Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins
Abstract
1. Introduction
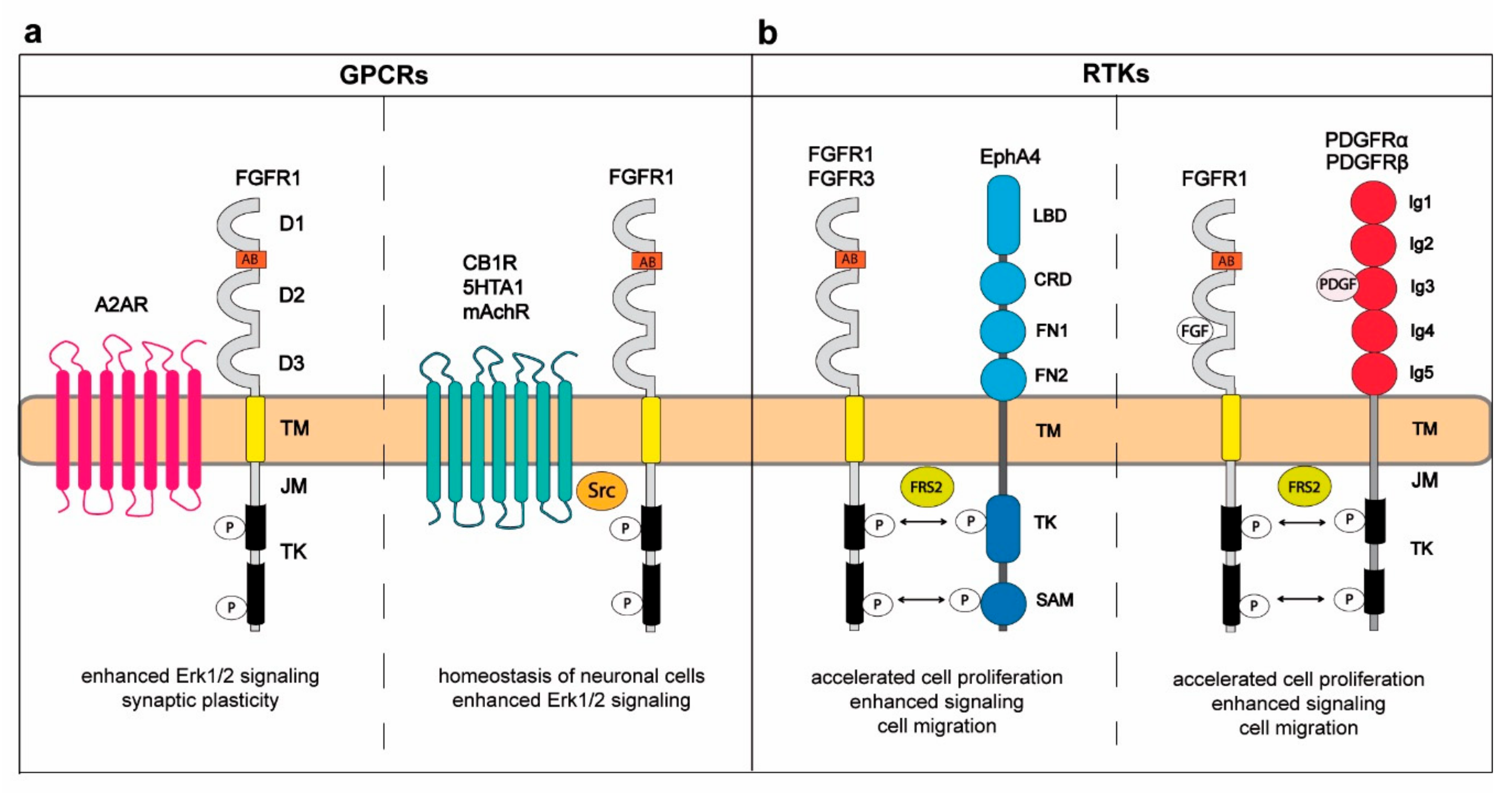
2. Cross-Talk between FGFRs and G-Protein-Coupled Receptors in Regulation of the Central Nervous System
3. Interplay between FGFRs and Other RTKs
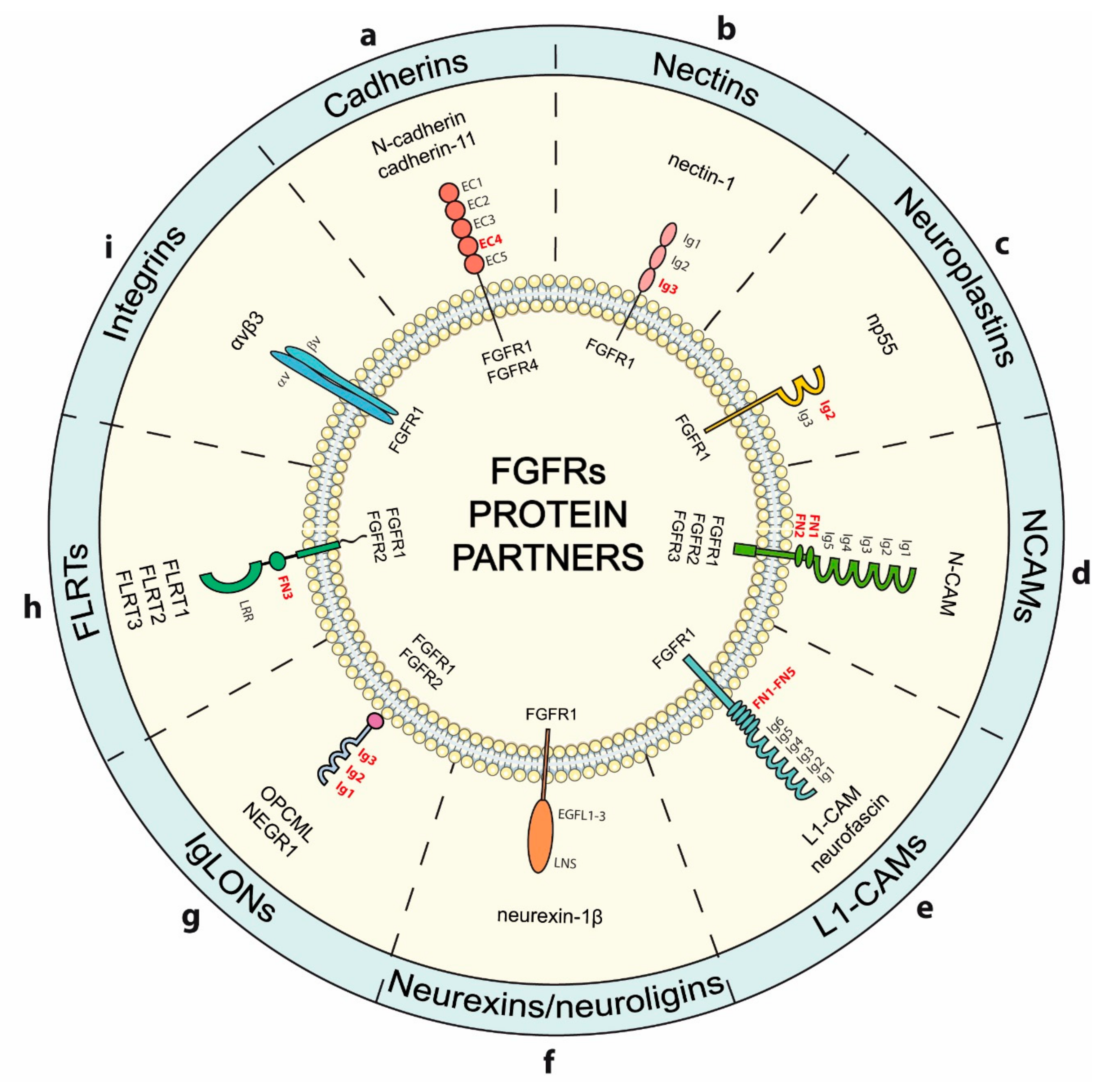
4. Modulation of FGFRs Activity by Cell-Surface Proteins Involved in Adhesion
4.1. Cadherins
4.2. Nectins
4.3. Neuroplastins
4.4. N-CAMs
4.5. L1-CAMs
4.6. Neurexins
4.7. IgLONs
4.8. FLRTs
4.9. Integrins
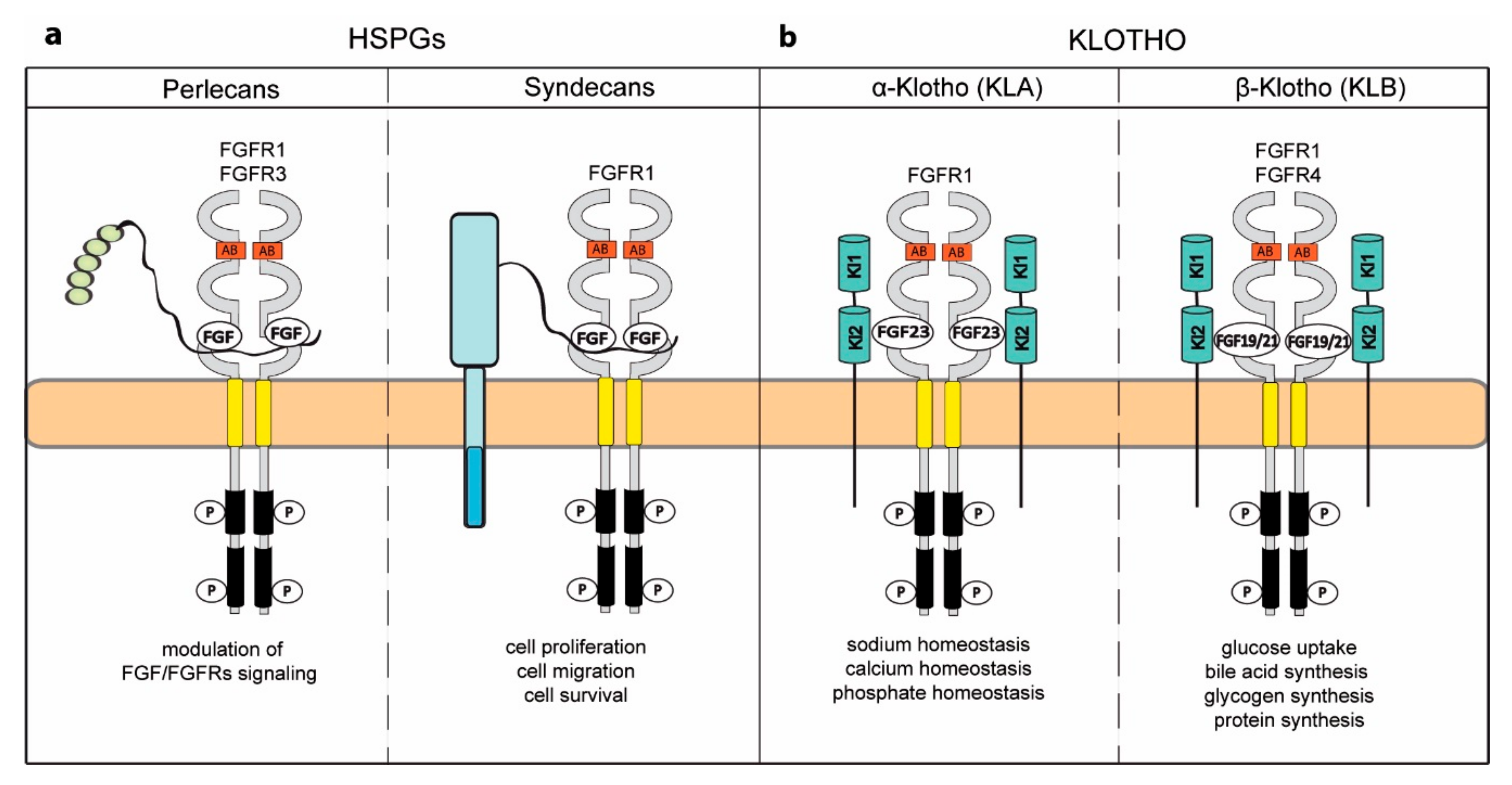
5. Novel Activities Acquired by FGFRs upon Binding to Specific Coreceptors
5.1. Heparan Sulfate Proteoglycans
5.2. Klotho Coreceptors
6. Modulation of FGFRs by Other Cell Surface Proteins
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 5-HT1A | 5-hydroksytriptamine receptor 1A |
| 8-OH-DPAT | 7(Dipropylamino)-5,6,7,8-tetrahydronaphthalen-1-ol |
| A2AR | adenosine receptor |
| AJ | adherens junction |
| AKT | protein kinase B |
| ATP | adenosine triphosphate |
| BRET | bioluminescence resonance energy transfer |
| CAMs | cell adhesion molecules |
| CB1R | cannabinoid receptor 1 |
| CNS | central nervous system |
| Dlg-1 | disks large homolog 1 |
| EGFs | epidermal growth factors |
| EGFRs | epidermal growth factor receptors |
| EMT | epithelial to mesenchymal transition |
| Eph | ephrin |
| ERK1/2 | extracellular regulated kinases 1/2 |
| FGFs | fibroblast growth factors |
| FGFRs | fibroblast growth factor receptors |
| FGFRL1 | fibroblast growth factor receptor like 1 |
| FLRTs | fibronectin leucine-rich transmembranes |
| FN3 | fibronectin type III |
| FRET | Förster Resonance Energy Transfer |
| FRS2 | fibroblast growth factor receptor substrate 2 |
| GAG | glysocaminoglycan |
| GPCRs | G-protein-coupled receptors |
| GPI | glycosylphosphatidylinositol |
| HS | heparan sulfate |
| HSPGs | heparan sulfate proteoglycans |
| IGFR | insulin-like growth factor receptor |
| JM | juxtamembrane |
| KLA | α-klotho |
| KLB | β-klotho |
| L1-CAM | L1 cell adhesion molecule |
| LNS | laminin, neurexin, sex hormone binding globulin |
| LPR | leucine-rich repeat domain |
| mAChR | muscarinic acetylcholine receptor |
| MAPK | mitogen-activated protein kinase |
| MOR | mu-opioid receptor |
| mTOR | mammalian target of rapamycin |
| N-CAMs | neural cell adhesion molecules |
| NEGR1 | neuronal growth regulator 1 |
| NFs | neurofascins |
| Np55 | neuroplastin 55 |
| OPCML | opioid binding protein cell adhesion molecule |
| PDGFs | platelet-derived growth factors |
| PDGFRs | platelet-derived growth factor receptors |
| PI13K | phosphoinositide 3-kinase |
| PKC | protein kinase C |
| PLA | proximity ligation assay |
| PLCɣ | phospholipase Cɣ |
| RTKs | receptor tyrosine kinases |
| SAM | sterile alpha motif |
| Sef | similar expression to fgf genes |
| SPA | solid-phase assay |
| SPR | surface plasmon resonance |
| STAT | signal transducer and activator of transcription |
| TGFs | transforming growth factors |
| TGFBRs | transforming growth factor receptors |
| Y2H | yeast two-hybrid |
References
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Hallinan, N.; Finn, S.; Cuffe, S.; Rafee, S.; O’Byrne, K.; Gately, K. Targeting the fibroblast growth factor receptor family in cancer. Cancer Treat. Rev. 2016, 46, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Marie, P.J. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015, 29, 1463–1486. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Olsen, S.K.; Ibrahimi, O.A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005, 16, 107–137. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.N.; Hubbard, S.R.; Schlessinger, J.; Mohammadi, M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 2000, 101, 413–424. [Google Scholar] [CrossRef]
- Olsen, S.K.; Ibrahimi, O.A.; Raucci, A.; Zhang, F.; Eliseenkova, A.V.; Yayon, A.; Basilico, C.; Linhardt, R.J.; Schlessinger, J.; Mohammadi, M. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc. Natl. Acad. Sci. USA 2004, 101, 935–940. [Google Scholar] [CrossRef]
- Kalinina, J.; Dutta, K.; Ilghari, D.; Beenken, A.; Goetz, R.; Eliseenkova, A.V.; Cowburn, D.; Mohammadi, M. The alternatively spliced acid box region plays a key role in FGF receptor autoinhibition. Structure 2012, 20, 77–88. [Google Scholar] [CrossRef]
- Kiselyov, V.V.; Kochoyan, A.; Poulsen, F.M.; Bock, E.; Berezin, V. Elucidation of the mechanism of the regulatory function of the Ig1 module of the fibroblast growth factor receptor 1. Protein Sci. 2006, 15, 2318–2322. [Google Scholar] [CrossRef] [PubMed]
- Opalinski, L.; Szczepara, M.; Sokolowska-Wedzina, A.; Zakrzewska, M.; Otlewski, J. The autoinhibitory function of D1 domain of FGFR1 goes beyond the inhibition of ligand binding. Int. J. Biochem. Cell Biol. 2017, 89, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.C.; Lin, X.; Torres, J. The strong dimerization of the transmembrane domain of the fibroblast growth factor receptor (FGFR) is modulated by C-terminal juxtamembrane residues. Protein Sci. 2009, 18, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Xu, J.; Ischenko, I.; Ornitz, D.M.; Halegoua, S.; Hayman, M.J. Identification of the cytoplasmic regions of fibroblast growth factor (FGF) receptor 1 which play important roles in induction of neurite outgrowth in PC12 cells by FGF-1. Mol. Cell Biol. 1998, 18, 3762–3770. [Google Scholar] [CrossRef]
- Burgar, H.R.; Burns, H.D.; Elsden, J.L.; Lalioti, M.D.; Heath, J.K. Association of the signaling adaptor FRS2 with fibroblast growth factor receptor 1 (Fgfr1) is mediated by alternative splicing of the juxtamembrane domain. J. Biol. Chem. 2002, 277, 4018–4023. [Google Scholar] [CrossRef] [PubMed]
- Sarabipour, S.; Hristova, K. FGFR3 unliganded dimer stabilization by the juxtamembrane domain. J. Mol. Biol. 2015, 427, 1705–1714. [Google Scholar] [CrossRef]
- Miki, T.; Bottaro, D.P.; Fleming, T.P.; Smith, C.L.; Burgess, W.H.; Chan, A.M.; Aaronson, S.A. Determination of ligand-binding specificity by alternative splicing: Two distinct growth factor receptors encoded by a single gene. Proc. Natl. Acad. Sci. USA 1992, 89, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Chellaiah, A.T.; McEwen, D.G.; Werner, S.; Xu, J.; Ornitz, D.M. Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J. Biol. Chem. 1994, 269, 11620–11627. [Google Scholar] [PubMed]
- Gong, S.G. Isoforms of receptors of fibroblast growth factors. J. Cell. Physiol. 2014, 229, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, M.; Trueb, B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics 2000, 69, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Trueb, B.; Zhuang, L.; Taeschler, S.; Wiedemann, M. Characterization of FGFRL1, a novel fibroblast growth factor (FGF) receptor preferentially expressed in skeletal tissues. J. Biol. Chem. 2003, 278, 33857–33865. [Google Scholar] [CrossRef]
- Goetz, R.; Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 2013, 14, 166–180. [Google Scholar] [CrossRef]
- Furdui, C.M.; Lew, E.D.; Schlessinger, J.; Anderson, K.S. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 2006, 21, 711–717. [Google Scholar] [CrossRef]
- Zinkle, A.; Mohammadi, M. A threshold model for receptor tyrosine kinase signaling specificity and cell fate determination. F1000Researcharch 2018, 7. [Google Scholar] [CrossRef]
- Sarabipour, S.; Hristova, K. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 2016, 7, 10262. [Google Scholar] [CrossRef]
- Duchesne, L.; Tissot, B.; Rudd, T.R.; Dell, A.; Fernig, D.G. N-glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J. Biol. Chem. 2006, 281, 27178–27189. [Google Scholar] [CrossRef] [PubMed]
