Purification of Functional Human TRP Channels Recombinantly Produced in Yeast
Abstract
:1. Introduction
2. Materials and Methods
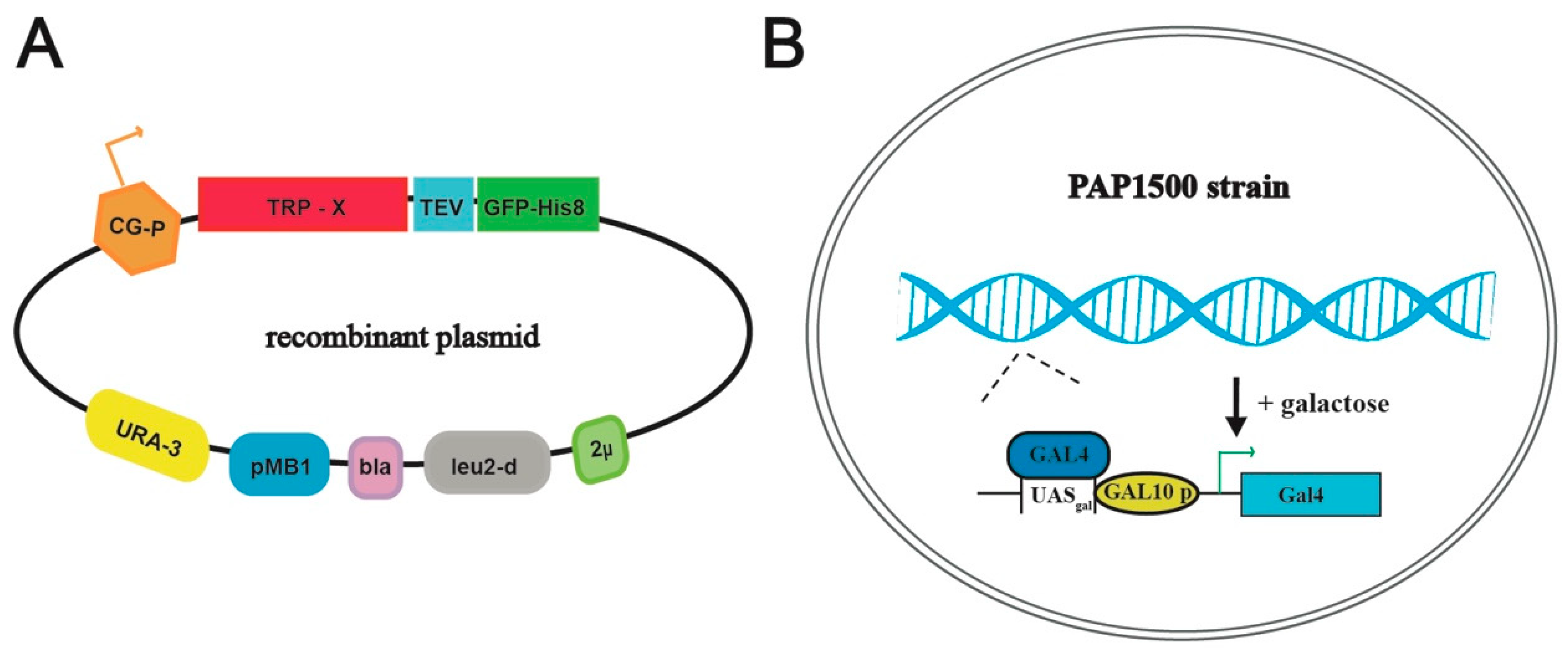
2.1. Cloning and Construction of Plasmids
2.2. Small-Scale Expression of TRP Channels and Live Cell Bioimaging
2.3. Membrane Preparation, Detergent Screens and F-SEC
2.4. Large-Scale Protein Production, TEV Protease Cleavage and SEC
2.5. Measurements of Single Channel Ion Conductance
3. Results
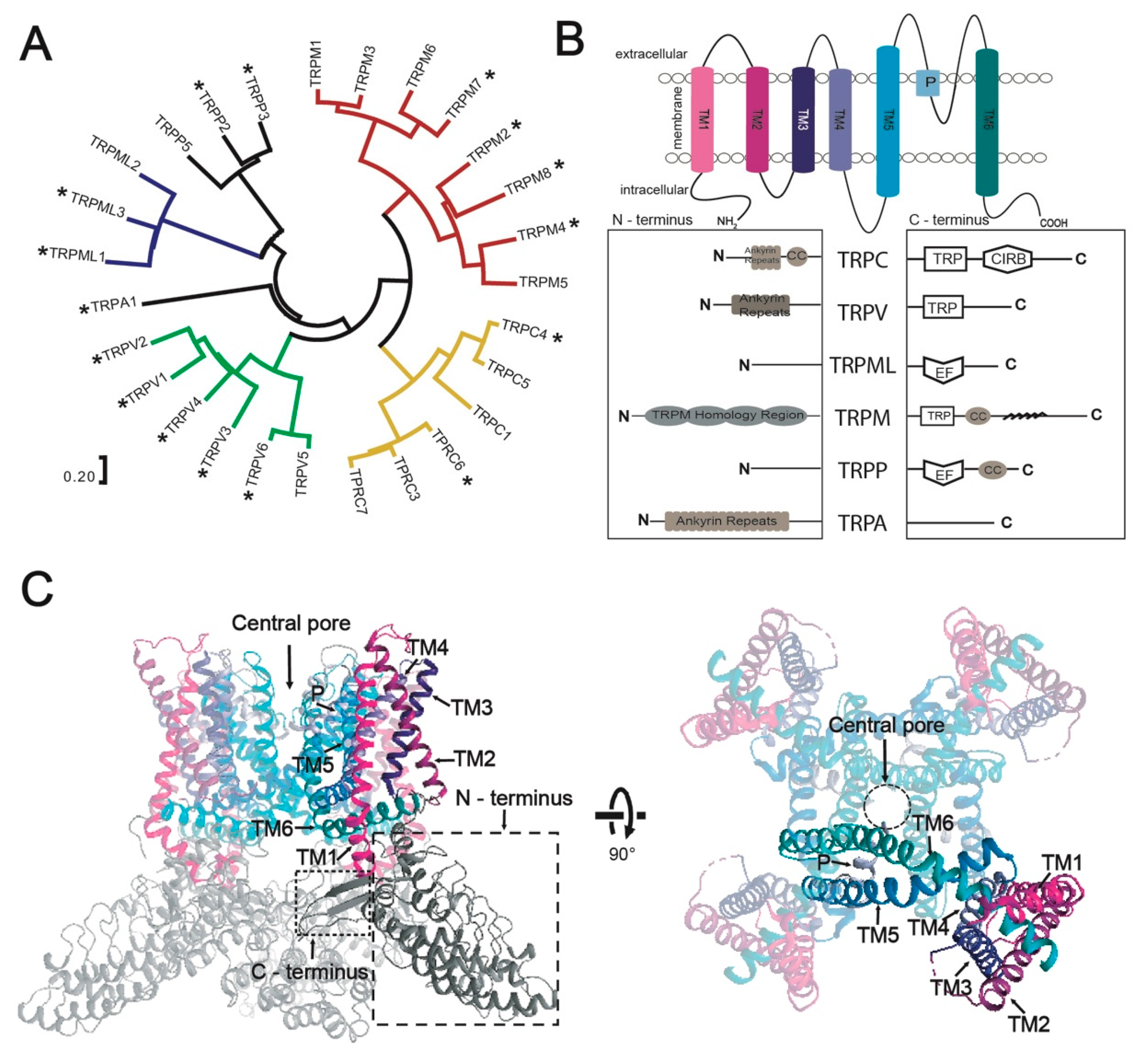
3.1. Selected TRP-Channel Targets and The Expression System
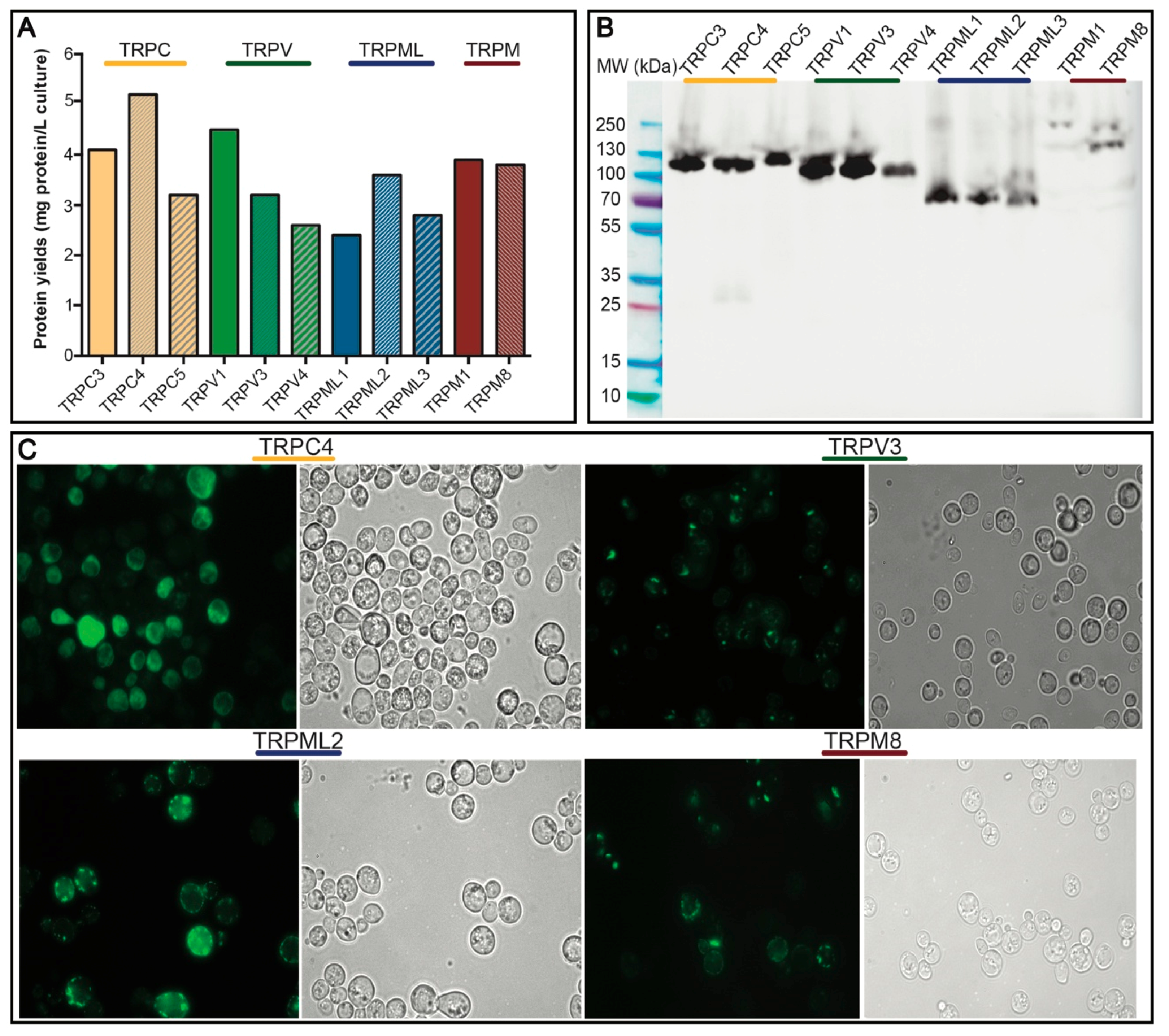
3.2. Small-Scale Production and Localization in S. cerevisiae
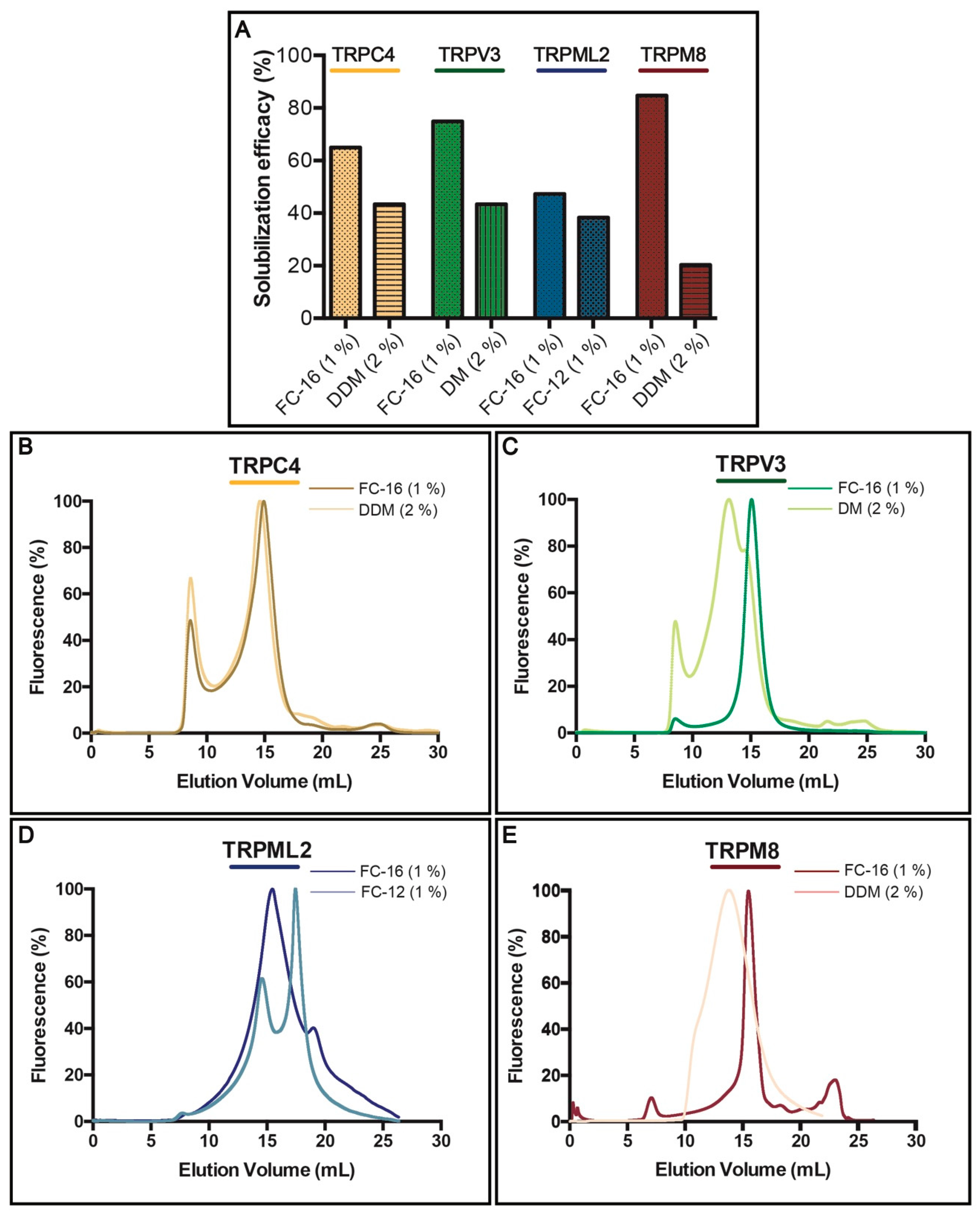
3.3. Solubilization Screen and F-SEC
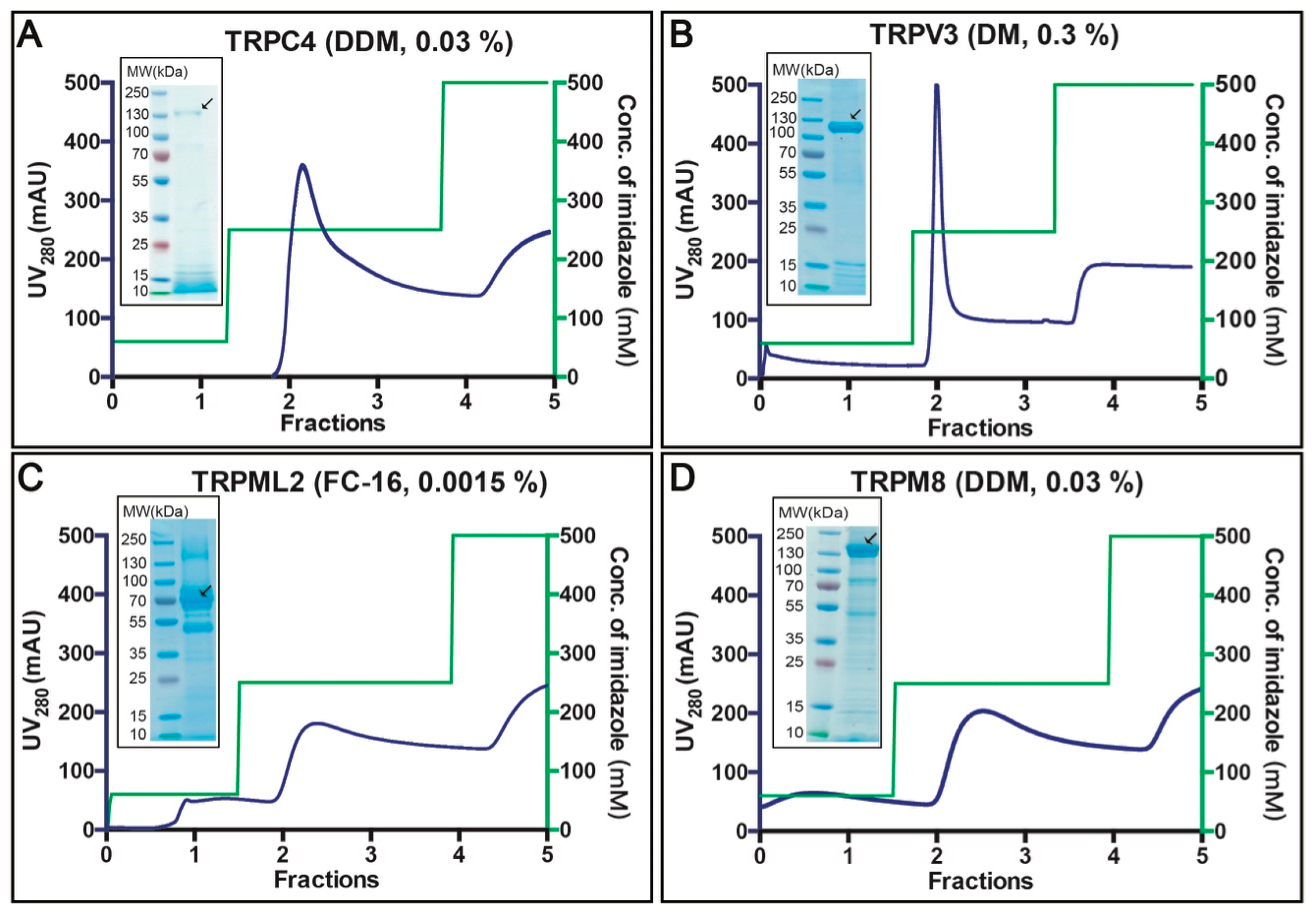
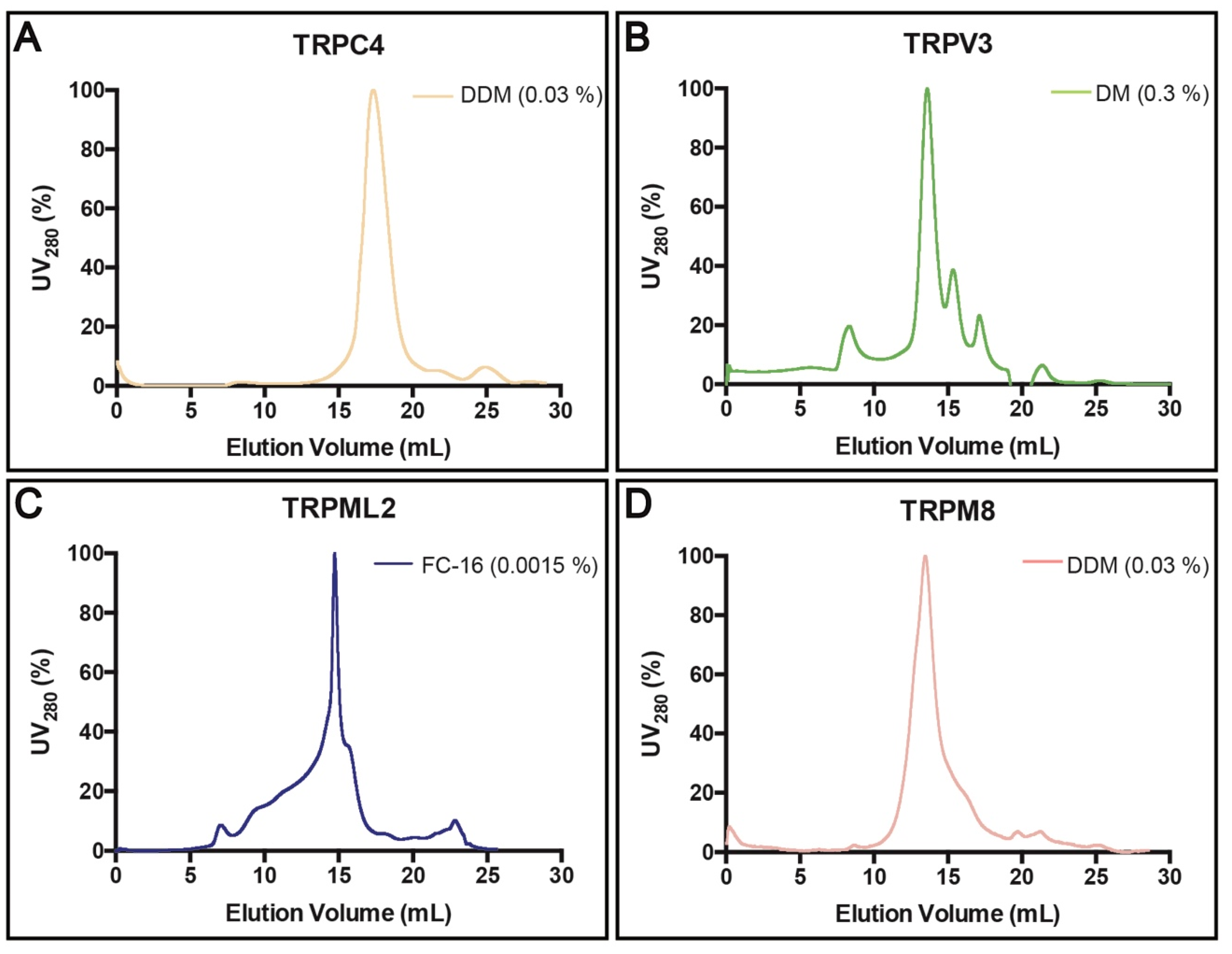
3.4. Purification
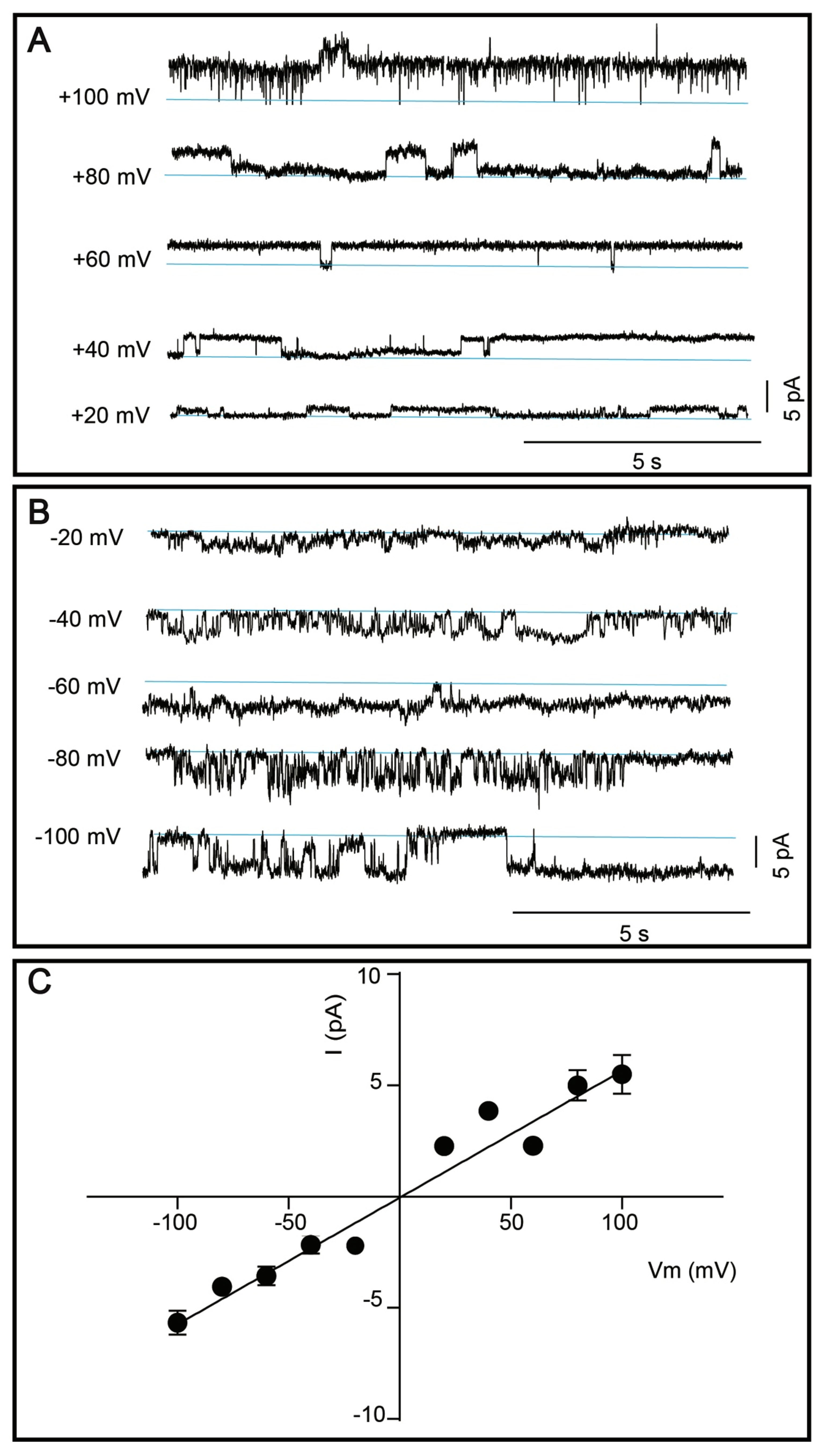
3.5. Single Channel Current Recordings of DDM-Solubilized TRPM8
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernandes, E.S.; Fernandes, M.A.; Keeble, J.E. The functions of TRPA1 and TRPV1: Moving away from sensory nerves. Br. J. Pharmacol. 2012, 166, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Gees, M.; Colsoul, B.; Nilius, B. The Role of Transient Receptor Potential Cation Channels in Ca2+ Signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a003962. [Google Scholar] [CrossRef] [PubMed]
- Brauchi, S. A Hot-Sensing Cold Receptor: C-Terminal Domain Determines Thermosensation in Transient Receptor Potential Channels. J. Neurosci. 2006, 26, 4830–4840. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.D.; Alessandri-Haber, N. TRP channels: Targets for the relief of pain. Biochim. Biophys. Acta Mol. Basis Dis. 2007, 1772, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Philippaert, K.; Pironet, A.; Mesuere, M.; Sones, W.; Vermeiren, L.; Kerselaers, S.; Pinto, S.; Segal, A.; Antoine, N.; Gysemans, C.; et al. Steviol glycosides enhance pancreatic beta-cell function and taste sensation by potentiation of TRPM5 channel activity. Nat. Commun. 2017. [Google Scholar] [CrossRef] [PubMed]
- Eccles, R. Nasal physiology and disease with reference to asthma. Agents Actions Suppl. 1989, 28, 249–261. [Google Scholar] [PubMed]
- Nilius, B. TRP channels in disease. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2007, 1772, 805–812. [Google Scholar] [CrossRef]
