmic growth stage; Figure S2: The micrographs of the cells of CRN and CRN/PPN1 strains of S. cerevisiae stained with 1,2,3-dihydrorhodamine; Table S1: The S. cerevisiae strains used in the study; Table S2: The Pi and polyP content in S. cerevisiae cells grown in control YPD medium and in the presence of 5mM MnSO4; Table S3: Gene counts estimated from the RNA-Seq data; Table S4: Differential gene expression data.
Author Contributions
Conceptualization—T.K.; methodology—T.K., A.P., A.V., and I.V.K.; software—I.V.K.; formal analysis—T.K., L.T., I.V.K., and I.E.; investigation—L.T., A.A.P., A.Z., A.V., I.A.E., and V.O.; writing—T.K., and I.V.K.; supervision—T.K.; visualization—A.Z., V.O., and I.V.K.
Funding
This work was supported by the Russian Foundation for Basic Research [primary experimental analysis: grant 17-04-00822 to T.K.; analysis of raw sequencing data: grant 18-34-20024 to I.V.K.]; Russian Science Foundation [analysis of differential gene expression: grant 17-74-10179 to I.A.E.]; Russian Academy of Sciences Projects [high-throughput sequencing: 0053-2019-0005; other bioinformatics analysis: 0112-2019-0001].
Acknowledgments
We personally thank Eugenia V. Serebrova and Elena Makeeva for valuable comments and suggestions.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Jiménez, J.; Bru, S.; Ribeiro, M.P.; Clotet, J. Phosphate: from stardust to eukaryotic cell cycle control. Int. Microbiol. 2016, 19, 133–141. [Google Scholar]
- Rao, N.N.; Gómez-García, M.R.; Kornberg, A. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. [Google Scholar] [CrossRef] [PubMed]
- Holmström, K.M.; Marina, N.; Baev, A.Y.; Wood, N.W.; Gourine, A.V.; Abramov, A.Y. Signalling properties of inorganic polyphosphate in the mammalian brain. Nat. Commun. 2013, 4, 1362. [Google Scholar] [CrossRef] [PubMed]
- Angelova, P.R.; Iversen, K.Z.; Teschemacher, A.G.; Kasparov, S.; Gourine, A.V.; Abramov, A.Y. Signal transduction in astrocytes: Localization and release of inorganic polyphosphate. Glia 2018, 66, 2126–2136. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, E.; Aschar-Sobbi, R.; Campanella, M.; Turner, R.J.; Gómez-García, M.R.; Abramov, A.Y. Inorganic polyphosphate and energy metabolism in mammalian cells. J. Biol. Chem. 2010, 285, 9420–9428. [Google Scholar] [CrossRef] [PubMed]
- Seidlmayer, L.K.; Gomez-Garcia, M.R.; Blatter, L.A.; Pavlov, E.; Dedkova, E.N. Inorganic polyphosphate is a potent activator of the mitochondrial permeability transition pore in cardiac myocytes. J. Gen. Physiol. 2012, 139, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Seidlmayer, L.K.; Juettner, V.V.; Kettlewell, S.; Pavlov, E.V.; Blatter, L.A.; Dedkova, E.N. Distinct mPTP activation mechanisms in ischaemia-reperfusion: contributions of Ca2+, ROS, pH, and inorganic polyphosphate. Cardiovasc. Res. 2015, 106, 237–248. [Google Scholar] [CrossRef]
- Dedkova, E.N. Inorganic polyphosphate in cardiac myocytes: from bioenergetics to the permeability transition pore and cell survival. Biochem. Soc. Trans. 2016, 44, 25–34. [Google Scholar] [CrossRef]
- Omelon, S.; Georgiou, J.; Henneman, Z.J.; Wise, L.M.; Sukhu, B.; Hunt, T.; Wynnyckyj, C.; Holmyard, D.; Bielecki, R.; Grynpas, M.D. Control of vertebrate skeletal mineralization by polyphosphates. PLoS ONE 2009, 4, e5634. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.G.; Wang, X.; Diehl-Seifert, B.; Kropf, K.; Schlossmacher, U.; Lieberwirth, I.; Glasser, G.; Wiens, M.; Schröder, H.C. Inorganic polymeric phosphate/polyphosphate as an inducer of alkaline phosphatase and a modulator of intracellular Ca2+ level in osteoblasts (SaOS-2 cells) in vitro. Acta Biomater. 2011, 7, 2661–2671. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, K.; Saito, N.; Kawazoe, Y.; Ooi, H.-K.; Shiba, T. Morphogenetic Study on the Maturation of Osteoblastic Cell as Induced by Inorganic Polyphosphate. PLoS ONE 2014, 9, e86834. [Google Scholar] [CrossRef] [PubMed]