- Polanska, U.M.; Duchesne, L.; Harries, J.C.; Fernig, D.G.; Kinnunen, T.K. N-Glycosylation regulates fibroblast growth factor receptor/EGL-15 activity in Caenorhabditis elegans in vivo. J. Biol. Chem. 2009, 284, 33030–33039. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.N.; Kilgour, E.; Smith, P.D. Molecular pathways: Fibroblast growth factor signaling: A new therapeutic opportunity in cancer. Clin. Cancer Res. 2012, 18, 1855–1862. [Google Scholar] [CrossRef] [PubMed]
- Haugsten, E.M.; Malecki, J.; Bjorklund, S.M.; Olsnes, S.; Wesche, J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol. Biol. Cell 2008, 19, 3390–3403. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, M.; Haugsten, E.M.; Nadratowska-Wesolowska, B.; Oppelt, A.; Hausott, B.; Jin, Y.; Otlewski, J.; Wesche, J.; Wiedlocha, A. ERK-mediated phosphorylation of fibroblast growth factor receptor 1 on Ser777 inhibits signaling. Sci. Signal. 2013, 6, ra11. [Google Scholar] [CrossRef]
- Porebska, N.; Latko, M.; Kucinska, M.; Zakrzewska, M.; Otlewski, J.; Opalinski, L. Targeting Cellular Trafficking of Fibroblast Growth Factor Receptors as a Strategy for Selective Cancer Treatment. J. Clin. Med. 2018, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Fan, J.; Chung, T.W.; Lythgoe, K.; Wang, X.; Xie, J.; Ge, Q.; Gu, T.L.; Polakiewicz, R.D.; Roesel, J.L.; et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol. Cell 2011, 44, 864–877. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, M.K.; Maher, P.A.; Stachowiak, E.K. Integrative nuclear signaling in cell development--a role for FGF receptor-1. DNA Cell Biol. 2007, 26, 811–826. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Ohta, H.; Konishi, M. Endocrine FGFs: Evolution, Physiology, Pathophysiology, and Pharmacotherapy. Front. Endocrinol. (Lausanne) 2015, 6, 154. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, A.; Cooper, H.J.; Trim, K.J.; Akbarzadeh, S.; Heath, J.K.; Wheldon, L.M. Protein partners in the life history of activated fibroblast growth factor receptors. Proteomics 2007, 7, 4565–4578. [Google Scholar] [CrossRef] [PubMed]
- Balek, L.; Nemec, P.; Konik, P.; Kunova Bosakova, M.; Varecha, M.; Gudernova, I.; Medalova, J.; Krakow, D.; Krejci, P. Proteomic analyses of signalling complexes associated with receptor tyrosine kinase identify novel members of fibroblast growth factor receptor 3 interactome. Cell Signal. 2018, 42, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Kostas, M.; Haugsten, E.M.; Zhen, Y.; Sorensen, V.; Szybowska, P.; Fiorito, E.; Lorenz, S.; Jones, N.; de Souza, G.A.; Wiedlocha, A.; et al. Protein Tyrosine Phosphatase Receptor Type G (PTPRG) Controls Fibroblast Growth Factor Receptor (FGFR) 1 Activity and Influences Sensitivity to FGFR Kinase Inhibitors. Mol. Cell Proteom. 2018, 17, 850–870. [Google Scholar] [CrossRef] [PubMed]
- Calebiro, D.; Koszegi, Z. The subcellular dynamics of GPCR signaling. Mol. Cell Endocrinol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Calebiro, D.; Jobin, M.L. Hot spots for GPCR signaling: Lessons from single-molecule microscopy. Curr. Opin. Cell Biol. 2018, 57, 57–63. [Google Scholar] [CrossRef]
- Milligan, G.; Ward, R.J.; Marsango, S. GPCR homo-oligomerization. Curr. Opin. Cell. Biol. 2018, 57, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Thal, D.M.; Glukhova, A.; Sexton, P.M.; Christopoulos, A. Structural insights into G-protein-coupled receptor allostery. Nature 2018, 559, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, J.P.; Sunahara, R.K. Mechanistic insights into GPCR-G protein interactions. Curr. Opin. Struct. Biol. 2016, 41, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [PubMed]
- Di Liberto, V.; Mudo, G.; Belluardo, N. Crosstalk between receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCR) in the brain: Focus on heteroreceptor complexes and related functional neurotrophic effects. Neuropharmacology 2018. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.C.Y.; Wong, Y.H. Role of G Protein-Coupled Receptors in the Regulation of Structural Plasticity and Cognitive Function. Molecules 2017, 22, 1239. [Google Scholar] [CrossRef] [PubMed]
- Belluardo, N.; Wu, G.; Mudo, G.; Hansson, A.C.; Pettersson, R.; Fuxe, K. Comparative localization of fibroblast growth factor receptor-1, -2, and -3 mRNAs in the rat brain: In situ hybridization analysis. J. Comp. Neurol. 1997, 379, 226–246. [Google Scholar] [CrossRef]
- Gonzalez, A.M.; Berry, M.; Maher, P.A.; Logan, A.; Baird, A. A comprehensive analysis of the distribution of FGF-2 and FGFR1 in the rat brain. Brain Res. 1995, 701, 201–226. [Google Scholar] [CrossRef]
- Ford-Perriss, M.; Abud, H.; Murphy, M. Fibroblast growth factors in the developing central nervous system. Clin. Exp. Pharm. Physiol. 2001, 28, 493–503. [Google Scholar] [CrossRef]
- Choubey, L.; Collette, J.C.; Smith, K.M. Quantitative assessment of fibroblast growth factor receptor 1 expression in neurons and glia. PeerJ. 2017, 5, e3173. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Yazaki, N.; Tagashira, S.; Miyake, A.; Ozaki, K.; Minami, M.; Satoh, M.; Ohta, M.; Kawasaki, T. Rat FGF receptor-4 mRNA in the brain is expressed preferentially in the medial habenular nucleus. Brain Res. Mol. Brain Res. 1994, 21, 344–348. [Google Scholar] [CrossRef]
- Turner, C.A.; Akil, H.; Watson, S.J.; Evans, S.J. The fibroblast growth factor system and mood disorders. Biol. Psychiatry 2006, 59, 1128–1135. [Google Scholar] [CrossRef]
- Flajolet, M.; Wang, Z.; Futter, M.; Shen, W.; Nuangchamnong, N.; Bendor, J.; Wallach, I.; Nairn, A.C.; Surmeier, D.J.; Greengard, P. FGF acts as a co-transmitter through adenosine A(2A) receptor to regulate synaptic plasticity. Nat. Neurosci. 2008, 11, 1402–1409. [Google Scholar] [CrossRef]
- Howlett, A.C.; Barth, F.; Bonner, T.I.; Cabral, G.; Casellas, P.; Devane, W.A.; Felder, C.C.; Herkenham, M.; Mackie, K.; Martin, B.R.; et al. International Union of Pharmacology. XXVII. Classification of cannabinoid receptors. Pharm. Rev. 2002, 54, 161–202. [Google Scholar] [CrossRef] [PubMed]
- Asimaki, O.; Leondaritis, G.; Lois, G.; Sakellaridis, N.; Mangoura, D. Cannabinoid 1 receptor-dependent transactivation of fibroblast growth factor receptor 1 emanates from lipid rafts and amplifies extracellular signal-regulated kinase 1/2 activation in embryonic cortical neurons. J. Neurochem. 2011, 116, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Di Liberto, V.; Borroto-Escuela, D.O.; Frinchi, M.; Verdi, V.; Fuxe, K.; Belluardo, N.; Mudo, G. Existence of muscarinic acetylcholine receptor (mAChR) and fibroblast growth factor receptor (FGFR) heteroreceptor complexes and their enhancement of neurite outgrowth in neural hippocampal cultures. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 235–245. [Google Scholar] [CrossRef]
- Borroto-Escuela, D.O.; Narvaez, M.; Perez-Alea, M.; Tarakanov, A.O.; Jimenez-Beristain, A.; Mudo, G.; Agnati, L.F.; Ciruela, F.; Belluardo, N.; Fuxe, K. Evidence for the existence of FGFR1-5-HT1A heteroreceptor complexes in the midbrain raphe 5-HT system. Biochem. Biophys. Res. Commun. 2015, 456, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; Romero-Fernandez, W.; Mudo, G.; Perez-Alea, M.; Ciruela, F.; Tarakanov, A.O.; Narvaez, M.; Di Liberto, V.; Agnati, L.F.; Belluardo, N.; et al. Fibroblast growth factor receptor 1- 5-hydroxytryptamine 1A heteroreceptor complexes and their enhancement of hippocampal plasticity. Biol. Psychiatry 2012, 71, 84–91. [Google Scholar] [CrossRef]