- Moran, M.M. TRP Channels as Potential Drug Targets. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 309–330. [Google Scholar] [CrossRef]
- Hellmich, U.A.; Gaudet, R. Structural biology of TRP channels. Handb. Exp. Pharmacol. 2014, 223, 963–990. [Google Scholar] [CrossRef]
- Vriens, J.; Owsianik, G.; Voets, T.; Droogmans, G.; Nilius, B. Invertebrate TRP proteins as functional models for mammalian channels. Pflugers Arch. Eur. J. Physiol. 2004, 449, 213–226. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Armache, J.P.; Gao, Y.; Cheng, Y.; Julius, D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 2015, 520, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, M.; Madej, M.G.; Kreuter, L.; Rhinow, D.; Heinz, V.; De Sanctis, S.; Ruppel, S.; Richter, R.M.; Joos, F.; Grieben, M.; et al. Molecular insights into lipid-assisted Ca2+ regulation of the TRP channel Polycystin-2. Nat. Struct. Mol. Biol. 2017, 24, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Hulse, R.E.; Li, Z.; Huang, R.K.; Zhang, J.; Clapham, D.E. Cryo-EM structure of the polycystin 2-l1 ion channel. eLife 2018, 7, e36931. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cao, E.; Julius, D.; Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 2016, 534, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Huynh, K.W.; Cohen, M.R.; Jiang, J.; Samanta, A.; Lodowski, D.T.; Zhou, Z.H.; Moiseenkova-Bell, V.Y. Structure of the full-length TRPV2 channel by cryo-EM. Nat. Commun. 2016, 7, 11130. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; McGoldrick, L.L.; Sobolevsky, A.I. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat. Struct. Mol. Biol. 2018, 25, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Paknejad, N.; Maksaev, G.; Sala-Rabanal, M.; Nichols, C.G.; Hite, R.K.; Yuan, P. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat. Struct. Mol. Biol. 2018, 25, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.E.T.; Pumroy, R.A.; Yazici, A.T.; Kasimova, M.A.; Fluck, E.C.; Huynh, K.W.; Samanta, A.; Molugu, S.K.; Zhou, Z.H.; Carnevale, V.; et al. Structural insights on TRPV5 gating by endogenous modulators. Nat. Commun. 2018, 9, 4198. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Saotome, K.; McGoldrick, L.L.; Sobolevsky, A.I. Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB. Nat. Commun. 2018, 9, 2465. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Choi, W.; Sun, W.; Du, J.; Lu, W. Structure of the human lipid-gated cation channel TRPC3. eLife 2018, 7, e36852. