Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors
Abstract
1. Introduction
2. Materials and Methods
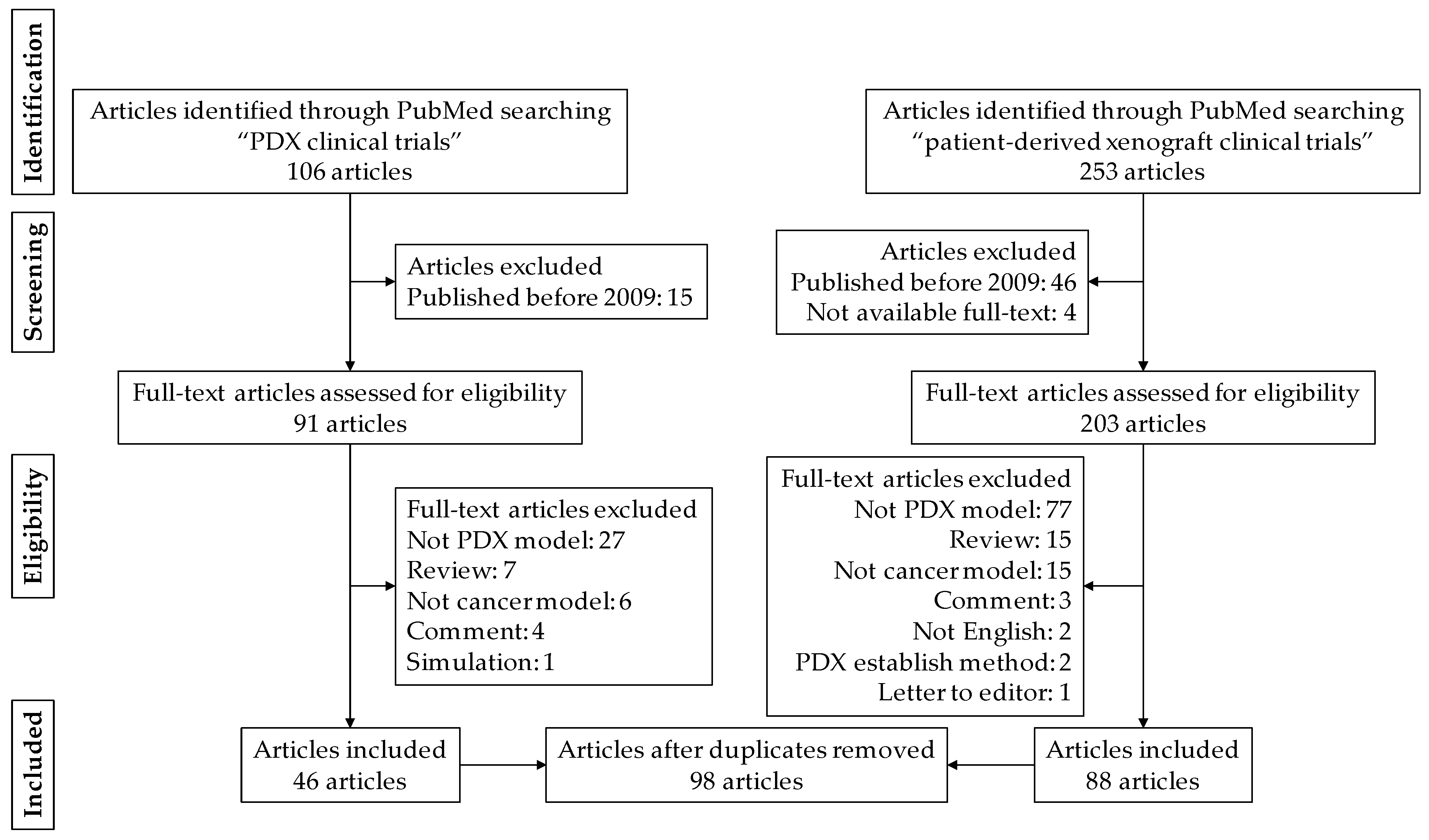
2.1. Article Search Strategy
2.2. Eligibility Criteria
2.3. Study Selection and Data Collection
3. Results
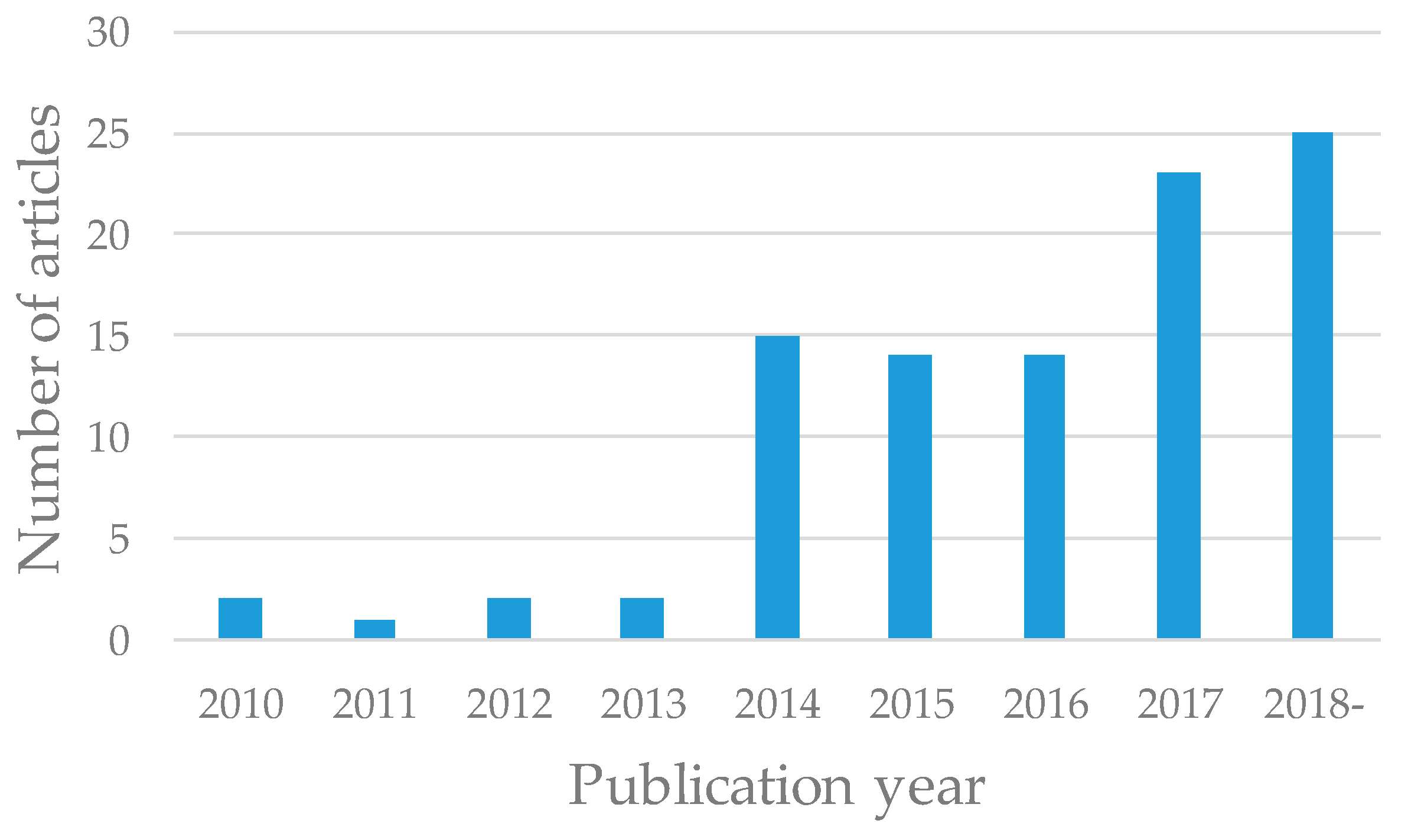
3.1. Publication Year
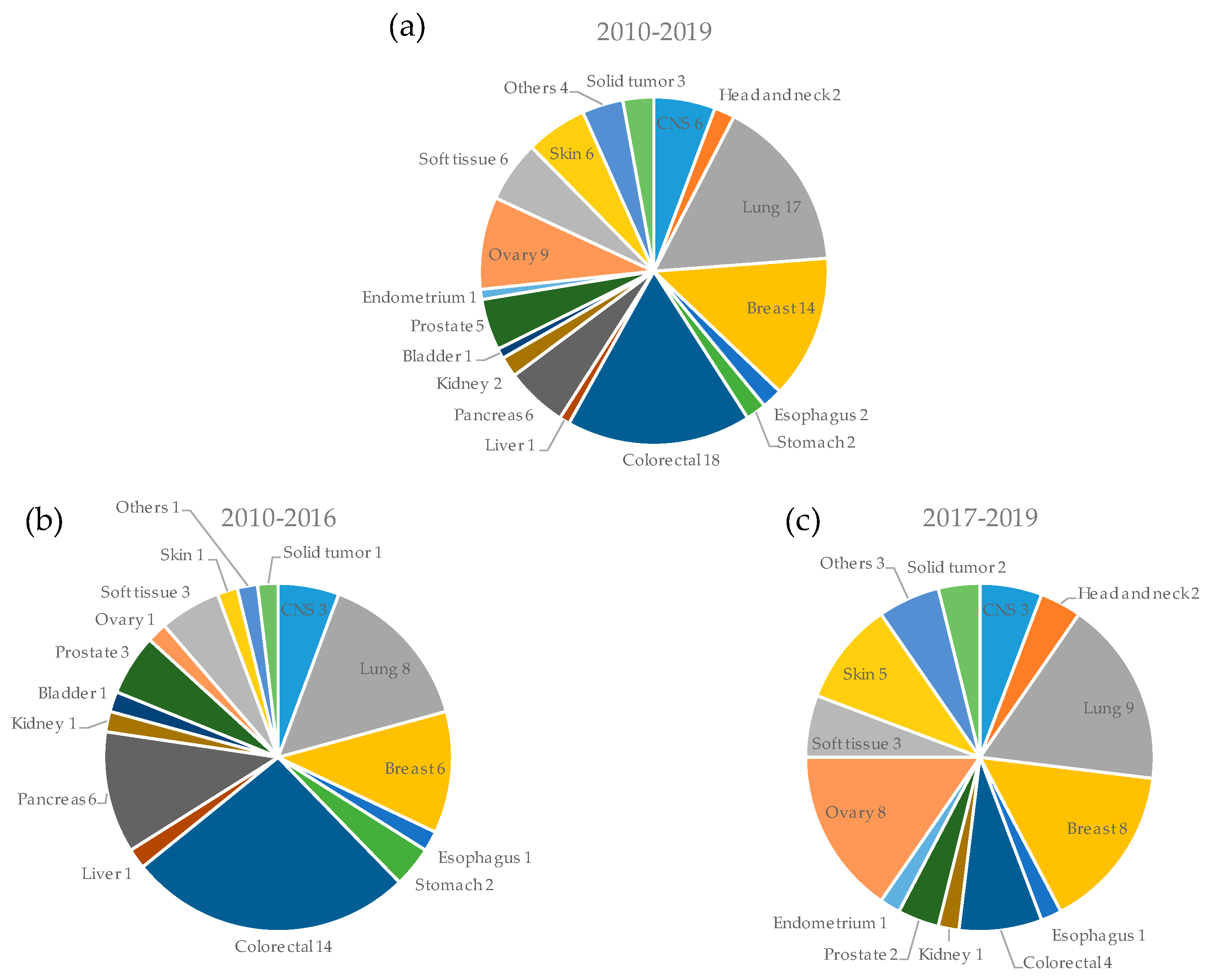
3.2. Types of Cancers Used for PDX Models
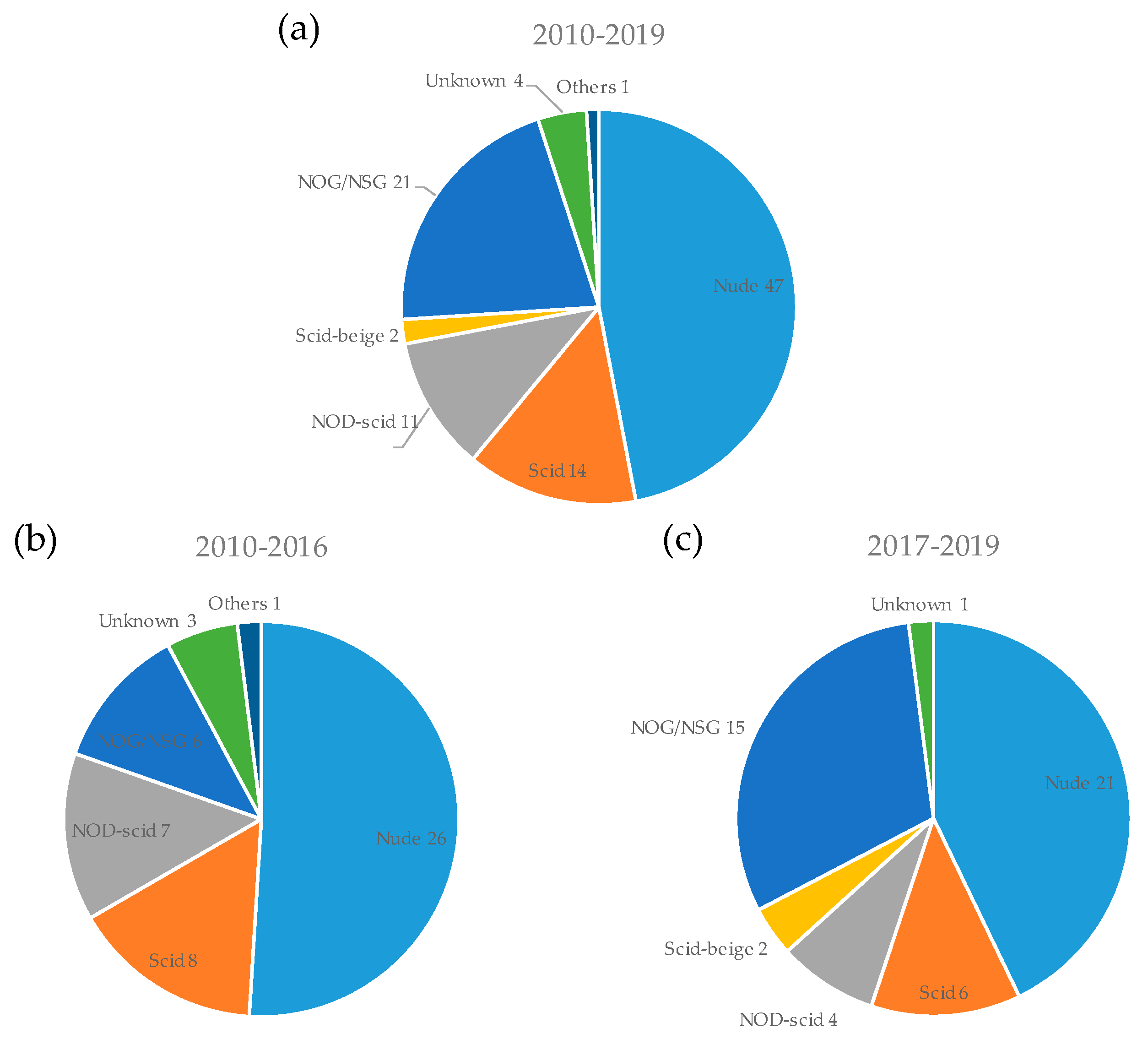
3.3. Mouse Strain and Implantation Site for PDX Model
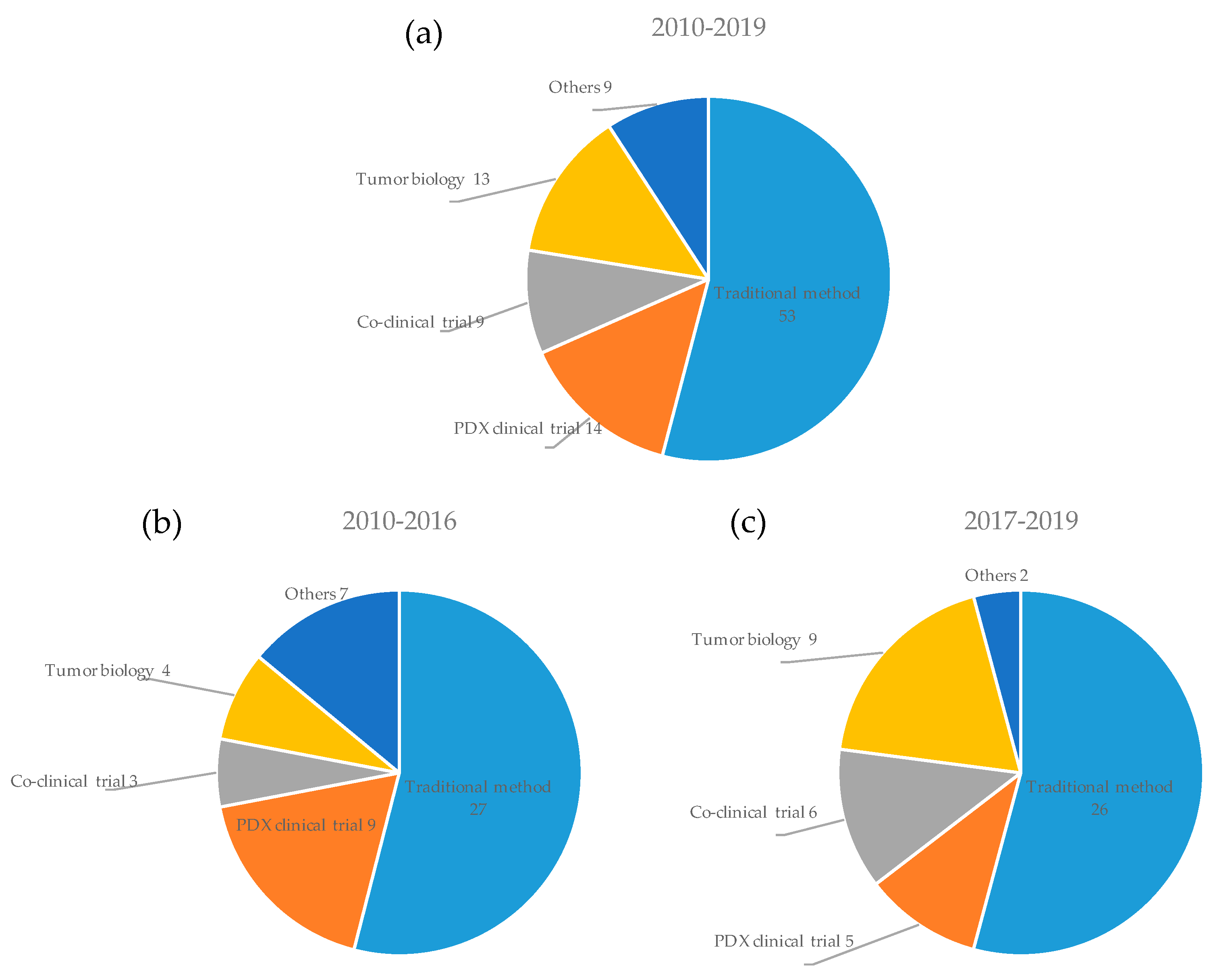
3.4. Applications of PDX Models in Cancer Research
3.5. PDX Clinical Trials
3.6. Co-clinical Trials
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cunningham, D.; Humblet, Y.; Siena, S.; Khayat, D.; Bleiberg, H.; Santoro, A.; Bets, D.; Mueser, M.; Harstrick, A.; Verslype, C.; et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N. Engl. J. Med. 2004, 351, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Procter, M.; Gelber, R.D.; Guillaume, S.; Feyereislova, A.; Dowsett, M.; Goldhirsch, A.; Untch, M.; Mariani, G.; Baselga, J.; et al. 2-year follow-up of trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer: A randomised controlled trial. Lancet 2007, 369, 29–36. [Google Scholar] [CrossRef]
- Hutchinson, L.; Kirk, R. High drug attrition rates--where are we going wrong? Nat. Rev. Clin. Oncol. 2011, 8, 189–190. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Maishi, N.; Hida, K. Tumor endothelial cells accelerate tumor metastasis. Cancer Sci. 2017, 108, 1921–1926. [Google Scholar] [CrossRef]
- Ishii, G.; Ochiai, A.; Neri, S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv. Drug Deliv. Rev. 2016, 99, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Takeya, M. Cafs and tams: Maestros of the tumour microenvironment. J. Pathol. 2017, 241, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Izumchenko, E.; Paz, K.; Ciznadija, D.; Sloma, I.; Katz, A.; Vasquez-Dunddel, D.; Ben-Zvi, I.; Stebbing, J.; McGuire, W.; Harris, W.; et al. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann. Oncol. 2017, 28, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. Us cancer institute to overhaul tumour cell lines. Nature 2016, 530, 391. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Clohessy, J.G.; Pandolfi, P.P. Mouse hospital and co-clinical trial project--from bench to bedside. Nat. Rev. Clin. Oncol. 2015, 12, 491–498. [Google Scholar] [CrossRef]
- Clohessy, J.G.; Pandolfi, P.P. The mouse hospital and its integration in ultra-precision approaches to cancer care. Front. Oncol. 2018, 8, 340. [Google Scholar] [CrossRef]
- Zhang, X.; Claerhout, S.; Prat, A.; Dobrolecki, L.E.; Petrovic, I.; Lai, Q.; Landis, M.D.; Wiechmann, L.; Schiff, R.; Giuliano, M.; et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013, 73, 4885–4897. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.L.; Mascaux, C.; Pham, N.A.; Sakashita, S.; Sykes, J.; Kim, L.; Yanagawa, N.; Allo, G.; Ishizawa, K.; Wang, D.; et al. Clinical utility of patient-derived xenografts to determine biomarkers of prognosis and map resistance pathways in EGFR-mutant lung adenocarcinoma. J. Clin. Oncol. 2015, 33, 2472–2480. [Google Scholar] [CrossRef] [PubMed]
- Vargas, R.; Gopal, P.; Kuzmishin, G.B.; DeBernardo, R.; Koyfman, S.A.; Jha, B.K.; Mian, O.Y.; Scott, J.; Adams, D.J.; Peacock, C.D.; et al. Case study: Patient-derived clear cell adenocarcinoma xenograft model longitudinally predicts treatment response. Npj Precis. Oncol. 2018, 2, 14. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The prisma statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- Rygaard, J.; Povlsen, C.O. Heterotransplantation of a human malignant tumour to "nude" mice. Acta Pathol. Microbiol. Scand. 1969, 77, 758–760. [Google Scholar] [CrossRef]
- Hammer, S.; Sommer, A.; Fichtner, I.; Becker, M.; Rolff, J.; Merk, J.; Klar, U.; Hoffmann, J. Comparative profiling of the novel epothilone, sagopilone, in xenografts derived from primary non-small cell lung cancer. Clin. Cancer Res. 2010, 16, 1452–1465. [Google Scholar] [CrossRef]
- Tentler, J.J.; Nallapareddy, S.; Tan, A.C.; Spreafico, A.; Pitts, T.M.; Morelli, M.P.; Selby, H.M.; Kachaeva, M.I.; Flanigan, S.A.; Kulikowski, G.N.; et al. Identification of predictive markers of response to the MEK1/2 inhibitor selumetinib (AZD6244) in K-ras-mutated colorectal cancer. Mol. Cancer Ther. 2010, 9, 3351–3362. [Google Scholar] [CrossRef]