- Borroto-Escuela, D.O.; Perez-Alea, M.; Narvaez, M.; Tarakanov, A.O.; Mudo, G.; Jimenez-Beristain, A.; Agnati, L.F.; Ciruela, F.; Belluardo, N.; Fuxe, K. Enhancement of the FGFR1 signaling in the FGFR1-5-HT1A heteroreceptor complex in midbrain raphe 5-HT neuron systems. Relevance for neuroplasticity and depression. Biochem. Biophys. Res. Commun. 2015, 463, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; DuPont, C.M.; Li, X.; Savelli, D.; Lattanzi, D.; Srivastava, I.; Narvaez, M.; Di Palma, M.; Barbieri, E.; Andrade-Talavera, Y.; et al. Disturbances in the FGFR1-5-HT1A Heteroreceptor Complexes in the Raphe-Hippocampal 5-HT System Develop in a Genetic Rat Model of Depression. Front. Cell Neurosci. 2017, 11, 309. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; Tarakanov, A.O.; Fuxe, K. FGFR1-5-HT1A Heteroreceptor Complexes: Implications for Understanding and Treating Major Depression. Trends Neurosci. 2016, 39, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Borroto-Escuela, D.O.; Carlsson, J.; Ambrogini, P.; Narvaez, M.; Wydra, K.; Tarakanov, A.O.; Li, X.; Millon, C.; Ferraro, L.; Cuppini, R.; et al. Understanding the Role of GPCR Heteroreceptor Complexes in Modulating the Brain Networks in Health and Disease. Front. Cell Neurosci. 2017, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Belcheva, M.M.; Haas, P.D.; Tan, Y.; Heaton, V.M.; Coscia, C.J. The fibroblast growth factor receptor is at the site of convergence between mu-opioid receptor and growth factor signaling pathways in rat C6 glioma cells. J. Pharm. Exp. 2002, 303, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Volinsky, N.; Kholodenko, B.N. Complexity of receptor tyrosine kinase signal processing. Cold Spring Harb. Perspect. Biol. 2013, 5, a009043. [Google Scholar] [CrossRef] [PubMed]
- Saha, N.; Robev, D.; Mason, E.O.; Himanen, J.P.; Nikolov, D.B. Therapeutic potential of targeting the Eph/ephrin signaling complex. Int. J. Biochem. Cell Biol. 2018, 105, 123–133. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005, 6, 462–475. [Google Scholar] [CrossRef]
- Lisabeth, E.M.; Falivelli, G.; Pasquale, E.B. Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Shiuan, E.; Chen, J. Eph Receptor Tyrosine Kinases in Tumor Immunity. Cancer Res. 2016, 76, 6452–6457. [Google Scholar] [CrossRef] [PubMed]
- Himanen, J.P.; Rajashankar, K.R.; Lackmann, M.; Cowan, C.A.; Henkemeyer, M.; Nikolov, D.B. Crystal structure of an Eph receptor-ephrin complex. Nature 2001, 414, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Kalo, M.S.; Pasquale, E.B. Signal transfer by Eph receptors. Cell Tissue Res. 1999, 298, 1–9. [Google Scholar] [CrossRef]
- Janes, P.W.; Nievergall, E.; Lackmann, M. Concepts and consequences of Eph receptor clustering. Semin. Cell Dev. Biol. 2012, 23, 43–50. [Google Scholar] [CrossRef]
- Himanen, J.P.; Yermekbayeva, L.; Janes, P.W.; Walker, J.R.; Xu, K.; Atapattu, L.; Rajashankar, K.R.; Mensinga, A.; Lackmann, M.; Nikolov, D.B.; et al. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. USA 2010, 107, 10860–10865. [Google Scholar] [CrossRef] [PubMed]
- Schmucker, D.; Zipursky, S.L. Signaling downstream of Eph receptors and ephrin ligands. Cell 2001, 105, 701–704. [Google Scholar] [CrossRef]
- Yokote, H.; Fujita, K.; Jing, X.; Sawada, T.; Liang, S.; Yao, L.; Yan, X.; Zhang, Y.; Schlessinger, J.; Sakaguchi, K. Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proc. Natl. Acad. Sci. USA 2005, 102, 18866–18871. [Google Scholar] [CrossRef] [PubMed]
- Fukai, J.; Yokote, H.; Yamanaka, R.; Arao, T.; Nishio, K.; Itakura, T. EphA4 promotes cell proliferation and migration through a novel EphA4-FGFR1 signaling pathway in the human glioma U251 cell line. Mol. Cancer 2008, 7, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008, 99, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Sawada, T.; Jing, X.; Zhang, Y.; Shimada, E.; Yokote, H.; Miyajima, M.; Sakaguchi, K. Ternary complex formation of EphA4, FGFR and FRS2alpha plays an important role in the proliferation of embryonic neural stem/progenitor cells. Genes Cells 2010, 15, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Sawada, T.; Arai, D.; Jing, X.; Furushima, K.; Chen, Q.; Kawakami, K.; Yokote, H.; Miyajima, M.; Sakaguchi, K. Trans-Activation between EphA and FGFR Regulates Self-Renewal and Differentiation of Mouse Embryonic Neural Stem/Progenitor Cells via Differential Activation of FRS2alpha. PLoS ONE 2015, 10, e0128826. [Google Scholar] [CrossRef]
- Zhang, Y.; Sawada, T.; Jing, X.; Yokote, H.; Yan, X.; Sakaguchi, K. Regulation of ephexin1, a guanine nucleotide exchange factor of Rho family GTPases, by fibroblast growth factor receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 2007, 282, 31103–31112. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Shatadal, S.; Griep, A.E. Dlg-1 Interacts With and Regulates the Activities of Fibroblast Growth Factor Receptors and EphA2 in the Mouse Lens. Invest. Ophthalmol. Vis. Sci. 2016, 57, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Griep, A.E. Loss of Dlg-1 in the mouse lens impairs fibroblast growth factor receptor signaling. PLoS ONE 2014, 9, e97470. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Multifarious functions of PDGFs and PDGFRs in tumor growth and metastasis. Trends Mol. Med. 2013, 19, 460–473. [Google Scholar] [CrossRef]
- Papadopoulos, N.; Lennartsson, J. The PDGF/PDGFR pathway as a drug target. Mol. Asp. Med. 2018, 62, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Shim, A.H.; Liu, H.; Focia, P.J.; Chen, X.; Lin, P.C.; He, X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc. Natl. Acad. Sci. USA 2010, 107, 11307–11312. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, K.; Backstrom, G.; Leppanen, O.; Persson, C.; Wernstedt, C.; Hellman, U.; Heldin, C.H.; Ostman, A. Role of immunoglobulin-like domains 2-4 of the platelet-derived growth factor alpha-receptor in ligand-receptor complex assembly. J. Biol. Chem. 1998, 273, 25495–25502. [Google Scholar] [CrossRef]
- Faraone, D.; Aguzzi, M.S.; Ragone, G.; Russo, K.; Capogrossi, M.C.; Facchiano, A. Heterodimerization of FGF-receptor 1 and PDGF-receptor-alpha: A novel mechanism underlying the inhibitory effect of PDGF-BB on FGF-2 in human cells. Blood 2006, 107, 1896–1902. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, P.Y.; Simons, M.; Friesel, R. FRS2 via fibroblast growth factor receptor 1 is required for platelet-derived growth factor receptor beta-mediated regulation of vascular smooth muscle marker gene expression. J. Biol. Chem. 2009, 284, 15980–15992. [Google Scholar] [CrossRef] [PubMed]
- Russo, K.; Ragone, R.; Facchiano, A.M.; Capogrossi, M.C.; Facchiano, A. Platelet-derived growth factor-BB and basic fibroblast growth factor directly interact in vitro with high affinity. J. Biol. Chem. 2002, 277, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- De Marchis, F.; Ribatti, D.; Giampietri, C.; Lentini, A.; Faraone, D.; Scoccianti, M.; Capogrossi, M.C.; Facchiano, A. Platelet-derived growth factor inhibits basic fibroblast growth factor angiogenic properties in vitro and in vivo through its alpha receptor. Blood 2002, 99, 2045–2053. [Google Scholar] [CrossRef]
- Facchiano, A.; De Marchis, F.; Turchetti, E.; Facchiano, F.; Guglielmi, M.; Denaro, A.; Palumbo, R.; Scoccianti, M.; Capogrossi, M.C. The chemotactic and mitogenic effects of platelet-derived growth factor-BB on rat aorta smooth muscle cells are inhibited by basic fibroblast growth factor. J. Cell Sci. 2000, 113 Pt 16, 2855–2863. [Google Scholar]
- Kono, S.A.; Heasley, L.E.; Doebele, R.C.; Camidge, D.R. Adding to the mix: Fibroblast growth factor and platelet-derived growth factor receptor pathways as targets in non-small cell lung cancer. Curr. Cancer Drug Targets 2012, 12, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 1995, 7, 619–627. [Google Scholar] [CrossRef]
- Kourtidis, A.; Lu, R.; Pence, L.J.; Anastasiadis, P.Z. A central role for cadherin signaling in cancer. Exp. Cell Res. 2017, 358, 78–85. [Google Scholar] [CrossRef]
- Gloushankova, N.A.; Rubtsova, S.N.; Zhitnyak, I.Y. Cadherin-mediated cell-cell interactions in normal and cancer cells. Tissue Barriers 2017, 5, e1356900. [Google Scholar] [CrossRef] [PubMed]
- Fontenete, S.; Pena-Jimenez, D.; Perez-Moreno, M. Heterocellular cadherin connections: Coordinating adhesive cues in homeostasis and cancer. F1000Research 2017, 6, 1010. [Google Scholar] [CrossRef]