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Guo, W.; Zheng, L.; Wu, J.X.; Liu, M.; Zhou, X.; Zhang, X.; Chen, L. Structure of the receptor-activated human TRPC6 and TRPC3 ion channels. Cell Res. 2018, 28, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Li, J.; Zeng, B.; Chen, G.L.; Peng, X.; Zhang, Y.; Wang, J.; Clapham, D.E.; Li, Z.; Zhang, J. Structure of the mouse TRPC4 ion channel. Nat. Commun. 2018, 9, 3102. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Li, J.; Chen, G.-L.; Zeng, B.; Clapham, D.; Li, Z.; Zhang, J. Cryo-EM structure of the receptor-activated TRPC5 ion channel at 2.9 angstrom resolution. bioRxiv 2018, 467969. [Google Scholar] [CrossRef]
- Azumaya, C.M.; Sierra-Valdez, F.; Cordero-Morales, J.F.; Nakagawa, T. Cryo-EM structure of the cytoplasmic domain of murine transient receptor potential cation channel subfamily C member 6 (TRPC6). J. Biol. Chem. 2018, 293, 10381–10391. [Google Scholar] [CrossRef] [PubMed]
- Schmiege, P.; Fine, M.; Blobel, G.; Li, X. Human TRPML1 channel structures in open and closed conformations. Nature 2017, 550, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, M.; Herzik, M.A.; Wie, J.; Suo, Y.; Borschel, W.F.; Ren, D.; Lander, G.C.; Lee, S.Y. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature 2017, 550, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tóth, B.; Szollosi, A.; Chen, J.; Csanády, L. Structure of a TRPM2 channel in complex with Ca2+explains unique gating regulation. eLife 2018, 7, e36409. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; She, J.; Zeng, W.; Chen, Q.; Bai, X.C.; Jiang, Y. Structures of the calcium-activated, non-selective cation channel TRPM4. Nature 2017, 552, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Li, Z.; Li, J.; Hulse, R.E.; Santa-Cruz, A.; Valinsky, W.C.; Abiria, S.A.; Krapivinsky, G.; Zhang, J.; Clapham, D.E. Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Proc. Natl. Acad. Sci. USA 2018, 115, E8201–E8210. [Google Scholar] [CrossRef]
- Yin, Y.; Wu, M.; Zubcevic, L.; Borschel, W.F.; Lander, G.C.; Lee, S.-Y. Structure of the cold- and menthol-sensing ion channel TRPM8. Science 2018, 359, 237–241. [Google Scholar] [CrossRef]