- Bertotti, A.; Migliardi, G.; Galimi, F.; Sassi, F.; Torti, D.; Isella, C.; Cora, D.; Di Nicolantonio, F.; Buscarino, M.; Petti, C.; et al. A molecularly annotated platform of patient-derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011, 1, 508–523. [Google Scholar] [CrossRef]
- Julien, S.; Merino-Trigo, A.; Lacroix, L.; Pocard, M.; Goere, D.; Mariani, P.; Landron, S.; Bigot, L.; Nemati, F.; Dartigues, P.; et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin. Cancer Res. 2012, 18, 5314–5328. [Google Scholar] [CrossRef]
- Laheru, D.; Shah, P.; Rajeshkumar, N.V.; McAllister, F.; Taylor, G.; Goldsweig, H.; Le, D.T.; Donehower, R.; Jimeno, A.; Linden, S.; et al. Integrated preclinical and clinical development of S-trans, trans-farnesylthiosalicylic acid (FTS, Salirasib) in pancreatic cancer. Invest. New Drugs 2012, 30, 2391–2399. [Google Scholar] [CrossRef]
- Kreahling, J.M.; Foroutan, P.; Reed, D.; Martinez, G.; Razabdouski, T.; Bui, M.M.; Raghavan, M.; Letson, D.; Gillies, R.J.; Altiok, S. Wee1 inhibition by mk-1775 leads to tumor inhibition and enhances efficacy of gemcitabine in human sarcomas. PLoS ONE 2013, 8, e57523. [Google Scholar] [CrossRef]
- Rodriguez-Pascual, J.; Sha, P.; Garcia-Garcia, E.; Rajeshkumar, N.V.; De Vicente, E.; Quijano, Y.; Cubillo, A.; Angulo, B.; Hernando, O.; Hidalgo, M. A preclinical and clinical study of mycophenolate mofetil in pancreatic cancer. Invest. New Drugs 2013, 31, 14–19. [Google Scholar] [CrossRef]
- Amendt, C.; Staub, E.; Friese-Hamim, M.; Storkel, S.; Stroh, C. Association of EGFR expression level and cetuximab activity in patient-derived xenograft models of human non-small cell lung cancer. Clin. Cancer Res. 2014, 20, 4478–4487. [Google Scholar] [CrossRef]
- Barbie, T.U.; Alexe, G.; Aref, A.R.; Li, S.; Zhu, Z.; Zhang, X.; Imamura, Y.; Thai, T.C.; Huang, Y.; Bowden, M.; et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J. Clin. Invest. 2014, 124, 5411–5423. [Google Scholar] [CrossRef]
- Burkhart, C.; Fleyshman, D.; Kohrn, R.; Commane, M.; Garrigan, J.; Kurbatov, V.; Toshkov, I.; Ramachandran, R.; Martello, L.; Gurova, K.V. Curaxin cbl0137 eradicates drug resistant cancer stem cells and potentiates efficacy of gemcitabine in preclinical models of pancreatic cancer. Oncotarget 2014, 5, 11038–11053. [Google Scholar] [CrossRef]
- du Manoir, S.; Orsetti, B.; Bras-Goncalves, R.; Nguyen, T.T.; Lasorsa, L.; Boissiere, F.; Massemin, B.; Colombo, P.E.; Bibeau, F.; Jacot, W.; et al. Breast tumor pdxs are genetically plastic and correspond to a subset of aggressive cancers prone to relapse. Mol. Oncol. 2014, 8, 431–443. [Google Scholar] [CrossRef]
- Ducker, G.S.; Atreya, C.E.; Simko, J.P.; Hom, Y.K.; Matli, M.R.; Benes, C.H.; Hann, B.; Nakakura, E.K.; Bergsland, E.K.; Donner, D.B.; et al. Incomplete inhibition of phosphorylation of 4E-BP1 as a mechanism of primary resistance to ATP-competitive mTOR inhibitors. Oncogene 2014, 33, 1590–1600. [Google Scholar] [CrossRef]
- Ingels, A.; Zhao, H.; Thong, A.E.; Saar, M.; Valta, M.P.; Nolley, R.; Santos, J.; Peehl, D.M. Preclinical trial of a new dual mTOR inhibitor, mln0128, using renal cell carcinoma tumorgrafts. Int. J. Cancer 2014, 134, 2322–2329. [Google Scholar] [CrossRef]
- Kobelt, D.; Aumann, J.; Schmidt, M.; Wittig, B.; Fichtner, I.; Behrens, D.; Lemm, M.; Freundt, G.; Schlag, P.M.; Walther, W. Preclinical study on combined chemo- and nonviral gene therapy for sensitization of melanoma using a human TNF-alpha expressing MIDGE DNA vector. Mol. Oncol. 2014, 8, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wozniak, A.; Sciot, R.; Cornillie, J.; Wellens, J.; Van Looy, T.; Vanleeuw, U.; Stas, M.; Hompes, D.; Debiec-Rychter, M.; et al. Pazopanib, a receptor tyrosine kinase inhibitor, suppresses tumor growth through angiogenesis in dedifferentiated liposarcoma xenograft models. Transl. Oncol. 2014, 7, 665–671. [Google Scholar] [CrossRef]
- Liu, Y.J.; Shen, D.; Yin, X.; Gavine, P.; Zhang, T.; Su, X.; Zhan, P.; Xu, Y.; Lv, J.; Qian, J.; et al. HER2, MET and FGFR2 oncogenic driver alterations define distinct molecular segments for targeted therapies in gastric carcinoma. Br. J. Cancer 2014, 110, 1169–1178. [Google Scholar] [CrossRef]
- Qu, S.; Wang, K.; Xue, H.; Wang, Y.; Wu, R.; Liu, C.; Gao, A.C.; Gout, P.W.; Collins, C.C.; Wang, Y. Enhanced anticancer activity of a combination of docetaxel and aneustat (OMN54) in a patient-derived, advanced prostate cancer tissue xenograft model. Mol. Oncol. 2014, 8, 311–322. [Google Scholar] [CrossRef]