- Nguyen, T.; Mege, R.M. N-Cadherin and Fibroblast Growth Factor Receptors crosstalk in the control of developmental and cancer cell migrations. Eur. J. Cell Biol. 2016, 95, 415–426. [Google Scholar] [CrossRef]
- Williams, E.J.; Furness, J.; Walsh, F.S.; Doherty, P. Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron 1994, 13, 583–594. [Google Scholar] [CrossRef]
- Saffell, J.L.; Williams, E.J.; Mason, I.J.; Walsh, F.S.; Doherty, P. Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron 1997, 18, 231–242. [Google Scholar] [CrossRef]
- Ronn, L.C.; Doherty, P.; Holm, A.; Berezin, V.; Bock, E. Neurite outgrowth induced by a synthetic peptide ligand of neural cell adhesion molecule requires fibroblast growth factor receptor activation. J. Neurochem. 2000, 75, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Heras, E.; Howell, F.V.; Williams, G.; Doherty, P. The fibroblast growth factor receptor acid box is essential for interactions with N-cadherin and all of the major isoforms of neural cell adhesion molecule. J. Biol. Chem. 2006, 281, 35208–35216. [Google Scholar] [CrossRef]
- Suyama, K.; Shapiro, I.; Guttman, M.; Hazan, R.B. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2002, 2, 301–314. [Google Scholar] [CrossRef]
- Hulit, J.; Suyama, K.; Chung, S.; Keren, R.; Agiostratidou, G.; Shan, W.; Dong, X.; Williams, T.M.; Lisanti, M.P.; Knudsen, K.; et al. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007, 67, 3106–3116. [Google Scholar] [CrossRef]
- Qian, X.; Anzovino, A.; Kim, S.; Suyama, K.; Yao, J.; Hulit, J.; Agiostratidou, G.; Chandiramani, N.; McDaid, H.M.; Nagi, C.; et al. N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene 2014, 33, 3411–3421. [Google Scholar] [CrossRef]
- Takehara, T.; Teramura, T.; Onodera, Y.; Frampton, J.; Fukuda, K. Cdh2 stabilizes FGFR1 and contributes to primed-state pluripotency in mouse epiblast stem cells. Sci. Rep. 2015, 5, 14722. [Google Scholar] [CrossRef] [PubMed]
- Cavallaro, U.; Niedermeyer, J.; Fuxa, M.; Christofori, G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat. Cell Biol. 2001, 3, 650–657. [Google Scholar] [CrossRef]
- Quintanal-Villalonga, A.; Ojeda-Marquez, L.; Marrugal, A.; Yague, P.; Ponce-Aix, S.; Salinas, A.; Carnero, A.; Ferrer, I.; Molina-Pinelo, S.; Paz-Ares, L. The FGFR4-388arg Variant Promotes Lung Cancer Progression by N-Cadherin Induction. Sci. Rep. 2018, 8, 2394. [Google Scholar] [CrossRef]
- Kimura, Y.; Matsunami, H.; Inoue, T.; Shimamura, K.; Uchida, N.; Ueno, T.; Miyazaki, T.; Takeichi, M. Cadherin-11 expressed in association with mesenchymal morphogenesis in the head, somite, and limb bud of early mouse embryos. Dev. Biol. 1995, 169, 347–358. [Google Scholar] [CrossRef]
- Sfikakis, P.P.; Vlachogiannis, N.I.; Christopoulos, P.F. Cadherin-11 as a therapeutic target in chronic, inflammatory rheumatic diseases. Clin. Immunol. 2017, 176, 107–113. [Google Scholar] [CrossRef]
- Birtolo, C.; Pham, H.; Morvaridi, S.; Chheda, C.; Go, V.L.; Ptasznik, A.; Edderkaoui, M.; Weisman, M.H.; Noss, E.; Brenner, M.B.; et al. Cadherin-11 Is a Cell Surface Marker Up-Regulated in Activated Pancreatic Stellate Cells and Is Involved in Pancreatic Cancer Cell Migration. Am. J. Pathol. 2017, 187, 146–155. [Google Scholar] [CrossRef]
- Ortiz, A.; Lee, Y.C.; Yu, G.; Liu, H.C.; Lin, S.C.; Bilen, M.A.; Cho, H.; Yu-Lee, L.Y.; Lin, S.H. Angiomotin is a novel component of cadherin-11/beta-catenin/p120 complex and is critical for cadherin-11-mediated cell migration. FASEB J. 2015, 29, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Choi, S.H.; Lee, T.R.; Lee, C.H.; Lee, A.Y. Cadherin 11, a miR-675 target, induces N-cadherin expression and epithelial-mesenchymal transition in melasma. J. Invest. Derm. 2014, 134, 2967–2976. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.; Cheng, C.J.; Ye, X.; Lee, Y.C.; Zurita, A.J.; Chen, D.T.; Yu-Lee, L.Y.; Zhang, S.; Yeh, E.T.; Hu, M.C.; et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol. Cancer Res. 2008, 6, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Boscher, C.; Mege, R.M. Cadherin-11 interacts with the FGF receptor and induces neurite outgrowth through associated downstream signalling. Cell Signal. 2008, 20, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, K.; Takai, Y. Nectin spot: A novel type of nectin-mediated cell adhesion apparatus. Biochem. J. 2016, 473, 2691–2715. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Lui, W.Y. Nectins and nectin-like molecules (Necls): Recent findings and their role and regulation in spermatogenesis. Semin. Cell Dev. Biol. 2016, 59, 54–61. [Google Scholar] [CrossRef]
- Bojesen, K.B.; Clausen, O.; Rohde, K.; Christensen, C.; Zhang, L.; Li, S.; Kohler, L.; Nielbo, S.; Nielsen, J.; Gjorlund, M.D.; et al. Nectin-1 binds and signals through the fibroblast growth factor receptor. J. Biol. Chem. 2012, 287, 37420–37433. [Google Scholar] [CrossRef]
- Owczarek, S.; Berezin, V. Neuroplastin: Cell adhesion molecule and signaling receptor. Int. J. Biochem. Cell Biol. 2012, 44, 1–5. [Google Scholar] [CrossRef]
- Langnaese, K.; Mummery, R.; Gundelfinger, E.D.; Beesley, P.W. Immunoglobulin superfamily members gp65 and gp55: Tissue distribution of glycoforms. FEBS Lett. 1998, 429, 284–288. [Google Scholar] [CrossRef]
- Langnaese, K.; Beesley, P.W.; Gundelfinger, E.D. Synaptic membrane glycoproteins gp65 and gp55 are new members of the immunoglobulin superfamily. J. Biol. Chem. 1997, 272, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Owczarek, S.; Kiryushko, D.; Larsen, M.H.; Kastrup, J.S.; Gajhede, M.; Sandi, C.; Berezin, V.; Bock, E.; Soroka, V. Neuroplastin-55 binds to and signals through the fibroblast growth factor receptor. FASEB J. 2010, 24, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Colombo, F.; Meldolesi, J. L1-CAM and N-CAM: From Adhesion Proteins to Pharmacological Targets. Trends Pharm. Sci. 2015, 36, 769–781. [Google Scholar] [CrossRef]
- Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily Regulate Synapse Formation, Maintenance, and Function. Trends Neurosci. 2017, 40, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Aonurm-Helm, A.; Jaako, K.; Jurgenson, M.; Zharkovsky, A. Pharmacological approach for targeting dysfunctional brain plasticity: Focus on neural cell adhesion molecule (NCAM). Pharm. Res. 2016, 113, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, C.; Cattaneo, P.; Berezin, V.; Bock, E.; Ami, D.; de Marco, A.; Christofori, G.; Cavallaro, U. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J. Cell Biol. 2009, 187, 1101–1116. [Google Scholar] [CrossRef]
- Christensen, C.; Lauridsen, J.B.; Berezin, V.; Bock, E.; Kiselyov, V.V. The neural cell adhesion molecule binds to fibroblast growth factor receptor 2. FEBS Lett. 2006, 580, 3386–3390. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, C.; Loeffler, S.; Piccini, D.; Kren, A.; Christofori, G.; Cavallaro, U. Neural cell adhesion molecule regulates the cellular response to fibroblast growth factor. J. Cell Sci. 2007, 120, 4388–4394. [Google Scholar] [CrossRef] [PubMed]
- Hansen, S.M.; Li, S.; Bock, E.; Berezin, V. Synthetic NCAM-derived ligands of the fibroblast growth factor receptor. Adv. Exp. Med. Biol. 2010, 663, 355–372. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Ohuchi, H.; Tanabe, H.; Imanaka, K.; Asano, H.; Kato, M.; Yokote, Y.; Kyo, S. Aortic root remodeling and coronary artery bypass grafting for acute type A aortic dissection involving the left main coronary artery; report of a case. Kyobu Geka 2005, 58, 897–901. [Google Scholar]
- Kiselyov, V.V.; Soroka, V.; Berezin, V.; Bock, E. Structural biology of NCAM homophilic binding and activation of FGFR. J. Neurochem. 2005, 94, 1169–1179. [Google Scholar] [CrossRef]