- Madej, M.G.; Ziegler, C.M. Dawning of a new era in TRP channel structural biology by cryo-electron microscopy. Pflug. Arch. Eur. J. Physiol. 2018, 470, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, V.; Rohacs, T. TRPV1: A target for rational drug design. Pharmaceuticals 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Kol, S.; Braun, C.; Thiel, G.; Doyle, D.A.; Sundström, M.; Gourdon, P.; Nissen, P. Heterologous expression and purification of an active human TRPV3 ion channel. FEBS J. 2013, 280, 6010–6021. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6, 160196. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Shin, K.; Patterson, R.E.; Liu, X.-Q.; Rainey, J.K. Current strategies for protein production and purification enabling membrane protein structural biology. Biochem. Cell Biol. 2016, 94, 507–527. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.R.; Byregowda, S.M.; Veeregowda, B.M.; Vinayagamurthy, B. An Overview of Heterologous Expression Host Systems for the Production of Recombinant Proteins. Adv. Anim. Vet. Sci. 2016, 52, 85–105. [Google Scholar] [CrossRef]
- Liu, Z.; Tyo, K.E.J.; Martínez, J.L.; Petranovic, D.; Nielsen, J. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Tyo, K.E.J.; Liu, Z.; Petranovic, D.; Nielsen, J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 491–510. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Ling, J.X.; Chen, M.; Johnson, R.D.; Tominaga, M.; Wang, C.Y.; Gu, J. TRPM8 mechanism of autonomic nerve response to cold in respiratory airway. Mol. Pain 2008, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Barritt, G.J. Evidence that TRPM8 is an androgen-dependent Ca2+channel required for the survival of prostate cancer cells. Cancer Res. 2004, 64, 8365–8373. [Google Scholar] [CrossRef] [PubMed]
- Bahk, J.D.; Kioka, N.; Sakai, H.; Komano, T. A runaway-replication plasmid pSY343 contains two ssi signals. Plasmid 1988, 20, 266–270. [Google Scholar] [CrossRef]
- Cesareni, G.; Murray, J.A.H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. In Genetic Engineering; Springer: Boston, MA, USA, 1987; pp. 135–154. [Google Scholar]
- Pedersen, P.A.; Rasmussen, J.H.; Jørgensen, P.L. Expression in high yield of pig α1β1 Na,K-ATPase and inactive mutants D369N and D807N in Saccharomyces cerevisiae. J. Biol. Chem. 1996. [Google Scholar] [CrossRef]
- Sherman, F. Getting Started with Yeast. Methods Enzymol. 1991, 194, 3–21. [Google Scholar] [CrossRef]
- Scharff-Poulsen, P.; Pedersen, P.A. Saccharomyces cerevisiae-Based Platform for Rapid Production and Evaluation of Eukaryotic Nutrient Transporters and Transceptors for Biochemical Studies and Crystallography. PLoS ONE 2013, 8, e76851. [Google Scholar] [CrossRef]