- Seguin, L.; Kato, S.; Franovic, A.; Camargo, M.F.; Lesperance, J.; Elliott, K.C.; Yebra, M.; Mielgo, A.; Lowy, A.M.; Husain, H.; et al. An integrin beta(3)-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nat. Cell Boil. 2014, 16, 457–468. [Google Scholar] [CrossRef]
- Stacchiotti, S.; Tortoreto, M.; Baldi, G.G.; Grignani, G.; Toss, A.; Badalamenti, G.; Cominetti, D.; Morosi, C.; Dei Tos, A.P.; Festinese, F.; et al. Preclinical and clinical evidence of activity of pazopanib in solitary fibrous tumour. Eur. J. Cancer 2014, 50, 3021–3028. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Paz, K.; Schwartz, G.K.; Wexler, L.H.; Maki, R.; Pollock, R.E.; Morris, R.; Cohen, R.; Shankar, A.; Blackman, G.; et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer 2014, 120, 2006–2015. [Google Scholar] [CrossRef]
- Wang, Z.; Da Silva, T.G.; Jin, K.; Han, X.; Ranganathan, P.; Zhu, X.; Sanchez-Mejias, A.; Bai, F.; Li, B.; Fei, D.L.; et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res. 2014, 74, 6364–6374. [Google Scholar] [CrossRef]
- Yang, A.; Rajeshkumar, N.V.; Wang, X.; Yabuuchi, S.; Alexander, B.M.; Chu, G.C.; Von Hoff, D.D.; Maitra, A.; Kimmelman, A.C. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014, 4, 905–913. [Google Scholar] [CrossRef]
- Bertolini, G.; D’Amico, L.; Moro, M.; Landoni, E.; Perego, P.; Miceli, R.; Gatti, L.; Andriani, F.; Wong, D.; Caserini, R.; et al. Microenvironment-modulated metastatic CD133+/CXCR4+/EPCAM− lung cancer-initiating cells sustain tumor dissemination and correlate with poor prognosis. Cancer Res. 2015, 75, 3636–3649. [Google Scholar] [CrossRef]
- Chen, D.; Huang, X.; Cai, J.; Guo, S.; Qian, W.; Wery, J.P.; Li, Q.X. A set of defined oncogenic mutation alleles seems to better predict the response to cetuximab in CRC patient-derived xenograft than KRAS 12/13 mutations. Oncotarget 2015, 6, 40815–40821. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Atreya, C.E.; Falchook, G.S.; Kwak, E.L.; Ryan, D.P.; Bendell, J.C.; Hamid, O.; Messersmith, W.A.; Daud, A.; Kurzrock, R.; et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J. Clin. Oncol. 2015, 33, 4023–4031. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Park, D.; Li, R.; Xie, M.; Owonikoko, T.K.; Zhang, G.; Sica, G.L.; Ding, C.; Zhou, J.; Magis, A.T.; et al. Small-molecule Bcl2 BH4 antagonist for lung cancer therapy. Cancer Cell 2015, 27, 852–863. [Google Scholar] [CrossRef]
- Hayes, G.M.; Chinn, L.; Cantor, J.M.; Cairns, B.; Levashova, Z.; Tran, H.; Velilla, T.; Duey, D.; Lippincott, J.; Zachwieja, J.; et al. Antitumor activity of an anti-CD98 antibody. Int. J. Cancer 2015, 137, 710–720. [Google Scholar] [CrossRef]
- Hu, C.; Dadon, T.; Chenna, V.; Yabuuchi, S.; Bannerji, R.; Booher, R.; Strack, P.; Azad, N.; Nelkin, B.D.; Maitra, A. Combined inhibition of cyclin-dependent kinases (dinaciclib) and AKT (MK-2206) blocks pancreatic tumor growth and metastases in patient-derived xenograft models. Mol. Cancer Ther. 2015, 14, 1532–1539. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- James, M.I.; Iwuji, C.; Irving, G.; Karmokar, A.; Higgins, J.A.; Griffin-Teal, N.; Thomas, A.; Greaves, P.; Cai, H.; Patel, S.R.; et al. Curcumin inhibits cancer stem cell phenotypes in ex vivo models of colorectal liver metastases, and is clinically safe and tolerable in combination with FOLFOX chemotherapy. Cancer Lett. 2015, 364, 135–141. [Google Scholar] [CrossRef]
- Kopetz, S.; Desai, J.; Chan, E.; Hecht, J.R.; O’Dwyer, P.J.; Maru, D.; Morris, V.; Janku, F.; Dasari, A.; Chung, W.; et al. Phase ii pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J. Clin. Oncol. 2015, 33, 4032–4038. [Google Scholar] [CrossRef]
- Meetze, K.; Vincent, S.; Tyler, S.; Mazsa, E.K.; Delpero, A.R.; Bottega, S.; McIntosh, D.; Nicoletti, R.; Winston, W.M.; Weiler, S.; et al. Neuregulin 1 expression is a predictive biomarker for response to AV-203, an ERBB3 inhibitory antibody, in human tumor models. Clin. Cancer Res. 2015, 21, 1106–1114. [Google Scholar] [CrossRef]
- Pan, C.X.; Zhang, H.; Tepper, C.G.; Lin, T.Y.; Davis, R.R.; Keck, J.; Ghosh, P.M.; Gill, P.; Airhart, S.; Bult, C.; et al. Development and characterization of bladder cancer patient-derived xenografts for molecularly guided targeted therapy. Plos ONE 2015, 10, e0134346. [Google Scholar] [CrossRef]