- Christensen, C.; Berezin, V.; Bock, E. Neural cell adhesion molecule differentially interacts with isoforms of the fibroblast growth factor receptor. Neuroreport 2011, 22, 727–732. [Google Scholar] [CrossRef]
- Zecchini, S.; Bombardelli, L.; Decio, A.; Bianchi, M.; Mazzarol, G.; Sanguineti, F.; Aletti, G.; Maddaluno, L.; Berezin, V.; Bock, E.; et al. The adhesion molecule NCAM promotes ovarian cancer progression via FGFR signalling. EMBO Mol. Med. 2011, 3, 480–494. [Google Scholar] [CrossRef]
- Colombo, N.; Cavallaro, U. The interplay between NCAM and FGFR signalling underlies ovarian cancer progression. Ecancermedicalscience 2011, 5, 226. [Google Scholar] [CrossRef]
- Zivotic, M.; Tampe, B.; Muller, G.; Muller, C.; Lipkovski, A.; Xu, X.; Nyamsuren, G.; Zeisberg, M.; Markovic-Lipkovski, J. Modulation of NCAM/FGFR1 signaling suppresses EMT program in human proximal tubular epithelial cells. PLoS ONE 2018, 13, e0206786. [Google Scholar] [CrossRef] [PubMed]
- Kulahin, N.; Li, S.; Hinsby, A.; Kiselyov, V.; Berezin, V.; Bock, E. Fibronectin type III (FN3) modules of the neuronal cell adhesion molecule L1 interact directly with the fibroblast growth factor (FGF) receptor. Mol. Cell Neurosci. 2008, 37, 528–536. [Google Scholar] [CrossRef]
- Riedle, S.; Kiefel, H.; Gast, D.; Bondong, S.; Wolterink, S.; Gutwein, P.; Altevogt, P. Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/gamma-secretase activity. Biochem. J. 2009, 420, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, V.; Temburni, M.K.; Kappes, J.C.; Galileo, D.S. L1CAM stimulates glioma cell motility and proliferation through the fibroblast growth factor receptor. Clin. Exp. Metastasis 2013, 30, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Martinez, D.; Kim, S.H.; Hu, Y.; Guimond, S.; Schofield, J.; Winyard, P.; Vannelli, G.B.; Turnbull, J.; Bouloux, P.M. Anosmin-1 modulates fibroblast growth factor receptor 1 signaling in human gonadotropin-releasing hormone olfactory neuroblasts through a heparan sulfate-dependent mechanism. J. Neurosci. 2004, 24, 10384–10392. [Google Scholar] [CrossRef]
- Bribian, A.; Barallobre, M.J.; Soussi-Yanicostas, N.; de Castro, F. Anosmin-1 modulates the FGF-2-dependent migration of oligodendrocyte precursors in the developing optic nerve. Mol. Cell Neurosci. 2006, 33, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalez, D.; Clemente, D.; Coelho, M.; Esteban, P.F.; Soussi-Yanicostas, N.; de Castro, F. Dynamic roles of FGF-2 and Anosmin-1 in the migration of neuronal precursors from the subventricular zone during pre- and postnatal development. Exp. Neurol. 2010, 222, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Murcia-Belmonte, V.; Esteban, P.F.; Garcia-Gonzalez, D.; De Castro, F. Biochemical dissection of Anosmin-1 interaction with FGFR1 and components of the extracellular matrix. J. Neurochem. 2010, 115, 1256–1265. [Google Scholar] [CrossRef]
- Hu, Y.; Guimond, S.E.; Travers, P.; Cadman, S.; Hohenester, E.; Turnbull, J.E.; Kim, S.H.; Bouloux, P.M. Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1. J. Biol. Chem. 2009, 284, 29905–29920. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Balzac, C.A.; Lazaro-Pena, M.I.; Ramos-Ortiz, G.A.; Bulow, H.E. The Adhesion Molecule KAL-1/anosmin-1 Regulates Neurite Branching through a SAX-7/L1CAM-EGL-15/FGFR Receptor Complex. Cell Rep. 2015, 11, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Kriebel, M.; Wuchter, J.; Trinks, S.; Volkmer, H. Neurofascin: A switch between neuronal plasticity and stability. Int. J. Biochem. Cell Biol. 2012, 44, 694–697. [Google Scholar] [CrossRef]
- Kirschbaum, K.; Kriebel, M.; Kranz, E.U.; Potz, O.; Volkmer, H. Analysis of non-canonical fibroblast growth factor receptor 1 (FGFR1) interaction reveals regulatory and activating domains of neurofascin. J. Biol. Chem. 2009, 284, 28533–28542. [Google Scholar] [CrossRef] [PubMed]
- Pruss, T.; Kranz, E.U.; Niere, M.; Volkmer, H. A regulated switch of chick neurofascin isoforms modulates ligand recognition and neurite extension. Mol. Cell Neurosci. 2006, 31, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Sudhof, T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 2008, 455, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, G. Neurexins - versatile molecular platforms in the synaptic cleft. Curr. Opin. Struct. Biol. 2019, 54, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Gjorlund, M.D.; Nielsen, J.; Pankratova, S.; Li, S.; Korshunova, I.; Bock, E.; Berezin, V. Neuroligin-1 induces neurite outgrowth through interaction with neurexin-1beta and activation of fibroblast growth factor receptor-1. FASEB J. 2012, 26, 4174–4186. [Google Scholar] [CrossRef]
- Kubick, N.; Brosamle, D.; Mickael, M.E. Molecular Evolution and Functional Divergence of the IgLON Family. Evol. Bioinform. Online 2018, 14, 1176934318775081. [Google Scholar] [CrossRef] [PubMed]
- Funatsu, N.; Miyata, S.; Kumanogoh, H.; Shigeta, M.; Hamada, K.; Endo, Y.; Sokawa, Y.; Maekawa, S. Characterization of a novel rat brain glycosylphosphatidylinositol-anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J. Biol. Chem. 1999, 274, 8224–8230. [Google Scholar] [CrossRef] [PubMed]
- Pischedda, F.; Piccoli, G. The IgLON Family Member Negr1 Promotes Neuronal Arborization Acting as Soluble Factor via FGFR2. Front. Mol. Neurosci. 2015, 8, 89. [Google Scholar] [CrossRef]
- Casey, J.P.; Magalhaes, T.; Conroy, J.M.; Regan, R.; Shah, N.; Anney, R.; Shields, D.C.; Abrahams, B.S.; Almeida, J.; Bacchelli, E.; et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum. Genet. 2012, 131, 565–579. [Google Scholar] [CrossRef] [PubMed]
- Szczurkowska, J.; Pischedda, F.; Pinto, B.; Manago, F.; Haas, C.A.; Summa, M.; Bertorelli, R.; Papaleo, F.; Schafer, M.K.; Piccoli, G.; et al. NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain 2018, 141, 2772–2794. [Google Scholar] [CrossRef]
- Sellar, G.C.; Watt, K.P.; Rabiasz, G.J.; Stronach, E.A.; Li, L.; Miller, E.P.; Massie, C.E.; Miller, J.; Contreras-Moreira, B.; Scott, D.; et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat. Genet. 2003, 34, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ye, F.; Zhang, J.; Lu, W.; Cheng, Q.; Xie, X. Loss of OPCML expression and the correlation with CpG island methylation and LOH in ovarian serous carcinoma. Eur. J. Gynaecol. Oncol. 2007, 28, 464–467. [Google Scholar]
- Reed, J.E.; Dunn, J.R.; du Plessis, D.G.; Shaw, E.J.; Reeves, P.; Gee, A.L.; Warnke, P.C.; Sellar, G.C.; Moss, D.J.; Walker, C. Expression of cellular adhesion molecule ’OPCML’ is down-regulated in gliomas and other brain tumours. Neuropathol. Appl. Neurobiol. 2007, 33, 77–85. [Google Scholar] [CrossRef]
- Cui, Y.; Ying, Y.; van Hasselt, A.; Ng, K.M.; Yu, J.; Zhang, Q.; Jin, J.; Liu, D.; Rhim, J.S.; Rha, S.Y.; et al. OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS ONE 2008, 3, e2990. [Google Scholar] [CrossRef]
- Zhang, N.; Xu, J.; Wang, Y.; Heng, X.; Yang, L.; Xing, X. Loss of opioid binding protein/cell adhesion molecule-like gene expression in gastric cancer. Oncol. Lett. 2018, 15, 9973–9977. [Google Scholar] [CrossRef] [PubMed]
- McKie, A.B.; Vaughan, S.; Zanini, E.; Okon, I.S.; Louis, L.; de Sousa, C.; Greene, M.I.; Wang, Q.; Agarwal, R.; Shaposhnikov, D.; et al. The OPCML tumor suppressor functions as a cell surface repressor-adaptor, negatively regulating receptor tyrosine kinases in epithelial ovarian cancer. Cancer Discov. 2012, 2, 156–171. [Google Scholar] [CrossRef]
- O’Sullivan, M.L.; de Wit, J.; Savas, J.N.; Comoletti, D.; Otto-Hitt, S.; Yates, J.R., 3rd; Ghosh, A. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron 2012, 73, 903–910. [Google Scholar] [CrossRef] [PubMed]
- Sando, R.; Jiang, X.; Sudhof, T.C. Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 2019, 363. [Google Scholar] [CrossRef] [PubMed]