- Mikami, Y.; Tsuda, H.; Akiyama, Y.; Honda, M.; Shimizu, N.; Suzuki, N.; Komiyama, K. Alkaline phosphatase determines polyphosphate-induced mineralization in a cell-type independent manner. J. Bone Miner. Metab. 2016, 34, 627–637. [Google Scholar] [CrossRef]
- Smith, S.A.; Morrissey, J.H. Polyphosphate as a general procoagulant agent. J. Thromb. Haemost. 2008, 6, 1750–1756. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Choi, S.H.; Davis-Harrison, R.; Huyck, J.; Boettcher, J.; Rienstra, C.M.; Reinstra, C.M.; Morrissey, J.H. Polyphosphate exerts differential effects on blood clotting, depending on polymer size. Blood 2010, 116, 4353–4359. [Google Scholar] [CrossRef] [PubMed]
- Lander, N.; Cordeiro, C.; Huang, G.; Docampo, R. Polyphosphate and acidocalcisomes. Biochem. Soc. Trans. 2016, 44, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hassanian, S.M.; Avan, A.; Ardeshirylajimi, A. Inorganic polyphosphate: a key modulator of inflammation. J. Thromb. Haemost. 2017, 15, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Chrysanthopoulou, A.; Kambas, K.; Stakos, D.; Mitroulis, I.; Mitsios, A.; Vidali, V.; Angelidou, I.; Bochenek, M.; Arelaki, S.; Arampatzioglou, A.; et al. Interferon lambda1/IL-29 and inorganic polyphosphate are novel regulators of neutrophil-driven thromboinflammation. J. Pathol. 2017, 243, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-M.; Bandyopadhyay, N.; Rifat, D.; Rubin, H.; Bader, J.S.; Karakousis, P.C. Deficiency of the Novel Exopolyphosphatase Rv1026/PPX2 Leads to Metabolic Downshift and Altered Cell Wall Permeability in Mycobacterium tuberculosis. MBio 2015, 6, e02428. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Gangaiah, D.; Torrelles, J.B.; Rajashekara, G. Polyphosphate and associated enzymes as global regulators of stress response and virulence in Campylobacter jejuni. World J. Gastroenterol. 2016, 22, 7402. [Google Scholar] [CrossRef]
- Albi, T.; Serrano, A. Inorganic polyphosphate in the microbial world. Emerging roles for a multifaceted biopolymer. World J. Microbiol. Biotechnol. 2016, 32, 27. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.; Bru, S.; Ribeiro, M.P.C.; Clotet, J. Polyphosphate: popping up from oblivion. Curr. Genet. 2017, 63, 15–18. [Google Scholar] [CrossRef]
- Kuroda, A. A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci. Biotechnol. Biochem. 2006, 70, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J. Inorganic Polyphosphate Accumulation in Escherichia coli Is Regulated by DksA but Not by (p)ppGpp. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Gray, M.J.; Jakob, U. Oxidative stress protection by polyphosphate—new roles for an old player. Curr. Opin. Microbiol. 2015, 24, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.J.; Wholey, W.-Y.; Wagner, N.O.; Cremers, C.M.; Mueller-Schickert, A.; Hock, N.T.; Krieger, A.G.; Smith, E.M.; Bender, R.A.; Bardwell, J.C.A.; et al. Polyphosphate is a primordial chaperone. Mol. Cell 2014, 53, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Culotta, V.C.; Daly, M.J. Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid. Redox Signal. 2013, 19, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Vagabov, V.M.; Trilisenko, L.V.; Kulaev, I.S. Dependence of inorganic polyphosphate chain length on the orthophosphate content in the culture medium of the yeast Saccharomyces cerevisiae. Biochemistry. (Mosc). 2000, 65, 349–354. [Google Scholar] [PubMed]
- Hothorn, M.; Neumann, H.; Lenherr, E.D.; Wehner, M.; Rybin, V.; Hassa, P.O.; Uttenweiler, A.; Reinhardt, M.; Schmidt, A.; Seiler, J.; et al. Catalytic Core of a Membrane-Associated Eukaryotic Polyphosphate Polymerase. Science 2009, 324, 513–516. [Google Scholar] [CrossRef]