- Song, E.K.; Tai, W.M.; Messersmith, W.A.; Bagby, S.; Purkey, A.; Quackenbush, K.S.; Pitts, T.M.; Wang, G.; Blatchford, P.; Yahn, R.; et al. Potent antitumor activity of cabozantinib, a c-MET and VEGFR2 inhibitor, in a colorectal cancer patient-derived tumor explant model. Int. J. Cancer 2015, 136, 1967–1975. [Google Scholar] [CrossRef]
- Zhu, Q.; Izumchenko, E.; Aliper, A.M.; Makarev, E.; Paz, K.; Buzdin, A.A.; Zhavoronkov, A.A.; Sidransky, D. Pathway activation strength is a novel independent prognostic biomarker for cetuximab sensitivity in colorectal cancer patients. Hum. Genome Var. 2015, 2, 15009. [Google Scholar] [CrossRef]
- Aparicio, A.M.; Shen, L.; Tapia, E.L.; Lu, J.F.; Chen, H.C.; Zhang, J.; Wu, G.; Wang, X.; Troncoso, P.; Corn, P.; et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin. Cancer Res. 2016, 22, 1520–1530. [Google Scholar] [CrossRef]
- Banerjee, S.; Modi, S.; McGinn, O.; Zhao, X.; Dudeja, V.; Ramakrishnan, S.; Saluja, A.K. Impaired synthesis of stromal components in response to minnelide improves vascular function, drug delivery, and survival in pancreatic cancer. Clin. Cancer Res. 2016, 22, 415–425. [Google Scholar] [CrossRef]
- Goncalves, A.; Bertucci, F.; Guille, A.; Garnier, S.; Adelaide, J.; Carbuccia, N.; Cabaud, O.; Finetti, P.; Brunelle, S.; Piana, G.; et al. Targeted NGS, array-CGH, and patient-derived tumor xenografts for precision medicine in advanced breast cancer: A single-center prospective study. Oncotarget 2016, 7, 79428–79441. [Google Scholar] [CrossRef]
- Guo, S.; Chen, D.; Huang, X.; Cai, J.; Wery, J.P.; Li, Q.X. Cetuximab response in CRC patient-derived xenografts seems predicted by an expression based RAS pathway signature. Oncotarget 2016, 7, 50575–50581. [Google Scholar] [CrossRef][Green Version]
- Gupta, S.K.; Kizilbash, S.H.; Carlson, B.L.; Mladek, A.C.; Boakye-Agyeman, F.; Bakken, K.K.; Pokorny, J.L.; Schroeder, M.A.; Decker, P.A.; Cen, L.; et al. Delineation of MGMT hypermethylation as a biomarker for veliparib-mediated temozolomide-sensitizing therapy of glioblastoma. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef]
- Kim, H.Y.; Kim, J.; Ha Thi, H.T.; Bang, O.S.; Lee, W.S.; Hong, S. Evaluation of anti-tumorigenic activity of BP3B against colon cancer with patient-derived tumor xenograft model. BMC Complement. Altern. Med. 2016, 16, 473. [Google Scholar] [CrossRef]
- Krytska, K.; Ryles, H.T.; Sano, R.; Raman, P.; Infarinato, N.R.; Hansel, T.D.; Makena, M.R.; Song, M.M.; Reynolds, C.P.; Mosse, Y.P. Crizotinib synergizes with chemotherapy in preclinical models of neuroblastoma. Clin. Cancer Res. 2016, 22, 948–960. [Google Scholar] [CrossRef]
- Lee, M.S.; Helms, T.L.; Feng, N.; Gay, J.; Chang, Q.E.; Tian, F.; Wu, J.Y.; Toniatti, C.; Heffernan, T.P.; Powis, G.; et al. Efficacy of the combination of MEK and CDK4/6 inhibitors in vitro and in vivo in KRAS mutant colorectal cancer models. Oncotarget 2016, 7, 39595–39608. [Google Scholar] [CrossRef]
- Ni, J.; Ramkissoon, S.H.; Xie, S.; Goel, S.; Stover, D.G.; Guo, H.; Luu, V.; Marco, E.; Ramkissoon, L.A.; Kang, Y.J.; et al. Combination inhibition of PI3K and MTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat. Med. 2016, 22, 723–726. [Google Scholar] [CrossRef]
- Nicolle, D.; Fabre, M.; Simon-Coma, M.; Gorse, A.; Kappler, R.; Nonell, L.; Mallo, M.; Haidar, H.; Deas, O.; Mussini, C.; et al. Patient-derived mouse xenografts from pediatric liver cancer predict tumor recurrence and advise clinical management. Hepatology 2016, 64, 1121–1135. [Google Scholar] [CrossRef]
- Nigim, F.; Esaki, S.; Hood, M.; Lelic, N.; James, M.F.; Ramesh, V.; Stemmer-Rachamimov, A.; Cahill, D.P.; Brastianos, P.K.; Rabkin, S.D.; et al. A new patient-derived orthotopic malignant meningioma model treated with oncolytic herpes simplex virus. Neuro-Oncol. 2016, 18, 1278–1287. [Google Scholar] [CrossRef]
- Owonikoko, T.K.; Zhang, G.; Kim, H.S.; Stinson, R.M.; Bechara, R.; Zhang, C.; Chen, Z.; Saba, N.F.; Pakkala, S.; Pillai, R.; et al. Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. J. Transl. Med. 2016, 14, 111. [Google Scholar] [CrossRef]
- Pearson, A.; Smyth, E.; Babina, I.S.; Herrera-Abreu, M.T.; Tarazona, N.; Peckitt, C.; Kilgour, E.; Smith, N.R.; Geh, C.; Rooney, C.; et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov 2016, 6, 838–851. [Google Scholar] [CrossRef]
- Varkaris, A.; Corn, P.G.; Parikh, N.U.; Efstathiou, E.; Song, J.H.; Lee, Y.C.; Aparicio, A.; Hoang, A.G.; Gaur, S.; Thorpe, L.; et al. Integrating murine and clinical trials with cabozantinib to understand roles of MET and VEGFR2 as targets for growth inhibition of prostate cancer. Clin. Cancer Res. 2016, 22, 107–121. [Google Scholar] [CrossRef]