- Del Toro, D.; Ruff, T.; Cederfjall, E.; Villalba, A.; Seyit-Bremer, G.; Borrell, V.; Klein, R. Regulation of Cerebral Cortex Folding by Controlling Neuronal Migration via FLRT Adhesion Molecules. Cell 2017, 169, 621–635.e616. [Google Scholar] [CrossRef] [PubMed]
- Jackson, V.A.; Mehmood, S.; Chavent, M.; Roversi, P.; Carrasquero, M.; Del Toro, D.; Seyit-Bremer, G.; Ranaivoson, F.M.; Comoletti, D.; Sansom, M.S.; et al. Super-complexes of adhesion GPCRs and neural guidance receptors. Nat. Commun. 2016, 7, 11184. [Google Scholar] [CrossRef] [PubMed]
- Lacy, S.E.; Bonnemann, C.G.; Buzney, E.A.; Kunkel, L.M. Identification of FLRT1, FLRT2, and FLRT3: A novel family of transmembrane leucine-rich repeat proteins. Genomics 1999, 62, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Karaulanov, E.E.; Bottcher, R.T.; Niehrs, C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO Rep. 2006, 7, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, R.T.; Pollet, N.; Delius, H.; Niehrs, C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat. Cell Biol. 2004, 6, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Xu, Y.; Tse, H.; Manolson, M.F.; Gong, S.G. Mouse FLRT2 interacts with the extracellular and intracellular regions of FGFR2. J. Dent. Res. 2011, 90, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Haines, B.P.; Wheldon, L.M.; Summerbell, D.; Heath, J.K.; Rigby, P.W. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev. Biol. 2006, 297, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Wheldon, L.M.; Haines, B.P.; Rajappa, R.; Mason, I.; Rigby, P.W.; Heath, J.K. Critical role of FLRT1 phosphorylation in the interdependent regulation of FLRT1 function and FGF receptor signalling. PLoS ONE 2010, 5, e10264. [Google Scholar] [CrossRef] [PubMed]
- Humphries, J.D.; Chastney, M.R.; Askari, J.A.; Humphries, M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019, 56, 14–21. [Google Scholar] [CrossRef]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Harburger, D.S.; Calderwood, D.A. Integrin signalling at a glance. J. Cell Sci. 2009, 122, 159–163. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef]
- Yamaji, S.; Saegusa, J.; Ieguchi, K.; Fujita, M.; Mori, S.; Takada, Y.K.; Takada, Y. A novel fibroblast growth factor-1 (FGF1) mutant that acts as an FGF antagonist. PLoS ONE 2010, 5, e10273. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Wu, C.Y.; Yamaji, S.; Saegusa, J.; Shi, B.; Ma, Z.; Kuwabara, Y.; Lam, K.S.; Isseroff, R.R.; Takada, Y.K.; et al. Direct binding of integrin alphavbeta3 to FGF1 plays a role in FGF1 signaling. J. Biol. Chem. 2008, 283, 18066–18075. [Google Scholar] [CrossRef]
- Mori, S.; Tran, V.; Nishikawa, K.; Kaneda, T.; Hamada, Y.; Kawaguchi, N.; Fujita, M.; Saegusa, J.; Takada, Y.K.; Matsuura, N.; et al. A dominant-negative FGF1 mutant (the R50E mutant) suppresses tumorigenesis and angiogenesis. PLoS ONE 2013, 8, e57927. [Google Scholar] [CrossRef]
- Mori, S.; Hatori, N.; Kawaguchi, N.; Hamada, Y.; Shih, T.C.; Wu, C.Y.; Lam, K.S.; Matsuura, N.; Yamamoto, H.; Takada, Y.K.; et al. The integrin-binding defective FGF2 mutants potently suppress FGF2 signalling and angiogenesis. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Yayon, A.; Klagsbrun, M.; Esko, J.D.; Leder, P.; Ornitz, D.M. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 1991, 64, 841–848. [Google Scholar] [CrossRef]
- Rapraeger, A.C.; Krufka, A.; Olwin, B.B. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 1991, 252, 1705–1708. [Google Scholar] [CrossRef]
- Pellegrini, L.; Burke, D.F.; von Delft, F.; Mulloy, B.; Blundell, T.L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 2000, 407, 1029–1034. [Google Scholar] [CrossRef]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, I.; Kimura-Yoshida, C. Extracellular modulation of Fibroblast Growth Factor signaling through heparan sulfate proteoglycans in mammalian development. Curr. Opin. Genet. Dev. 2013, 23, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.S.; Tang, F.; Rnjak-Kovacina, J.; Smith, J.G.W.; Melrose, J.; Whitelock, J.M. The multifaceted roles of perlecan in fibrosis. Matrix Biol. 2018, 68–69, 150–166. [Google Scholar] [CrossRef]
- Chuang, C.Y.; Lord, M.S.; Melrose, J.; Rees, M.D.; Knox, S.M.; Freeman, C.; Iozzo, R.V.; Whitelock, J.M. Heparan sulfate-dependent signaling of fibroblast growth factor 18 by chondrocyte-derived perlecan. Biochemistry 2010, 49, 5524–5532. [Google Scholar] [CrossRef] [PubMed]
- Knox, S.; Merry, C.; Stringer, S.; Melrose, J.; Whitelock, J. Not all perlecans are created equal: Interactions with fibroblast growth factor (FGF) 2 and FGF receptors. J. Biol. Chem. 2002, 277, 14657–14665. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; West, L.A.; Govindraj, P.; Zhang, X.; Ornitz, D.M.; Hassell, J.R. Heparan and chondroitin sulfate on growth plate perlecan mediate binding and delivery of FGF-2 to FGF receptors. Matrix Biol. 2007, 26, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; West, L.A.; Hassell, J.R. The core protein of growth plate perlecan binds FGF-18 and alters its mitogenic effect on chondrocytes. Arch. Biochem. Biophys. 2007, 468, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Aviezer, D.; Hecht, D.; Safran, M.; Eisinger, M.; David, G.; Yayon, A. Perlecan, basal lamina proteoglycan, promotes basic fibroblast growth factor-receptor binding, mitogenesis, and angiogenesis. Cell 1994, 79, 1005–1013. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Karamanos, N.K. Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biol. 2019, 75–76, 220–259. [Google Scholar] [CrossRef]
- Afratis, N.A.; Nikitovic, D.; Multhaupt, H.A.; Theocharis, A.D.; Couchman, J.R.; Karamanos, N.K. Syndecans—Key regulators of cell signaling and biological functions. FEBS J. 2017, 284, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Multhaupt, H.A.; Oh, E.S.; Couchman, J.R. Minireview: Syndecans and their crucial roles during tissue regeneration. FEBS Lett. 2016, 590, 2408–2417. [Google Scholar] [CrossRef]
- Bernfield, M.; Sanderson, R.D. Syndecan, a developmentally regulated cell surface proteoglycan that binds extracellular matrix and growth factors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1990, 327, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Olwin, B.B.; Rapraeger, A. Repression of myogenic differentiation by aFGF, bFGF, and K-FGF is dependent on cellular heparan sulfate. J. Cell Biol. 1992, 118, 631–639. [Google Scholar] [CrossRef]
- Clasper, S.; Vekemans, S.; Fiore, M.; Plebanski, M.; Wordsworth, P.; David, G.; Jackson, D.G. Inducible expression of the cell surface heparan sulfate proteoglycan syndecan-2 (fibroglycan) on human activated macrophages can regulate fibroblast growth factor action. J. Biol. Chem. 1999, 274, 24113–24123. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Kan, M.; Wang, F.; Jin, C.; Yu, C.; McKeehan, W.L. A rare premalignant prostate tumor epithelial cell syndecan-1 forms a fibroblast growth factor-binding complex with progression-promoting ectopic fibroblast growth factor receptor 1. Cancer Res. 2001, 61, 5295–5302. [Google Scholar] [PubMed]
- Iwabuchi, T.; Goetinck, P.F. Syndecan-4 dependent FGF stimulation of mouse vibrissae growth. Mech. Dev. 2006, 123, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Filla, M.S.; Dam, P.; Rapraeger, A.C. The cell surface proteoglycan syndecan-1 mediates fibroblast growth factor-2 binding and activity. J. Cell Physiol. 1998, 174, 310–321. [Google Scholar] [CrossRef]
- Jang, E.; Albadawi, H.; Watkins, M.T.; Edelman, E.R.; Baker, A.B. Syndecan-4 proteoliposomes enhance fibroblast growth factor-2 (FGF-2)-induced proliferation, migration, and neovascularization of ischemic muscle. Proc. Natl. Acad. Sci. USA 2012, 109, 1679–1684. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Nguyen, L.T.; Zhuang, Z.W.; Moodie, K.L.; Carmeliet, P.; Stan, R.V.; Simons, M. The FGF system has a key role in regulating vascular integrity. J. Clin. Invest. 2008, 118, 3355–3366. [Google Scholar] [CrossRef] [PubMed]