- Gerasimaitė, R.; Mayer, A. Ppn2, a novel Zn 2+ -dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J. Cell Sci. 2017, 130, 1625–1636. [Google Scholar] [CrossRef]
- Lichko, L.P.; Andreeva, N.A.; Kulakovskaya, T.V.; Kulaev, I.S. Exopolyphosphatases of the yeast Saccharomyces cerevisiae. FEMS Yeast Res. 2003, 3, 233–238. [Google Scholar] [CrossRef]
- Ortiz-Severín, J.; Varas, M.; Bravo-Toncio, C.; Guiliani, N.; Chávez, F.P. Multiple antibiotic susceptibility of polyphosphate kinase mutants (ppk1 and ppk2) from Pseudomonas aeruginosa PAO1 as revealed by global phenotypic analysis. Biol. Res. 2015, 48, 22. [Google Scholar] [CrossRef] [PubMed]
- Bashatwah, R.M.; Khanfar, M.A.; Bardaweel, S.K. Discovery of potent polyphosphate kinase 1 (PPK1) inhibitors using structure-based exploration of PPK1Pharmacophoric space coupled with docking analyses. J. Mol. Recognit. 2018, 31, e2726. [Google Scholar] [CrossRef] [PubMed]
- Desfougères, Y.; Gerasimaitė, R.U.; Jessen, H.J.; Mayer, A. Vtc5, a Novel Subunit of the Vacuolar Transporter Chaperone Complex, Regulates Polyphosphate Synthesis and Phosphate Homeostasis in Yeast. J. Biol. Chem. 2016, 291, 22262–22275. [Google Scholar] [CrossRef] [PubMed]
- Gerasimaite, R.; Sharma, S.; Desfougeres, Y.; Schmidt, A.; Mayer, A. Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 2014, 127, 5093–5104. [Google Scholar] [CrossRef] [PubMed]
- Trilisenko, L.; Tomashevsky, A.; Kulakovskaya, T.; Kulaev, I. V-ATPase dysfunction suppresses polyphosphate synthesis in Saccharomyces cerevisiae. Folia Microbiol. (Praha.) 2013, 58, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Müller, O.; Neumann, H.; Bayer, M.J.; Mayer, A. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 2003, 116, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Milgrom, E.; Diab, H.; Middleton, F.; Kane, P.M. Loss of vacuolar proton-translocating ATPase activity in yeast results in chronic oxidative stress. J. Biol. Chem. 2007, 282, 7125–7136. [Google Scholar] [CrossRef] [PubMed]
- Charoenbhakdi, S.; Dokpikul, T.; Burphan, T.; Techo, T.; Auesukaree, C. Vacuolar H + -ATPase Protects Saccharomyces cerevisiae Cells against Ethanol-Induced Oxidative and Cell Wall Stresses. Appl. Environ. Microbiol. 2016, 82, 3121–3130. [Google Scholar] [CrossRef]
- Nishikawa, H.; Miyazaki, T.; Nakayama, H.; Minematsu, A.; Yamauchi, S.; Yamashita, K.; Takazono, T.; Shimamura, S.; Nakamura, S.; Izumikawa, K.; et al. Roles of vacuolar H + -ATPase in the oxidative stress response of Candida glabrata. FEMS Yeast Res. 2016, 16, fow054. [Google Scholar] [CrossRef] [PubMed]
- Eldarov, M.A.; Baranov, M.V.; Dumina, M.V.; Shgun, A.A.; Andreeva, N.A.; Trilisenko, L.V.; Kulakovskaya, T.V.; Ryasanova, L.P.; Kulaev, I.S. Polyphosphates and exopolyphosphatase activities in the yeast Saccharomyces cerevisiae under overexpression of homologous and heterologous PPN1 genes. Biochem. 2013, 78, 946–953. [Google Scholar] [CrossRef]
- Kulakovskaya, T. Inorganic polyphosphates and heavy metal resistance in microorganisms. World J. Microbiol. Biotechnol. 2018, 34, 139. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, N.; Ryazanova, L.; Dmitriev, V.; Kulakovskaya, T.; Kulaev, I. Adaptation of Saccharomyces cerevisiae to toxic manganese concentration triggers changes in inorganic polyphosphates. FEMS Yeast Res. 2013, 13, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, R.; Park, J.; Karra, K.; Hitz, B.C.; Binkley, G.; Hong, E.L.; Sullivan, J.; Micklem, G.; Cherry, J.M. YeastMine-An integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database 2012, 2012, bar062. [Google Scholar] [CrossRef] [PubMed]