- Bialucha, C.U.; Collins, S.D.; Li, X.; Saxena, P.; Zhang, X.; Durr, C.; Lafont, B.; Prieur, P.; Shim, Y.; Mosher, R.; et al. Discovery and optimization of hkt288, a cadherin-6-targeting ADC for the treatment of ovarian and renal cancers. Cancer Discov. 2017, 7, 1030–1045. [Google Scholar] [CrossRef]
- Bonanno, L.; De Paoli, A.; Zulato, E.; Esposito, G.; Calabrese, F.; Favaretto, A.; Santo, A.; Conte, A.D.; Chilosi, M.; Oniga, F.; et al. LKB1 expression correlates with increased survival in patients with advanced non-small cell lung cancer treated with chemotherapy and bevacizumab. Clin. Cancer Res. 2017, 23, 3316–3324. [Google Scholar] [CrossRef]
- Calvo, E.; Soria, J.C.; Ma, W.W.; Wang, T.; Bahleda, R.; Tolcher, A.W.; Gernhardt, D.; O’Connell, J.; Millham, R.; Giri, N.; et al. A phase I clinical trial and independent patient-derived xenograft study of combined targeted treatment with dacomitinib and figitumumab in advanced solid tumors. Clin. Cancer Res. 2017, 23, 1177–1185. [Google Scholar] [CrossRef]
- Colon-Otero, G.; Weroha, S.J.; Foster, N.R.; Haluska, P.; Hou, X.; Wahner-Hendrickson, A.E.; Jatoi, A.; Block, M.S.; Dinh, T.A.; Robertson, M.W.; et al. Phase 2 trial of everolimus and letrozole in relapsed estrogen receptor-positive high-grade ovarian cancers. Gynecol. Oncol. 2017, 146, 64–68. [Google Scholar] [CrossRef]
- Damelin, M.; Bankovich, A.; Bernstein, J.; Lucas, J.; Chen, L.; Williams, S.; Park, A.; Aguilar, J.; Ernstoff, E.; Charati, M.; et al. A PTK7-targeted antibody-drug conjugate reduces tumor-initiating cells and induces sustained tumor regressions. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Eigenmann, M.J.; Frances, N.; Lave, T.; Walz, A.C. PKPD modeling of acquired resistance to anti-cancer drug treatment. J. Pharmacokinet. Pharmacodyn. 2017, 44, 617–630. [Google Scholar] [CrossRef]
- Engstrom, L.D.; Aranda, R.; Lee, M.; Tovar, E.A.; Essenburg, C.J.; Madaj, Z.; Chiang, H.; Briere, D.; Hallin, J.; Lopez-Casas, P.P.; et al. Glesatinib exhibits antitumor activity in lung cancer models and patients harboring MET exon 14 mutations and overcomes mutation-mediated resistance to type I MET inhibitors in nonclinical models. Clin. Cancer Res. 2017, 23, 6661–6672. [Google Scholar] [CrossRef]
- Eritja, N.; Chen, B.J.; Rodriguez-Barrueco, R.; Santacana, M.; Gatius, S.; Vidal, A.; Marti, M.D.; Ponce, J.; Bergada, L.; Yeramian, A.; et al. Autophagy orchestrates adaptive responses to targeted therapy in endometrial cancer. Autophagy 2017, 13, 608–624. [Google Scholar] [CrossRef]
- Francis, A.M.; Alexander, A.; Liu, Y.; Vijayaraghavan, S.; Low, K.H.; Yang, D.; Bui, T.; Somaiah, N.; Ravi, V.; Keyomarsi, K.; et al. CDK4/6 inhibitors sensitize Rb-positive sarcoma cells to Wee1 kinase inhibition through reversible cell-cycle arrest. Mol. Cancer Ther. 2017, 16, 1751–1764. [Google Scholar] [CrossRef]
- Frankel, A.E.; Eskiocak, U.; Gill, J.G.; Yuan, S.; Ramesh, V.; Froehlich, T.W.; Ahn, C.; Morrison, S.J. Digoxin plus trametinib therapy achieves disease control in BRAF wild-type metastatic melanoma patients. Neoplasia 2017, 19, 255–260. [Google Scholar] [CrossRef]
- Ionkina, A.A.; Tentler, J.J.; Kim, J.; Capasso, A.; Pitts, T.M.; Ryall, K.A.; Howison, R.R.; Kabos, P.; Sartorius, C.A.; Tan, A.C.; et al. Efficacy and molecular mechanisms of differentiated response to the aurora and angiogenic kinase inhibitor ENMD-2076 in preclinical models of p53-mutated triple-negative breast cancer. Front. Oncol. 2017, 7, 94. [Google Scholar] [CrossRef]
- Kim, H.R.; Kang, H.N.; Shim, H.S.; Kim, E.Y.; Kim, J.; Kim, D.J.; Lee, J.G.; Lee, C.Y.; Hong, M.H.; Kim, S.M.; et al. Co-clinical trials demonstrate predictive biomarkers for dovitinib, an FGFR inhibitor, in lung squamous cell carcinoma. Ann. Oncol. 2017, 28, 1250–1259. [Google Scholar] [CrossRef]
- Kirouac, D.C.; Schaefer, G.; Chan, J.; Merchant, M.; Orr, C.; Huang, S.A.; Moffat, J.; Liu, L.; Gadkar, K.; Ramanujan, S. Clinical responses to ERK inhibition in BRAF(V600E)-mutant colorectal cancer predicted using a computational model. Npj Syst. Biol. Appl. 2017, 3, 14. [Google Scholar] [CrossRef]
- Kondrashova, O.; Nguyen, M.; Shield-Artin, K.; Tinker, A.V.; Teng, N.N.H.; Harrell, M.I.; Kuiper, M.J.; Ho, G.Y.; Barker, H.; Jasin, M.; et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017, 7, 984–998. [Google Scholar] [CrossRef]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef]