- Elfenbein, A.; Lanahan, A.; Zhou, T.X.; Yamasaki, A.; Tkachenko, E.; Matsuda, M.; Simons, M. Syndecan 4 regulates FGFR1 signaling in endothelial cells by directing macropinocytosis. Sci. Signal. 2012, 5, ra36. [Google Scholar] [CrossRef] [PubMed]
- Fico, A.; Maina, F.; Dono, R. Fine-tuning of cell signaling by glypicans. Cell Mol. Life Sci. 2011, 68, 923–929. [Google Scholar] [CrossRef]
- Galli, A.; Roure, A.; Zeller, R.; Dono, R. Glypican 4 modulates FGF signalling and regulates dorsoventral forebrain patterning in Xenopus embryos. Development 2003, 130, 4919–4929. [Google Scholar] [CrossRef]
- Gutierrez, J.; Brandan, E. A novel mechanism of sequestering fibroblast growth factor 2 by glypican in lipid rafts, allowing skeletal muscle differentiation. Mol. Cell Biol. 2010, 30, 1634–1649. [Google Scholar] [CrossRef] [PubMed]
- Berman, B.; Ostrovsky, O.; Shlissel, M.; Lang, T.; Regan, D.; Vlodavsky, I.; Ishai-Michaeli, R.; Ron, D. Similarities and differences between the effects of heparin and glypican-1 on the bioactivity of acidic fibroblast growth factor and the keratinocyte growth factor. J. Biol. Chem. 1999, 274, 36132–36138. [Google Scholar] [CrossRef] [PubMed]
- Qiao, D.; Meyer, K.; Mundhenke, C.; Drew, S.A.; Friedl, A. Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 signaling in brain endothelial cells. Specific role for glypican-1 in glioma angiogenesis. J. Biol. Chem. 2003, 278, 16045–16053. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Meyer, K.; Nandini, C.D.; Qiao, D.; Salamat, S.; Friedl, A. Glypican-1 is frequently overexpressed in human gliomas and enhances FGF-2 signaling in glioma cells. Am. J. Pathol. 2006, 168, 2014–2026. [Google Scholar] [CrossRef]
- Itoh, N.; Nakayama, Y.; Konishi, M. Roles of FGFs As Paracrine or Endocrine Signals in Liver Development, Health, and Disease. Front. Cell Dev. Biol. 2016, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N. Hormone-like (endocrine) Fgfs: Their evolutionary history and roles in development, metabolism, and disease. Cell Tissue Res. 2010, 342, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Ibrahimi, O.A.; Goetz, R.; Zhang, F.; Davis, S.I.; Garringer, H.J.; Linhardt, R.J.; Ornitz, D.M.; Mohammadi, M.; White, K.E. Analysis of the biochemical mechanisms for the endocrine actions of fibroblast growth factor-23. Endocrinology 2005, 146, 4647–4656. [Google Scholar] [CrossRef]
- Yie, J.; Wang, W.; Deng, L.; Tam, L.T.; Stevens, J.; Chen, M.M.; Li, Y.; Xu, J.; Lindberg, R.; Hecht, R.; et al. Understanding the physical interactions in the FGF21/FGFR/beta-Klotho complex: Structural requirements and implications in FGF21 signaling. Chem. Biol. Drug Des. 2012, 79, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Goetz, R.; Ohnishi, M.; Kir, S.; Kurosu, H.; Wang, L.; Pastor, J.; Ma, J.; Gai, W.; Kuro-o, M.; Razzaque, M.S.; et al. Conversion of a paracrine fibroblast growth factor into an endocrine fibroblast growth factor. J. Biol. Chem. 2012, 287, 29134–29146. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.C.; Cheng, C.C.; Coskun, T.; Kharitonenkov, A. FGF21 requires betaklotho to act in vivo. PLoS ONE 2012, 7, e49977. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Boney-Montoya, J.; Owen, B.M.; Bookout, A.L.; Coate, K.C.; Mangelsdorf, D.J.; Kliewer, S.A. betaKlotho is required for fibroblast growth factor 21 effects on growth and metabolism. Cell Metab. 2012, 16, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Kurosu, H.; Ogawa, Y.; Miyoshi, M.; Yamamoto, M.; Nandi, A.; Rosenblatt, K.P.; Baum, M.G.; Schiavi, S.; Hu, M.C.; Moe, O.W.; et al. Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 2006, 281, 6120–6123. [Google Scholar] [CrossRef]
- Urakawa, I.; Yamazaki, Y.; Shimada, T.; Iijima, K.; Hasegawa, H.; Okawa, K.; Fujita, T.; Fukumoto, S.; Yamashita, T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 2006, 444, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Kharitonenkov, A.; Dunbar, J.D.; Bina, H.A.; Bright, S.; Moyers, J.S.; Zhang, C.; Ding, L.; Micanovic, R.; Mehrbod, S.F.; Knierman, M.D.; et al. FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. J. Cell Physiol. 2008, 215, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kuro-o, M.; Matsumura, Y.; Aizawa, H.; Kawaguchi, H.; Suga, T.; Utsugi, T.; Ohyama, Y.; Kurabayashi, M.; Kaname, T.; Kume, E.; et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 1997, 390, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Aizawa, H.; Shiraki-Iida, T.; Nagai, R.; Kuro-o, M.; Nabeshima, Y. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem. Biophys. Res. Commun. 1998, 242, 626–630. [Google Scholar] [CrossRef]
- Chen, G.; Liu, Y.; Goetz, R.; Fu, L.; Jayaraman, S.; Hu, M.C.; Moe, O.W.; Liang, G.; Li, X.; Mohammadi, M. alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 2018, 553, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Ye, S.; Li, X.; Lu, W. Emerging Structure-Function Paradigm of Endocrine FGFs in Metabolic Diseases. Trends Pharm. Sci. 2019, 40, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Takashi, Y.; Fukumoto, S. FGF23 beyond Phosphotropic Hormone. Trends Endocrinol. Metab. 2018, 29, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Farrow, E.G.; Davis, S.I.; Summers, L.J.; White, K.E. Initial FGF23-mediated signaling occurs in the distal convoluted tubule. J. Am. Soc. Nephrol. 2009, 20, 955–960. [Google Scholar] [CrossRef]
- Andrukhova, O.; Smorodchenko, A.; Egerbacher, M.; Streicher, C.; Zeitz, U.; Goetz, R.; Shalhoub, V.; Mohammadi, M.; Pohl, E.E.; Lanske, B.; et al. FGF23 promotes renal calcium reabsorption through the TRPV5 channel. EMBO J. 2014, 33, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Goetz, R.; Nakada, Y.; Hu, M.C.; Kurosu, H.; Wang, L.; Nakatani, T.; Shi, M.; Eliseenkova, A.V.; Razzaque, M.S.; Moe, O.W.; et al. Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23-FGFR-Klotho complex formation. Proc. Natl. Acad. Sci. USA 2010, 107, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Andrukhova, O.; Zeitz, U.; Goetz, R.; Mohammadi, M.; Lanske, B.; Erben, R.G. FGF23 acts directly on renal proximal tubules to induce phosphaturia through activation of the ERK1/2-SGK1 signaling pathway. Bone 2012, 51, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; David, V.; Quarles, L.D. Regulation and function of the FGF23/Klotho endocrine pathways. Physiol. Rev. 2012, 92, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, L.S.; Gonzalez, S.G.; Tomero, J.A.S.; Aguilera, A.; Junco, E.O. Bone mineral disorder in chronic kidney disease: Klotho and FGF23; cardiovascular implications. Nefrologia 2016, 36, 333–464. [Google Scholar] [CrossRef][Green Version]
- Lu, X.; Hu, M.C. Klotho/FGF23 Axis in Chronic Kidney Disease and Cardiovascular Disease. Kidney Dis. 2017, 3, 15–23. [Google Scholar] [CrossRef]
- Shiohama, A.; Sasaki, T.; Noda, S.; Minoshima, S.; Shimizu, N. Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 2003, 304, 184–190. [Google Scholar] [CrossRef]
- Kurosu, H.; Choi, M.; Ogawa, Y.; Dickson, A.S.; Goetz, R.; Eliseenkova, A.V.; Mohammadi, M.; Rosenblatt, K.P.; Kliewer, S.A.; Kuro-o, M. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 2007, 282, 26687–26695. [Google Scholar] [CrossRef]
- Ogawa, Y.; Kurosu, H.; Yamamoto, M.; Nandi, A.; Rosenblatt, K.P.; Goetz, R.; Eliseenkova, A.V.; Mohammadi, M.; Kuro-o, M. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proc. Natl. Acad. Sci. USA 2007, 104, 7432–7437. [Google Scholar] [CrossRef]