- Trilisenko, L.; Kulakovskaya, E.; Kulakovskaya, T. The cadmium tolerance in Saccharomyces cerevisiae depends on inorganic polyphosphate. J. Basic Microbiol. 2017, 57, 982–986. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.; Fernandes, E.; Lima, J.L.F.C. Fluorescence probes used for detection of reactive oxygen species. J. Biochem. Biophys. Methods 2005, 65, 45–80. [Google Scholar] [CrossRef]
- Andreeva, N.; Kulakovskaya, E.; Zvonarev, A.; Penin, A.; Eliseeva, I.; Teterina, A.; Lando, A.; Kulakovskiy, I.V.; Kulakovskaya, T. Transcriptome profile of yeast reveals the essential role of PMA2 and uncharacterized gene YBR056W-A (MNC1) in adaptation to toxic manganese concentration. Metallomics 2017, 9, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, N.; DeRisi, J.; Brown, P.O. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 2000, 11, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Wawrzycka, D.; Sobczak, I.; Bartosz, G.; Bocer, T.; Ułaszewski, S.; Goffeau, A. Vmr 1p is a novel vacuolar multidrug resistance ABC transporter in Saccharomyces cerevisiae. FEMS Yeast Res. 2010, 10, 828–838. [Google Scholar] [CrossRef]
- Sousa, C.A.; Hanselaer, S.; Soares, E.V. ABCC Subfamily Vacuolar Transporters are Involved in Pb (Lead) Detoxification in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2015, 175, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, R.; Tamás, M.J. How Saccharomyces cerevisiae copes with toxic metals and metalloids. FEMS Microbiol. Rev. 2010, 34, 925–951. [Google Scholar] [CrossRef] [PubMed]
- Reddi, A.R.; Jensen, L.T.; Culotta, V.C. Manganese homeostasis in Saccharomyces cerevisiae. Chem. Rev. 2009, 109, 4722–4732. [Google Scholar] [CrossRef] [PubMed]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2012, 40, D71–D75. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.-K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Pan, H. Functions of CaPhm7 in the regulation of ion homeostasis, drug tolerance, filamentation and virulence in Candida albicans. BMC Microbiol. 2018, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yang, Y. The putative transient receptor potential channel protein encoded by the orf19.4805 gene is involved in cation sensitivity, antifungal tolerance, and filamentation in Candida albicans. Can. J. Microbiol. 2018, 64, 727–731. [Google Scholar] [CrossRef]
- Tomar, P.; Sinha, H. Conservation of PHO pathway in ascomycetes and the role of Pho84. J. Biosci. 2014, 39, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Popova, Y.; Thayumanavan, P.; Lonati, E.; Agrochao, M.; Thevelein, J.M. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc. Natl. Acad. Sci. USA 2010, 107, 2890–2895. [Google Scholar] [CrossRef] [PubMed]
- Wykoff, D.D.; Rizvi, A.H.; Raser, J.M.; Margolin, B.; O’Shea, E.K. Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol. Cell 2007, 27, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.T.; Ajua-Alemanji, M.; Culotta, V.C. The Saccharomyces cerevisiae high affinity phosphate transporter encoded by PHO84 also functions in manganese homeostasis. J. Biol. Chem. 2003, 278, 42036–42040. [Google Scholar] [CrossRef] [PubMed]
- Ofiteru, A.M.; Ruta, L.L.; Rotaru, C.; Dumitru, I.; Ene, C.D.; Neagoe, A.; Farcasanu, I.C. Overexpression of the PHO84 gene causes heavy metal accumulation and induces Ire1p-dependent unfolded protein response in Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2012, 94, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.-N.; Uppuluri, P.; Broggi, A.; Besold, A.; Ryman, K.; Kambara, H.; Solis, N.; Lorenz, V.; Qi, W.; Acosta-Zaldívar, M.; et al. Intersection of phosphate transport, oxidative stress and TOR signalling in Candida albicans virulence. PLOS Pathog. 2018, 14, e1007076. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).