- Prabakaran, P.J.; Javaid, A.M.; Swick, A.D.; Werner, L.R.; Nickel, K.P.; Sampene, E.; Hu, R.; Ong, I.M.; Bruce, J.Y.; Hartig, G.K.; et al. Radiosensitization of adenoid cystic carcinoma with MDM2 inhibition. Clin. Cancer Res. 2017, 23, 6044–6053. [Google Scholar] [CrossRef]
- Schmidt, C.; Schubert, N.A.; Brabetz, S.; Mack, N.; Schwalm, B.; Chan, J.A.; Selt, F.; Herold-Mende, C.; Witt, O.; Milde, T.; et al. Preclinical drug screen reveals topotecan, actinomycin d, and volasertib as potential new therapeutic candidates for ETMR brain tumor patients. Neuro-Oncol. 2017, 19, 1607–1617. [Google Scholar] [CrossRef]
- Weeden, C.E.; Holik, A.Z.; Young, R.J.; Ma, S.B.; Garnier, J.M.; Fox, S.B.; Antippa, P.; Irving, L.B.; Steinfort, D.P.; Wright, G.M.; et al. Cisplatin increases sensitivity to FGFR inhibition in patient-derived xenograft models of lung squamous cell carcinoma. Mol Cancer Ther. 2017, 16, 1610–1622. [Google Scholar] [CrossRef]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef]
- Xu, Y.C.; Wang, X.; Chen, Y.; Chen, S.M.; Yang, X.Y.; Sun, Y.M.; Geng, M.Y.; Ding, J.; Meng, L.H. Integration of receptor tyrosine kinases determines sensitivity to pI3Kalpha-selective inhibitors in breast cancer. Theranostics 2017, 7, 974–986. [Google Scholar] [CrossRef]
- Yao, Y.M.; Donoho, G.P.; Iversen, P.W.; Zhang, Y.; Van Horn, R.D.; Forest, A.; Novosiadly, R.D.; Webster, Y.W.; Ebert, P.; Bray, S.; et al. Mouse PDX trial suggests synergy of concurrent inhibition of RAF and EGFR in colorectal cancer with BRAF or KRAS mutations. Clin. Cancer Res. 2017, 23, 5547–5560. [Google Scholar] [CrossRef]
- Yu, J.; Qin, B.; Moyer, A.M.; Sinnwell, J.P.; Thompson, K.J.; Copland, J.A., 3rd; Marlow, L.A.; Miller, J.L.; Yin, P.; Gao, B.; et al. Establishing and characterizing patient-derived xenografts using pre-chemotherapy percutaneous biopsy and post-chemotherapy surgical samples from a prospective neoadjuvant breast cancer study. Breast Cancer Res. 2017, 19, 130. [Google Scholar] [CrossRef]
- Yuan, B.; Ye, N.; Song, S.S.; Wang, Y.T.; Song, Z.; Chen, H.D.; Chen, C.H.; Huan, X.J.; Wang, Y.Q.; Su, Y.; et al. Poly(ADP-ribose)polymerase (PARP) inhibition and anticancer activity of simmiparib, a new inhibitor undergoing clinical trials. Cancer Lett. 2017, 386, 47–56. [Google Scholar] [CrossRef]
- Campbell, K.M.; Lin, T.; Zolkind, P.; Barnell, E.K.; Skidmore, Z.L.; Winkler, A.E.; Law, J.H.; Mardis, E.R.; Wartman, L.D.; Adkins, D.R.; et al. Oral cavity squamous cell carcinoma xenografts retain complex genotypes and intertumor molecular heterogeneity. Cell Rep. 2018, 24, 2167–2178. [Google Scholar] [CrossRef]
- Cazet, A.S.; Hui, M.N.; Elsworth, B.L.; Wu, S.Z.; Roden, D.; Chan, C.L.; Skhinas, J.N.; Collot, R.; Yang, J.; Harvey, K.; et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018, 9, 2897. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Zammarchi, F.; Williams, D.G.; Havenith, C.E.G.; Monks, N.R.; Tyrer, P.; D’Hooge, F.; Fleming, R.; Vashisht, K.; Dimasi, N.; et al. Antitumor activity of MEDI3726 (ADCT-401), a pyrrolobenzodiazepine antibody-drug conjugate targeting PSMA, in preclinical models of prostate cancer. Mol. Cancer Ther. 2018, 17, 2176–2186. [Google Scholar] [CrossRef]
- Couts, K.L.; Bemis, J.; Turner, J.A.; Bagby, S.M.; Murphy, D.; Christiansen, J.; Hintzsche, J.D.; Le, A.; Pitts, T.M.; Wells, K.; et al. ALK inhibitor response in melanomas expressing EML4-ALK fusions and alternate ALK isoforms. Mol. Cancer Ther. 2018, 17, 222–231. [Google Scholar] [CrossRef]
- Decaudin, D.; El Botty, R.; Diallo, B.; Massonnet, G.; Fleury, J.; Naguez, A.; Raymondie, C.; Davies, E.; Smith, A.; Wilson, J.; et al. Selumetinib-based therapy in uveal melanoma patient-derived xenografts. Oncotarget 2018, 9, 21674–21686. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Somwar, R.; Mangatt, B.P.; Edgren, H.; Desmeules, P.; Ruusulehto, A.; Smith, R.S.; Delasos, L.; Vojnic, M.; Plodkowski, A.J.; et al. Response to ERBB3-directed targeted therapy in NRG1-rearranged cancers. Cancer Discov. 2018, 8, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Einarsdottir, B.O.; Karlsson, J.; Soderberg, E.M.V.; Lindberg, M.F.; Funck-Brentano, E.; Jespersen, H.; Brynjolfsson, S.F.; Bagge, R.O.; Carstam, L.; Scobie, M.; et al. A patient-derived xenograft pre-clinical trial reveals treatment responses and a resistance mechanism to karonudib in metastatic melanoma. Cell Death Dis. 2018, 9, 810. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Chen, M.; Ouyang, Y.; Li, R.; Zhang, X.; Gao, X.; Lin, S.; Wang, X. Icaritin induces ovarian cancer cell apoptosis through activation of p53 and inhibition of Akt/mTOR pathway. Life Sci. 2018, 202, 188–194. [Google Scholar] [CrossRef]