- Klaerke, D.A.; Tejada, M.L.A.; Christensen, V.G.; Lassen, M.; Pedersen, P.A.; Calloe, K. Reconstitution and Electrophysiological Characterization of Ion Channels in Lipid Bilayers. Curr. Protoc. Pharmacol. 2018, 81, e37. [Google Scholar] [CrossRef]
- Erhart, E.; Hollenberg, C.P. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J. Bacteriol. 1983, 156, 625–635. [Google Scholar] [CrossRef]
- Drew, D.; Newstead, S.; Sonoda, Y.; Kim, H.; von Heijne, G.; Iwata, S. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 2008, 3, 784–798. [Google Scholar] [CrossRef]
- Molbaek, K.; Scharff-Poulsen, P.; Helix-Nielsen, C.; Klaerke, D.A.; Pedersen, A.A. High yield purification of full-length functional hERG K+channels produced in Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 15. [Google Scholar] [CrossRef]
- Ulăreanu, R.; Chiriţoiu, G.; Cojocaru, F.; Deftu, A.; Ristoiu, V.; Stănică, L.; Mihăilescu, D.F.; Cucu, D. N-glycosylation of the transient receptor potential melastatin 8 channel is altered in pancreatic cancer cells. Tumor Biol. 2017, 39, 1010428317720940. [Google Scholar] [CrossRef]
- Tang, H.; Wang, S.; Wang, J.; Song, M.; Xu, M.; Zhang, M.; Shen, Y.; Hou, J.; Bao, X. N-hypermannose glycosylation disruption enhances recombinant protein production by regulating secretory pathway and cell wall integrity in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 25654. [Google Scholar] [CrossRef]
- Moiseenkova, V.Y.; Hellmich, H.L.; Christensen, B.N. Overexpression and purification of the vanilloid receptor in yeast (Saccharomyces cerevisiae). Biochem. Biophys. Res. Commun. 2003, 310, 196–201. [Google Scholar] [CrossRef]
- Bomholt, J.; Hélix-Nielsen, C.; Scharff-Poulsen, P.; Pedersen, P.A. Recombinant Production of Human Aquaporin-1 to an Exceptional High Membrane Density in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e56431. [Google Scholar] [CrossRef]
- Arachea, B.T.; Sun, Z.; Potente, N.; Malik, R.; Isailovic, D.; Viola, R.E. Detergent selection for enhanced extraction of membrane proteins. Protein Expr. Purif. 2012, 86, 12–20. [Google Scholar] [CrossRef]
- Kawate, T.; Gouaux, E. Fluorescence-Detection Size-Exclusion Chromatography for Precrystallization Screening of Integral Membrane Proteins. Structure 2006, 14, 673–681. [Google Scholar] [CrossRef]
- Geertsma, E.R.; Groeneveld, M.; Slotboom, D.-J.; Poolman, B. Quality control of overexpressed membrane proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 5722–5727. [Google Scholar] [CrossRef]