- Adams, A.C.; Yang, C.; Coskun, T.; Cheng, C.C.; Gimeno, R.E.; Luo, Y.; Kharitonenkov, A. The breadth of FGF21’s metabolic actions are governed by FGFR1 in adipose tissue. Mol. Metab. 2012, 2, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, J.; Mohanty, J.; Sousa, L.P.; Tome, F.; Pardon, E.; Steyaert, J.; Lemmon, M.A.; Lax, I.; Schlessinger, J. Structures of beta-klotho reveal a ‘zip code’-like mechanism for endocrine FGF signalling. Nature 2018, 553, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.Y.; Lu, Y.W.; Richardson, J.; Min, X.; Weiszmann, J.; Richards, W.G.; Wang, Z.; Zhang, Z.; Zhang, J.; Li, Y. A systematic dissection of sequence elements determining beta-Klotho and FGF interaction and signaling. Sci. Rep. 2018, 8, 11045. [Google Scholar] [CrossRef] [PubMed]
- Kharitonenkov, A.; Shiyanova, T.L.; Koester, A.; Ford, A.M.; Micanovic, R.; Galbreath, E.J.; Sandusky, G.E.; Hammond, L.J.; Moyers, J.S.; Owens, R.A.; et al. FGF-21 as a novel metabolic regulator. J. Clin. Investig. 2005, 115, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xie, Y.; Berglund, E.D.; Coate, K.C.; He, T.T.; Katafuchi, T.; Xiao, G.; Potthoff, M.J.; Wei, W.; Wan, Y.; et al. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. eLife 2012, 1, e00065. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Regulation of longevity by FGF21: Interaction between energy metabolism and stress responses. Ageing Res. Rev. 2017, 37, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, T.; Choi, M.; Moschetta, A.; Peng, L.; Cummins, C.L.; McDonald, J.G.; Luo, G.; Jones, S.A.; Goodwin, B.; Richardson, J.A.; et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005, 2, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Wang, F.; Kan, M.; Jin, C.; Jones, R.B.; Weinstein, M.; Deng, C.X.; McKeehan, W.L. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 2000, 275, 15482–15489. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; John, L.M.; Adams, S.H.; Yu, X.X.; Tomlinson, E.; Renz, M.; Williams, P.M.; Soriano, R.; Corpuz, R.; Moffat, B.; et al. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology 2004, 145, 2594–2603. [Google Scholar] [CrossRef] [PubMed]
- Kir, S.; Beddow, S.A.; Samuel, V.T.; Miller, P.; Previs, S.F.; Suino-Powell, K.; Xu, H.E.; Shulman, G.I.; Kliewer, S.A.; Mangelsdorf, D.J. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 2011, 331, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Kuro, O.M. The Klotho proteins in health and disease. Nat. Rev. Nephrol. 2019, 15, 27–44. [Google Scholar] [CrossRef]
- Babaknejad, N.; Nayeri, H.; Hemmati, R.; Bahrami, S.; Esmaillzadeh, A. An Overview of FGF19 and FGF21: The Therapeutic Role in the Treatment of the Metabolic Disorders and Obesity. Horm. Metab. Res. 2018, 50, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sola, G.; Uriarte, I.; Latasa, M.U.; Urtasun, R.; Barcena-Varela, M.; Elizalde, M.; Jimenez, M.; Rodriguez-Ortigosa, C.M.; Corrales, F.J.; Fernandez-Barrena, M.G.; et al. Fibroblast Growth Factor 15/19 in Hepatocarcinogenesis. Dig. Dis. 2017, 35, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Casillas, F.; Cheifetz, S.; Doody, J.; Andres, J.L.; Lane, W.S.; Massague, J. Structure and expression of the membrane proteoglycan betaglycan, a component of the TGF-beta receptor system. Cell 1991, 67, 785–795. [Google Scholar] [CrossRef]
- Knelson, E.H.; Gaviglio, A.L.; Tewari, A.K.; Armstrong, M.B.; Mythreye, K.; Blobe, G.C. Type III TGF-beta receptor promotes FGF2-mediated neuronal differentiation in neuroblastoma. J. Clin. Investig. 2013, 123, 4786–4798. [Google Scholar] [CrossRef] [PubMed]
- Andres, J.L.; DeFalcis, D.; Noda, M.; Massague, J. Binding of two growth factor families to separate domains of the proteoglycan betaglycan. J. Biol. Chem. 1992, 267, 5927–5930. [Google Scholar] [PubMed]
- Furthauer, M.; Lin, W.; Ang, S.L.; Thisse, B.; Thisse, C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat. Cell Biol. 2002, 4, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Preger, E.; Ziv, I.; Shabtay, A.; Sher, I.; Tsang, M.; Dawid, I.B.; Altuvia, Y.; Ron, D. Alternative splicing generates an isoform of the human Sef gene with altered subcellular localization and specificity. Proc. Natl. Acad. Sci. USA 2004, 101, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Harduf, H.; Halperin, E.; Reshef, R.; Ron, D. Sef is synexpressed with FGFs during chick embryogenesis and its expression is differentially regulated by FGFs in the developing limb. Dev. Dyn. 2005, 233, 301–312. [Google Scholar] [CrossRef]
- Lin, W.; Furthauer, M.; Thisse, B.; Thisse, C.; Jing, N.; Ang, S.L. Cloning of the mouse Sef gene and comparative analysis of its expression with Fgf8 and Spry2 during embryogenesis. Mech. Dev. 2002, 113, 163–168. [Google Scholar] [CrossRef]
- Yang, R.B.; Ng, C.K.; Wasserman, S.M.; Komuves, L.G.; Gerritsen, M.E.; Topper, J.N. A novel interleukin-17 receptor-like protein identified in human umbilical vein endothelial cells antagonizes basic fibroblast growth factor-induced signaling. J. Biol. Chem. 2003, 278, 33232–33238. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, D.; Yang, X.; Nadeau, R.J.; Harkins, L.K.; Friesel, R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J. Biol. Chem. 2003, 278, 14087–14091. [Google Scholar] [CrossRef]
- Xiong, S.; Zhao, Q.; Rong, Z.; Huang, G.; Huang, Y.; Chen, P.; Zhang, S.; Liu, L.; Chang, Z. hSef inhibits PC-12 cell differentiation by interfering with Ras-mitogen-activated protein kinase MAPK signaling. J. Biol. Chem. 2003, 278, 50273–50282. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Ren, Y.; Cheng, L.; Li, Z.; Li, Y.; Sun, Y.; Li, H.; Xiong, S.; Chang, Z. Sef-S, an alternative splice isoform of sef gene, inhibits NIH3T3 cell proliferation via a mitogen-activated protein kinases p42 and p44 (ERK1/2)-independent mechanism. Cell Signal. 2007, 19, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Friesel, R.; Kudoh, T.; Dawid, I.B. Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 2002, 4, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Korsensky, L.; Ron, D. Regulation of FGF signaling: Recent insights from studying positive and negative modulators. Semin. Cell Dev. Biol. 2016, 53, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Ziv, I.; Fuchs, Y.; Preger, E.; Shabtay, A.; Harduf, H.; Zilpa, T.; Dym, N.; Ron, D. The human sef-a isoform utilizes different mechanisms to regulate receptor tyrosine kinase signaling pathways and subsequent cell fate. J. Biol. Chem. 2006, 281, 39225–39235. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.; Darby, S.; Mathers, M.E.; Gnanapragasam, V.J. Evidence for distinct alterations in the FGF axis in prostate cancer progression to an aggressive clinical phenotype. J. Pathol. 2010, 220, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Wadhwa, K.; Pisupati, V.; Zecchini, V.; Ramos-Montoya, A.; Warren, A.Y.; Neal, D.E.; Gnanapragasam, V.J. Loss of hSef promotes metastasis through upregulation of EMT in prostate cancer. Int. J. Cancer 2017, 140, 1881–1887. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 2019, 16, 105–122. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latko, M.; Czyrek, A.; Porębska, N.; Kucińska, M.; Otlewski, J.; Zakrzewska, M.; Opaliński, Ł. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 2019, 8, 455. https://doi.org/10.3390/cells8050455
Latko M, Czyrek A, Porębska N, Kucińska M, Otlewski J, Zakrzewska M, Opaliński Ł. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells. 2019; 8(5):455. https://doi.org/10.3390/cells8050455
Chicago/Turabian StyleLatko, Marta, Aleksandra Czyrek, Natalia Porębska, Marika Kucińska, Jacek Otlewski, Małgorzata Zakrzewska, and Łukasz Opaliński. 2019. "Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins" Cells 8, no. 5: 455. https://doi.org/10.3390/cells8050455
APA StyleLatko, M., Czyrek, A., Porębska, N., Kucińska, M., Otlewski, J., Zakrzewska, M., & Opaliński, Ł. (2019). Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells, 8(5), 455. https://doi.org/10.3390/cells8050455