- Harris, A.L.; Lee, S.E.; Dawson, L.K.; Marlow, L.A.; Edenfield, B.H.; Durham, W.F.; Flotte, T.J.; Thompson, M.; Small, D.L.; Synnott, A.J.; et al. Targeting the cyclin dependent kinase and retinoblastoma axis overcomes standard of care resistance in BRAF (V600E) -mutant melanoma. Oncotarget 2018, 9, 10905–10919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heisey, D.A.R.; Lochmann, T.L.; Floros, K.V.; Coon, C.M.; Powell, K.M.; Jacob, S.; Calbert, M.L.; Ghotra, M.S.; Stein, G.T.; Maves, Y.K.; et al. The ewing family of tumors relies on BCL-2 and BCL-XL to escape PARP inhibitor toxicity. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 3970. [Google Scholar] [CrossRef]
- Kurokawa, C.; Iankov, I.D.; Anderson, S.K.; Aderca, I.; Leontovich, A.A.; Maurer, M.J.; Oberg, A.L.; Schroeder, M.A.; Giannini, C.; Greiner, S.M.; et al. Constitutive interferon pathway activation in tumors as an efficacy determinant following oncolytic virotherapy. J. Natl. Cancer Inst. 2018, 110, 1123–1132. [Google Scholar] [CrossRef]
- Liu, X.L.; Xu, Y.C.; Wang, Y.X.; Chen, Y.; Wang, B.B.; Wang, Y.; Chen, Y.H.; Tan, C.; Hu, L.D.; Ma, Q.Y.; et al. Decrease in phosphorylated ERK indicates the therapeutic efficacy of a clinical PI3Kalpha-selective inhibitor CYH33 in breast cancer. Cancer Lett. 2018, 433, 273–282. [Google Scholar] [CrossRef]
- Phillips, A.C.; Boghaert, E.R.; Vaidya, K.S.; Falls, H.D.; Mitten, M.J.; DeVries, P.J.; Benatuil, L.; Hsieh, C.M.; Meulbroek, J.A.; Panchal, S.C.; et al. Characterization of ABBV-221, a tumor-selective EGFR-targeting antibody drug conjugate. Mol Cancer Ther. 2018, 17, 795–805. [Google Scholar] [CrossRef]
- Ponath, P.; Menezes, D.; Pan, C.; Chen, B.; Oyasu, M.; Strachan, D.; LeBlanc, H.; Sun, H.; Wang, X.T.; Rangan, V.S.; et al. A novel, fully human anti-fucosyl-GM1 antibody demonstrates potent in vitro and in vivo antitumor activity in preclinical models of small cell lung cancer. Clin. Cancer Res. 2018, 24, 5178–5189. [Google Scholar] [CrossRef]
- Qu, S.; Xue, H.; Dong, X.; Lin, D.; Wu, R.; Nabavi, N.; Collins, C.C.; Gleave, M.E.; Gout, P.W.; Wang, Y. Aneustat (OMN54) has aerobic glycolysis-inhibitory activity and also immunomodulatory activity as indicated by a first-generation PDX prostate cancer model. Int. J. Cancer 2018, 143, 419–429. [Google Scholar] [CrossRef]
- Robichaux, J.P.; Elamin, Y.Y.; Tan, Z.; Carter, B.W.; Zhang, S.; Liu, S.; Li, S.; Chen, T.; Poteete, A.; Estrada-Bernal, A.; et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat. Med. 2018, 24, 638–646. [Google Scholar] [CrossRef]
- Ruicci, K.M.; Meens, J.; Sun, R.X.; Rizzo, G.; Pinto, N.; Yoo, J.; Fung, K.; MacNeil, D.; Mymryk, J.S.; Barrett, J.W.; et al. A controlled trial of hnscc patient-derived xenografts reveals broad efficacy of PI3Kalpha inhibition in controlling tumor growth. Int. J. Cancer 2018. [Google Scholar] [CrossRef]
- Wagner, J.; Kline, C.L.; Zhou, L.; Khazak, V.; El-Deiry, W.S. Anti-tumor effects of onc201 in combination with VEGF-inhibitors significantly impacts colorectal cancer growth and survival in vivo through complementary non-overlapping mechanisms. J. Exp. Clin. Cancer Res. 2018, 37, 11. [Google Scholar] [CrossRef]
- Wang, F.; Li, H.; Markovsky, E.; Glass, R.; de Stanchina, E.; Powell, S.N.; Schwartz, G.K.; Haimovitz-Friedman, A. Pazopanib radio-sensitization of human sarcoma tumors. Oncotarget 2018, 9, 9311–9324. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, T.; Qin, Z.; Jiang, J.; Wang, Q.; Yang, S.; Rivard, C.; Gao, G.; Ng, T.L.; Tu, M.M.; et al. HER2 exon 20 insertions in non-small cell lung cancer are sensitive to the irreversible pan-HER receptor tyrosine kinase inhibitor pyrotinib. Ann. Oncol. 2018. [Google Scholar] [CrossRef]
- Zaytseva, Y.Y.; Rychahou, P.G.; Le, A.T.; Scott, T.L.; Flight, R.M.; Kim, J.T.; Harris, J.; Liu, J.; Wang, C.; Morris, A.J.; et al. Preclinical evaluation of novel fatty acid synthase inhibitors in primary colorectal cancer cells and a patient-derived xenograft model of colorectal cancer. Oncotarget 2018, 9, 24787–24800. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, C.; Wang, J.; Chen, D.; Deng, J.; Deng, J.; Fan, J.; Badakhshi, H.; Huang, X.; Zhang, L.; et al. A subset of esophageal squamous cell carcinoma patient-derived xenografts respond to cetuximab, which is predicted by high EGFR expression and amplification. J. Thorac. Dis. 2018, 10, 5328–5338. [Google Scholar] [CrossRef]