- Stetsenko, A.; Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 2017, 7, 197. [Google Scholar] [CrossRef]
- Almeida, J.G.; Preto, A.J.; Koukos, P.I.; Bonvin, A.M.J.J.; Moreira, I.S. Membrane proteins structures: A review on computational modeling tools. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2021–2039. [Google Scholar] [CrossRef]
- Audagnotto, M.; Dal Peraro, M. Protein post-translational modifications: In silico prediction tools and molecular modeling. Comput. Struct. Biotechnol. J. 2017, 15, 307–319. [Google Scholar] [CrossRef]
- Luby, C.J.; Coughlin, B.P.; MacE, C.R. Enrichment and Recovery of Mammalian Cells from Contaminated Cultures Using Aqueous Two-Phase Systems. Anal. Chem. 2018, 90, 2103–2110. [Google Scholar] [CrossRef]
- Bjørkskov, F.B.; Krabbe, S.L.; Nurup, C.N.; Missel, J.W.; Spulber, M.; Bomholt, J.; Molbaek, K.; Helix-Nielsen, C.; Gotfryd, K.; Gourdon, P.; et al. Purification and functional comparison of nine human Aquaporins produced in Saccharomyces cerevisiae for the purpose of biophysical characterization. Sci. Rep. 2017, 7, 1–21. [Google Scholar] [CrossRef]
- Gotfryd, K.; Mósca, A.F.; Missel, J.W.; Truelsen, S.F. Human adipose glycerol flux is regulated by a pH gate in AQP10. Nat. Commun. 2018, 9, 4749. [Google Scholar] [CrossRef]
- Edavettal, S.C.; Hunter, M.J.; Swanson, R.V. Genetic construct design and recombinant protein expression for structural biology. Methods Mol. Biol. 2012, 841, 29–47. [Google Scholar] [CrossRef]
- Lanza, A.M.; Curran, K.A.; Rey, L.G.; Alper, H.S. A condition-specific codon optimization approach for improved heterologous gene expression in Saccharomyces cerevisiae. BMC Syst. Biol. 2014, 8, 33. [Google Scholar] [CrossRef]
- Drew, D.; Lerch, M.; Kunji, E.; Slotboom, D.J.; de Gier, J.W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 2006, 3, 303–313. [Google Scholar] [CrossRef]
- Schultz, L.D.; Hofmann, K.J.; Mylin, L.M.; Montgomery, D.L.; Ellis, R.W.; Hopper, J.E. Regulated overproduction of the GAL4 gene product greatly increases expression from galactose-inducible promoters on multi-copy expression vectors in yeast. Gene 1987, 61, 123–133. [Google Scholar] [CrossRef]
- Wang, J.M.; Partoens, P.M.; Callebaut, D.P.; Coen, E.P.; Martin, J.J.; De Potter, W.P. Phenotype plasticity and immunocytochemical evidence for ChAT and DβH co-localization in fetal pig superior cervical ganglion cells. Dev. Brain Res. 1995, 90, 17–23. [Google Scholar] [CrossRef]
