Integration of Transcriptome, Proteome, and Metabolome Provides Insights into How Calcium Enhances the Mechanical Strength of Herbaceous Peony Inflorescence Stems
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatment
2.2. Morphological Indices and Photosynthetic Characteristics Measurements
2.3. Transcriptome with RNA-Seq and Data Analysis
2.4. Proteome with ITRAQ and Data Analysis
2.5. mRNA and Protein Correlation Analysis
2.6. Gene Expression Analysis with Quantitative Real-Time PCR (qRT-PCR)
2.7. Metabolome with LC-MS/MS and Data Analysis
2.8. Microstructures Observation and Histological Staining
2.9. Cell Wall Compositions Analysis
2.10. Statistical Analysis
3. Results
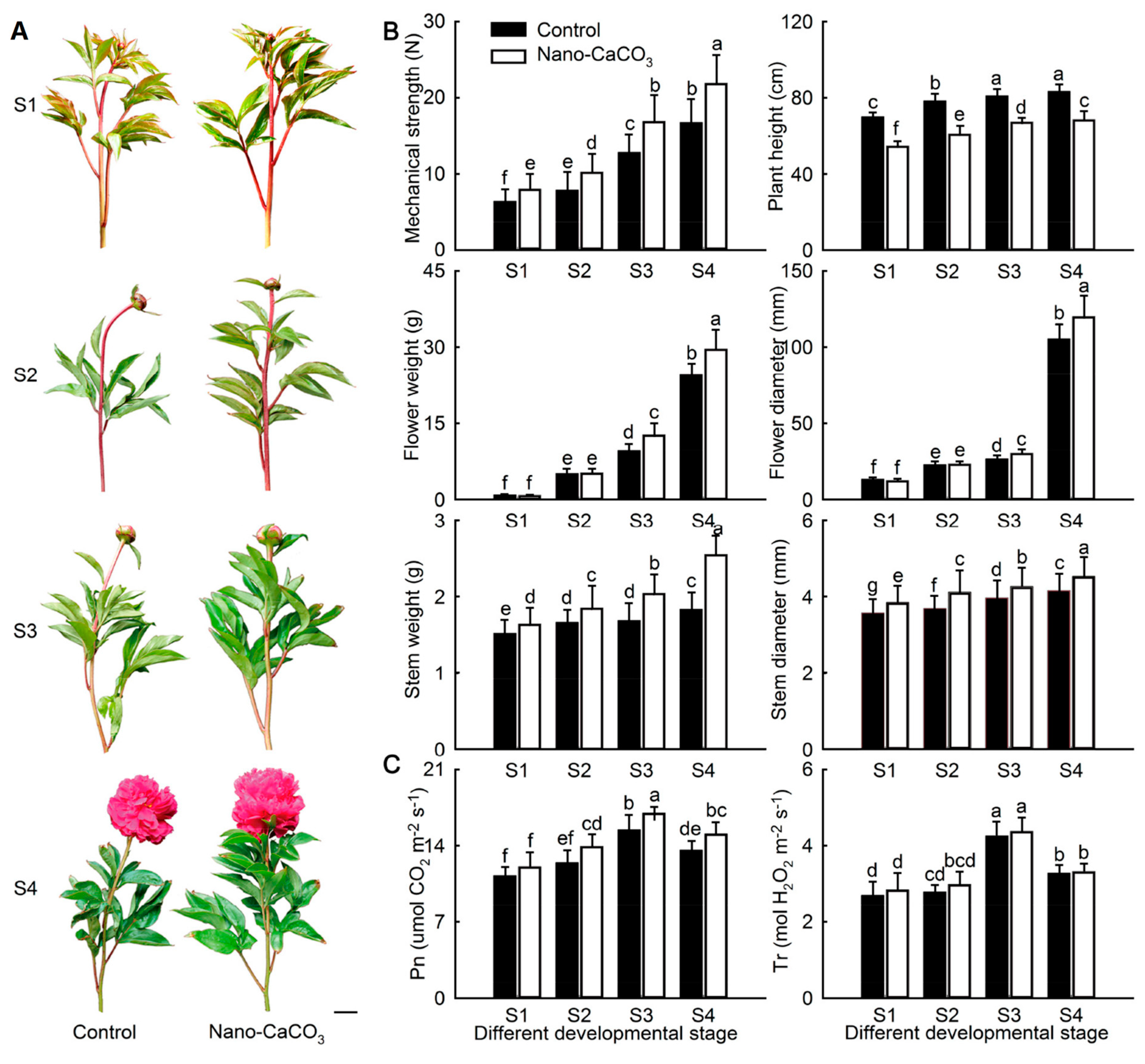