- Zhong, H.; Chen, C.; Tammali, R.; Breen, S.; Zhang, J.; Fazenbaker, C.; Kennedy, M.; Conway, J.; Higgs, B.W.; Holoweckyj, N.; et al. Improved therapeutic window in brca-mutant tumors with antibody-linked pyrrolobenzodiazepine dimers with and without PARP inhibition. Mol. Cancer Ther. 2019, 18, 89–99. [Google Scholar] [CrossRef]
- Cho, S.Y.; Kang, W.; Han, J.Y.; Min, S.; Kang, J.; Lee, A.; Kwon, J.Y.; Lee, C.; Park, H. An integrative approach to precision cancer medicine using patient-derived xenografts. Mol. Cells 2016, 39, 77–86. [Google Scholar] [CrossRef]
- Hoffman, R.M. Patient-derived orthotopic xenografts: Better mimic of metastasis than subcutaneous xenografts. Nat. Reviews. Cancer 2015, 15, 451–452. [Google Scholar] [CrossRef]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Reviews. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef]
- Braekeveldt, N.; Wigerup, C.; Gisselsson, D.; Mohlin, S.; Merselius, M.; Beckman, S.; Jonson, T.; Borjesson, A.; Backman, T.; Tadeo, I.; et al. Neuroblastoma patient-derived orthotopic xenografts retain metastatic patterns and geno- and phenotypes of patient tumours. Int. J. Cancer 2015, 136, E252–E261. [Google Scholar] [CrossRef]
- Braekeveldt, N.; Bexell, D. Patient-derived xenografts as preclinical neuroblastoma models. Cell Tissue Res. 2018, 372, 233–243. [Google Scholar] [CrossRef]
- Capasso, A.; Lang, J.; Pitts, T.M.; Jordan, K.R.; Lieu, C.H.; Davis, S.L.; Diamond, J.R.; Kopetz, S.; Barbee, J.; Peterson, J.; et al. Characterization of immune responses to anti-PD-1 mono and combination immunotherapy in hematopoietic humanized mice implanted with tumor xenografts. J. Immunother. Cancer 2019, 7, 37. [Google Scholar] [CrossRef]
- Meehan, T.F.; Conte, N.; Goldstein, T.; Inghirami, G.; Murakami, M.A.; Brabetz, S.; Gu, Z.; Wiser, J.A.; Dunn, P.; Begley, D.A.; et al. PDX-MI: Minimal information for patient-derived tumor xenograft models. Cancer Res. 2017, 77, e62–e66. [Google Scholar] [CrossRef]
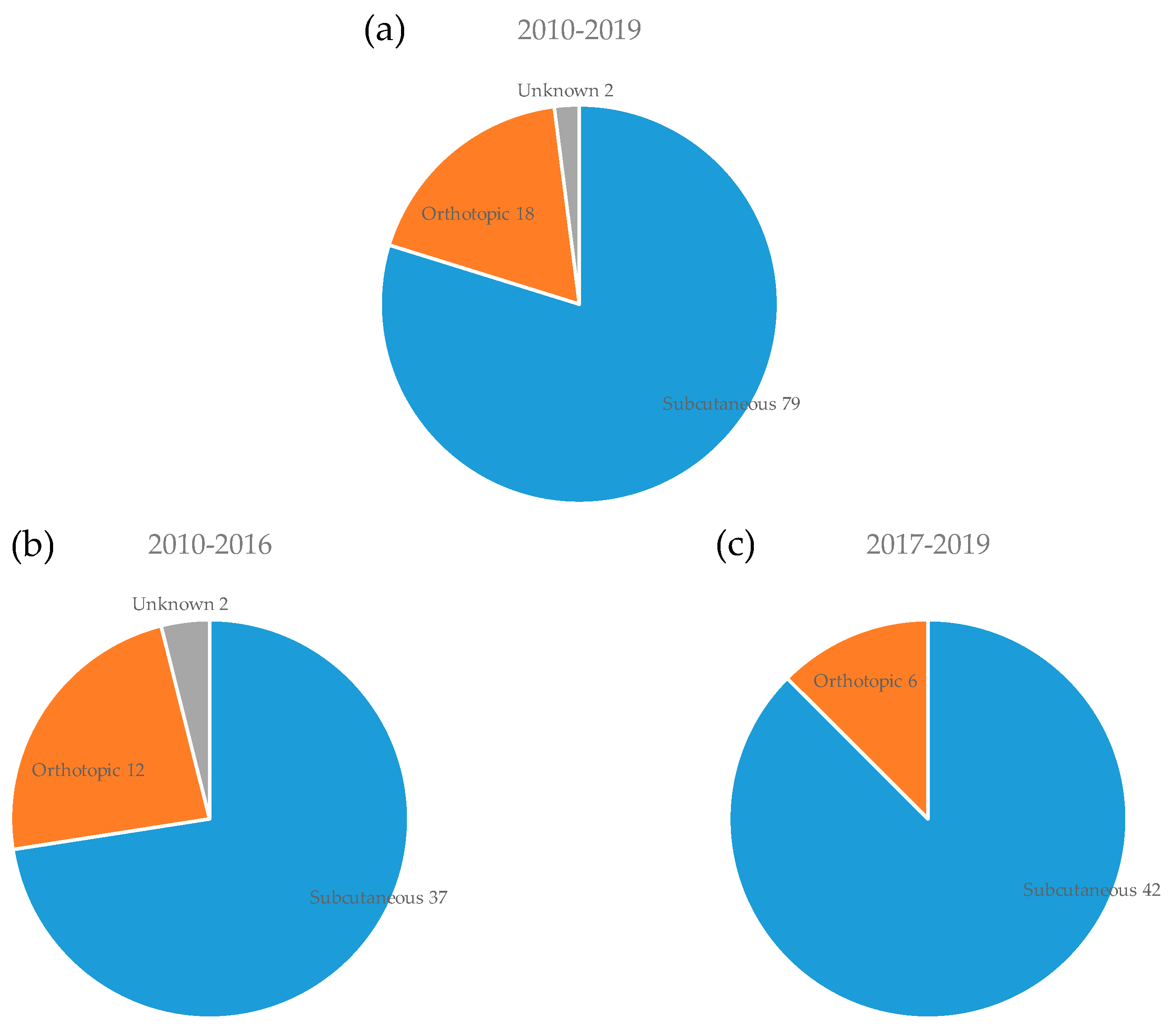
| Author | Year1 | Origin2 | Cases3 | Strain4 | Site5 | Animals6 |
|---|---|---|---|---|---|---|
| Hammer [18] | 2010 | NSCLC7 | 22 | Nude | sc12 | 6 |
| Bertotti [20] | 2011 | CRC8 | 85 | NOD-scid | sc12 | 6 |
| Laheru [22] | 2012 | Pancreas | 14 | Nude | sc12 | 5 |
| Amendt [25] | 2014 | NSCLC7 | 45 | Nude | sc12 | 10 |
| Chen [41] | 2015 | CRC8 | 27 | Nude | sc12 | 5 |
| Gao [10] | 2015 | Solid tumor | 1075 | Nude | sc12 | 1 |
| Pan [50] | 2015 | Bladder | 22 | NSG | sc12/ortho13 | 8 to 10 |
| Guo [56] | 2016 | CRC8 | 25 | NSG | sc12 | Unknown |
| Gupta [57] | 2016 | GBM9 | 28 | Nude | ortho13 | 8 to 10 |
| Bialucha [67] | 2017 | Ovary/RCC10 | 30 | Nude | sc12 | 1 |
| Yao [87] | 2017 | CRC | 79 | Nude | sc12 | 1 |
| Einarsdottir [96] | 2018 | Melanoma | 31 | NOG | sc12 | 1 |
| Ruicci [107] | 2018 | HNSCC11 | 20 | NSG | sc12 | 2 |
| Zhong [113] | 2019 | Breast/Ovary | 23 | Nude | sc12 | 2 to 3 |
| Author | Year1 | Origin2 | Cases3 | Strain4 | Site5 | Animals6 |
|---|---|---|---|---|---|---|
| Stebbing [37] | 2014 | Sarcoma | 16 | Nude | sc11 | Unknown |
| Kopetz [48] | 2015 | CRC7 | 1 | NSG | sc11 | 1 |
| Owonikoko [64] | 2016 | SCLC8 | 5 | Nude | sc11 | 3 to 6 |
| Frankel [76] | 2017 | Melanoma | 4 | NSG | sc11 | 7 to 10 |
| Kim [78] | 2017 | LSCC9 | 5 | NOG/Nude | sc11 | 6 to 7 |
| Pauli [81] | 2017 | Solid tumor | 19 | Nude | sc11 | 5 |
| Campbell [90] | 2018 | OCSCC10 | 1 | NSG | sc11 | 7 |
| Harris [98] | 2018 | Melanoma | 3 | Nude | sc11 | 8 to 10 |
| Vargas [15] | 2018 | Clear cell adenocarcinoma | 1 | NSG | sc11 | 3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koga, Y.; Ochiai, A. Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors. Cells 2019, 8, 418. https://doi.org/10.3390/cells8050418
Koga Y, Ochiai A. Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors. Cells. 2019; 8(5):418. https://doi.org/10.3390/cells8050418
Chicago/Turabian StyleKoga, Yoshikatsu, and Atsushi Ochiai. 2019. "Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors" Cells 8, no. 5: 418. https://doi.org/10.3390/cells8050418
APA StyleKoga, Y., & Ochiai, A. (2019). Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors. Cells, 8(5), 418. https://doi.org/10.3390/cells8050418