- Egan, T.J.; Acuna, M.A.; Zenobi-Wong, M.; Zeilhofer, H.U.; Urech, D. Effects of N-glycosylation of the human cation channel TRPA1 on agonist-sensitivity. Biosci. Rep. 2016. [Google Scholar] [CrossRef]
- Dietrich, A.; Mederos Y Schnitzler, M.; Emmel, J.; Kalwa, H.; Hofmann, T.; Gudermann, T. N-Linked Protein Glycosylation Is a Major Determinant for Basal TRPC3 and TRPC6 Channel Activity. J. Biol. Chem. 2003, 278, 47842–47852. [Google Scholar] [CrossRef]
- Pertusa, M.; Madrid, R.; Morenilla-Palao, C.; Belmonte, C.; Viana, F. N-glycosylation of TRPM8 ion channels modulates temperature sensitivity of cold thermoreceptor neurons. J. Biol. Chem. 2012, 287, 18218–18229. [Google Scholar] [CrossRef]
- Woo, S.K.; Kwon, M.S.; Ivanov, A.; Geng, Z.; Gerzanich, V.; Simard, J.M. Complex n-Glycosylation stabilizes surface expression of transient receptor potential melastatin 4b protein. J. Biol. Chem. 2013, 288, 36409–36417. [Google Scholar] [CrossRef]
- Dong, X.P.; Wang, X.; Xu, H. TRP channels of intracellular membranes. J. Neurochem. 2010, 113, 313–328. [Google Scholar] [CrossRef]
- Wen, W.; Que, K.; Zang, C.; Wen, J.; Sun, G.; Zhao, Z.; Li, Y. Expression and distribution of three transient receptor potential vanilloid(TRPV) channel proteins in human odontoblast-like cells. J. Mol. Histol. 2017, 48, 367–377. [Google Scholar] [CrossRef]
- Newstead, S.; Kim, H.; von Heijne, G.; Iwata, S.; Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2007, 104, 13936–13941. [Google Scholar] [CrossRef]
- Raddatz, N.; Castillo, J.P.; Gonzalez, C.; Alvarez, O.; Latorre, R. Temperature and voltage coupling to channel opening in transient receptor potential melastatin 8 (TRPM8). J. Biol. Chem. 2014, 289, 35438–35454. [Google Scholar] [CrossRef]
- Zakharian, E.; Thyagarajan, B.; French, R.J.; Pavlov, E.; Rohacs, T. Inorganic polyphosphate modulates TRPM8 channels. PLoS ONE 2009, 4, e5404. [Google Scholar] [CrossRef]
| Target | Organism | Expression System | Full Length | Truncation | Structure Determination Method | Resolution Å |
|---|---|---|---|---|---|---|
| TRPC3 | Homo sapiens | Homo sapiens (AD-HEK293) | x | cryo-EM | 4.4 | |
| TRPC4 | Mus musculus | Spodoptera frugiperda | x | cryo-EM | 3.3 | |
| TRPC5 | Mus musculus | Homo sapiens (HEK293S) | x | cryo-EM | 2.9 | |
| TRPC6 | Mus musculus | Spodoptera frugiperda | x | cryo-EM | 3.8 | |
| TRPC6 | Homo sapiens | Homo sapiens (AD-HEK293) | x | cryo-EM | 3.8 | |
| TRPV1 | Rattus norvegicus | Homo sapiens (HEK293S) | minimal function | cryo-EM | 4.2 | |
| TRPV2 | Rattus norvegicus | Saccharomyces cerevisiae | x | cryo-EM | 4.4 | |
| TRPV2 | Oryctolagus cuniculus | Spodoptera frugiperda | x | cryo-EM | 3.8 | |
| TRPV2 | Oryctolagus cuniculus | Spodoptera frugiperda | minimal function | X-ray | 3.9 | |
| TRPV3 | Mus musculus | Homo sapiens (HEK293S) | x | cryo-EM | 4.3 | |
| TRPV4 | Xenopus tropicalis | Pichia pastoris | x | cryo-EM | 3.8 | |
| TRPV5 | Oryctolagus cuniculus | Saccharomyces cerevisiae | x | cryo-EM | 3.9 | |
| TRPV6 | Rattus norvegicus | Homo sapiens (HEK293S) | x | X-ray | 3.2 | |
| TRPV6 | Homo sapiens | Homo sapiens (HEK293S) | x | cryo-EM | 3.6 | |
| TRPML1 | Homo sapiens | Homo sapiens (HEK293S) | x | cryo-EM | 3.7 | |
| TRPML3 | Callithrix jacchus | Spodoptera frugiperda | x | cryo-EM | 2.9 | |
| TRPM2 | Danio rerio | Homo sapiens (HEK293S) | x | cryo-EM | 3.3 | |
| TRPM4 | Homo sapiens | Homo sapiens (HEK293F) | x | cryo-EM | 3.2 | |
| TRPM7 | Mus musculus | Homo sapiens (HEK293S) | x | cryo-EM | 3.3 | |
| TRPM8 | Ficedula albicollis | Homo sapiens (HEK293S) | x | cryo-EM | 4.1 | |
| TRPA1 | Homo sapiens | Homo sapiens (HEK293) | x | cryo-EM | 4.2 | |
| TRPP2 | Homo sapiens | Homo sapiens (HEK293S) | x | cryo-EM | 4.2 | |
| TRPP3 | Homo sapiens | Homo sapiens (HEK293S) | x | cryo-EM | 3.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Wang, K.; Klaerke, D.A.; Calloe, K.; Lowrey, L.; Pedersen, P.A.; Gourdon, P.; Gotfryd, K. Purification of Functional Human TRP Channels Recombinantly Produced in Yeast. Cells 2019, 8, 148. https://doi.org/10.3390/cells8020148
Zhang L, Wang K, Klaerke DA, Calloe K, Lowrey L, Pedersen PA, Gourdon P, Gotfryd K. Purification of Functional Human TRP Channels Recombinantly Produced in Yeast. Cells. 2019; 8(2):148. https://doi.org/10.3390/cells8020148
Chicago/Turabian StyleZhang, Liying, Kaituo Wang, Dan Arne Klaerke, Kirstine Calloe, Lillian Lowrey, Per Amstrup Pedersen, Pontus Gourdon, and Kamil Gotfryd. 2019. "Purification of Functional Human TRP Channels Recombinantly Produced in Yeast" Cells 8, no. 2: 148. https://doi.org/10.3390/cells8020148
APA StyleZhang, L., Wang, K., Klaerke, D. A., Calloe, K., Lowrey, L., Pedersen, P. A., Gourdon, P., & Gotfryd, K. (2019). Purification of Functional Human TRP Channels Recombinantly Produced in Yeast. Cells, 8(2), 148. https://doi.org/10.3390/cells8020148