3.1. Morphological Indices and Photosynthetic Characteristics
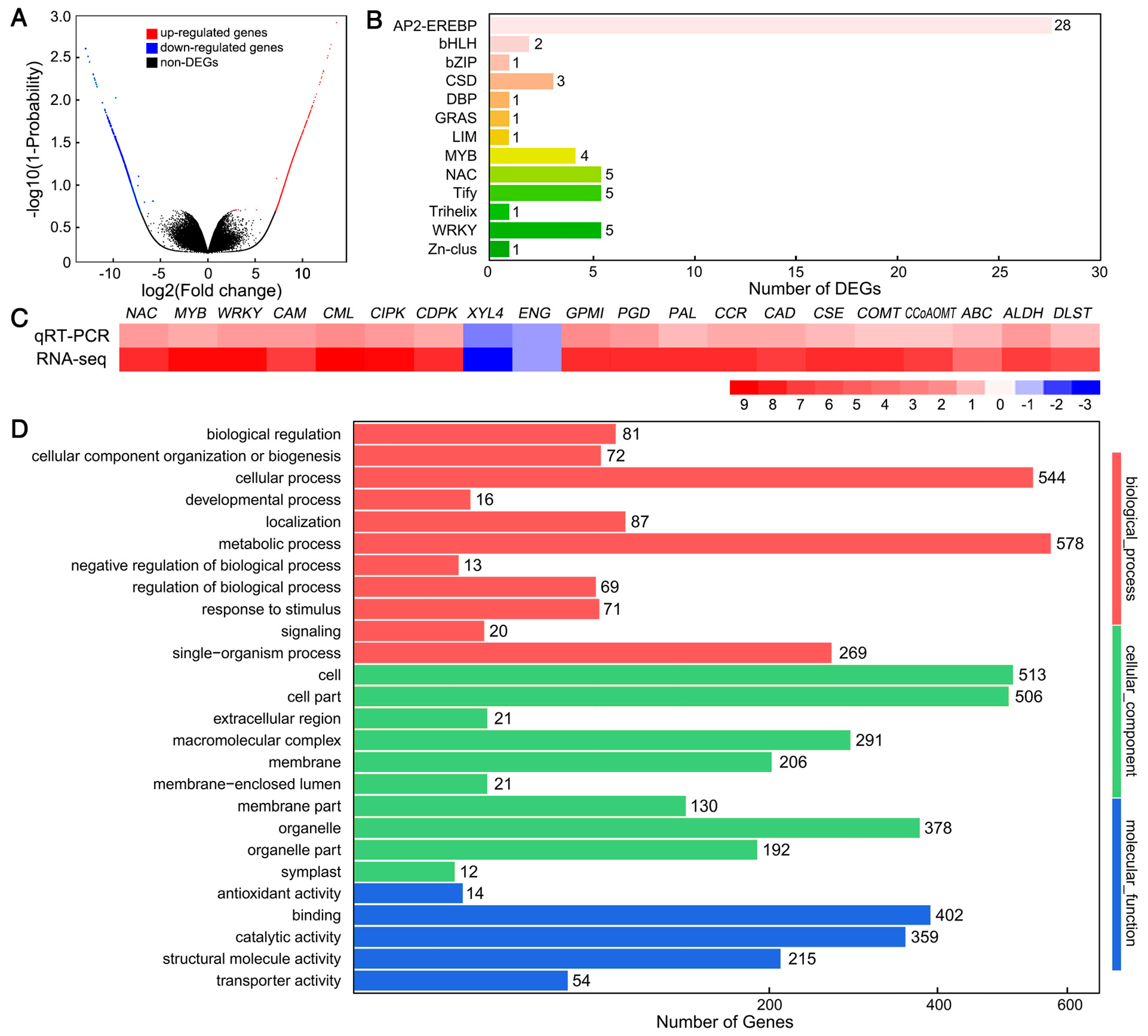
3.2. Transcriptome Analysis
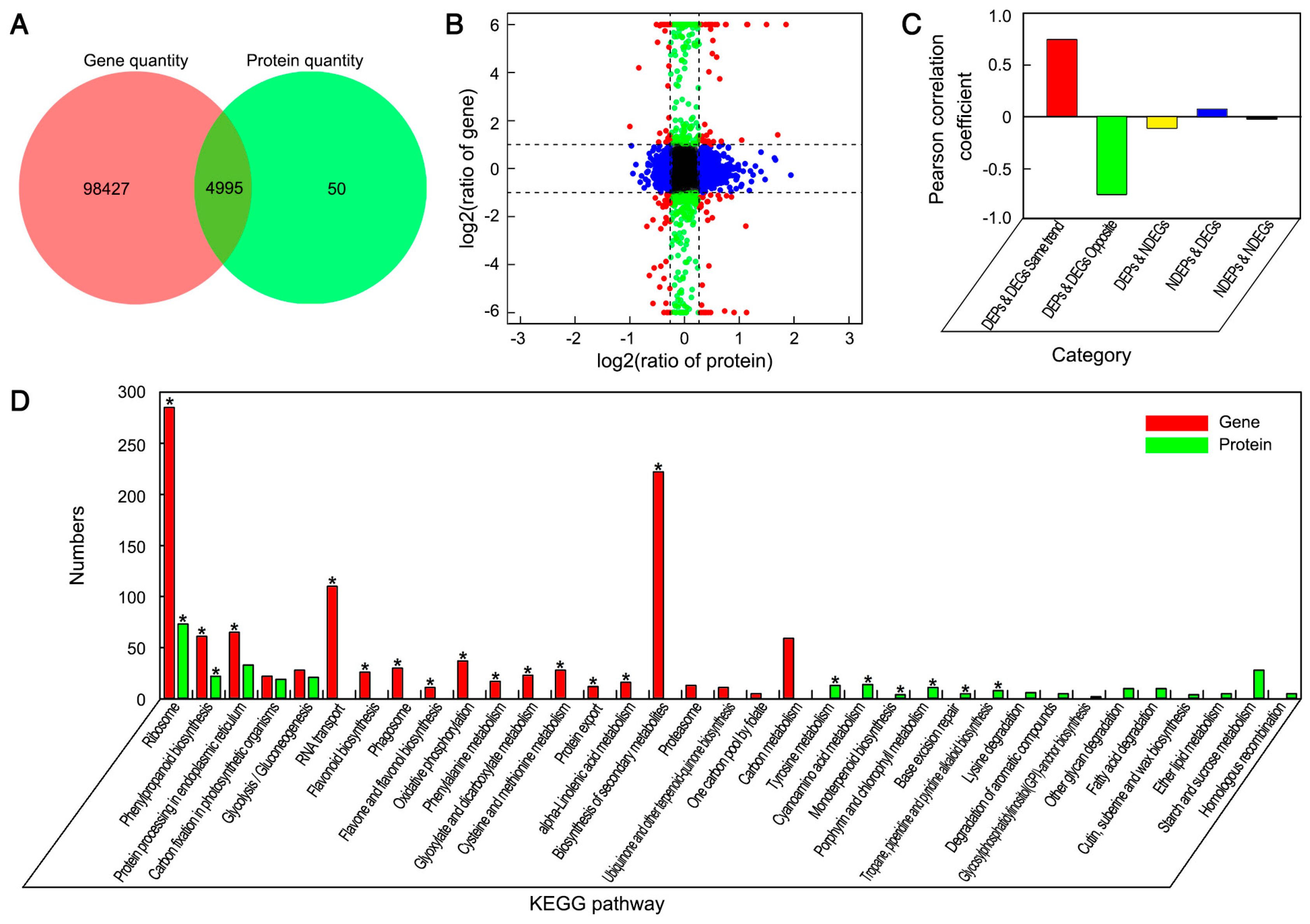
3.3. Integrative Analysis of Proteome and Transcriptome
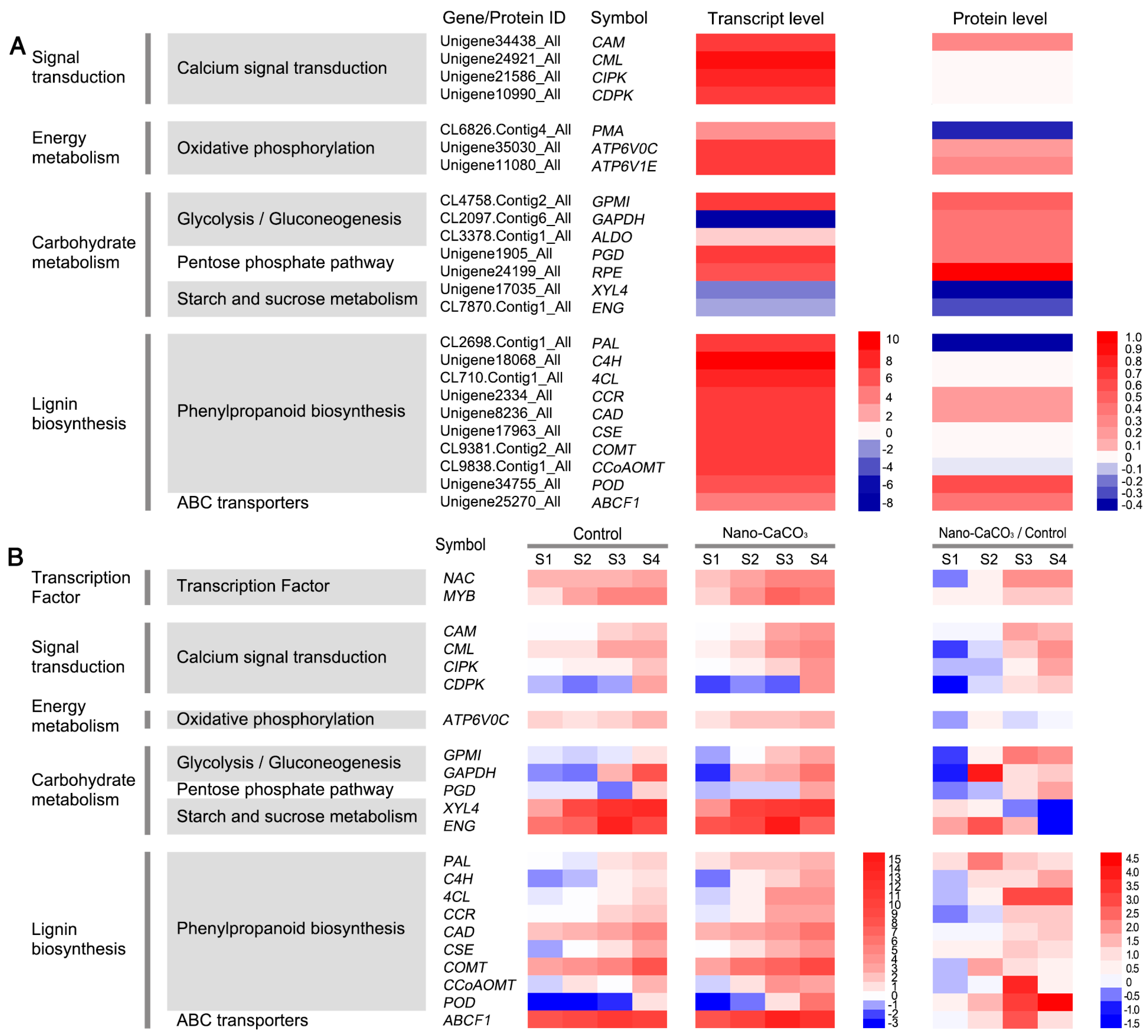
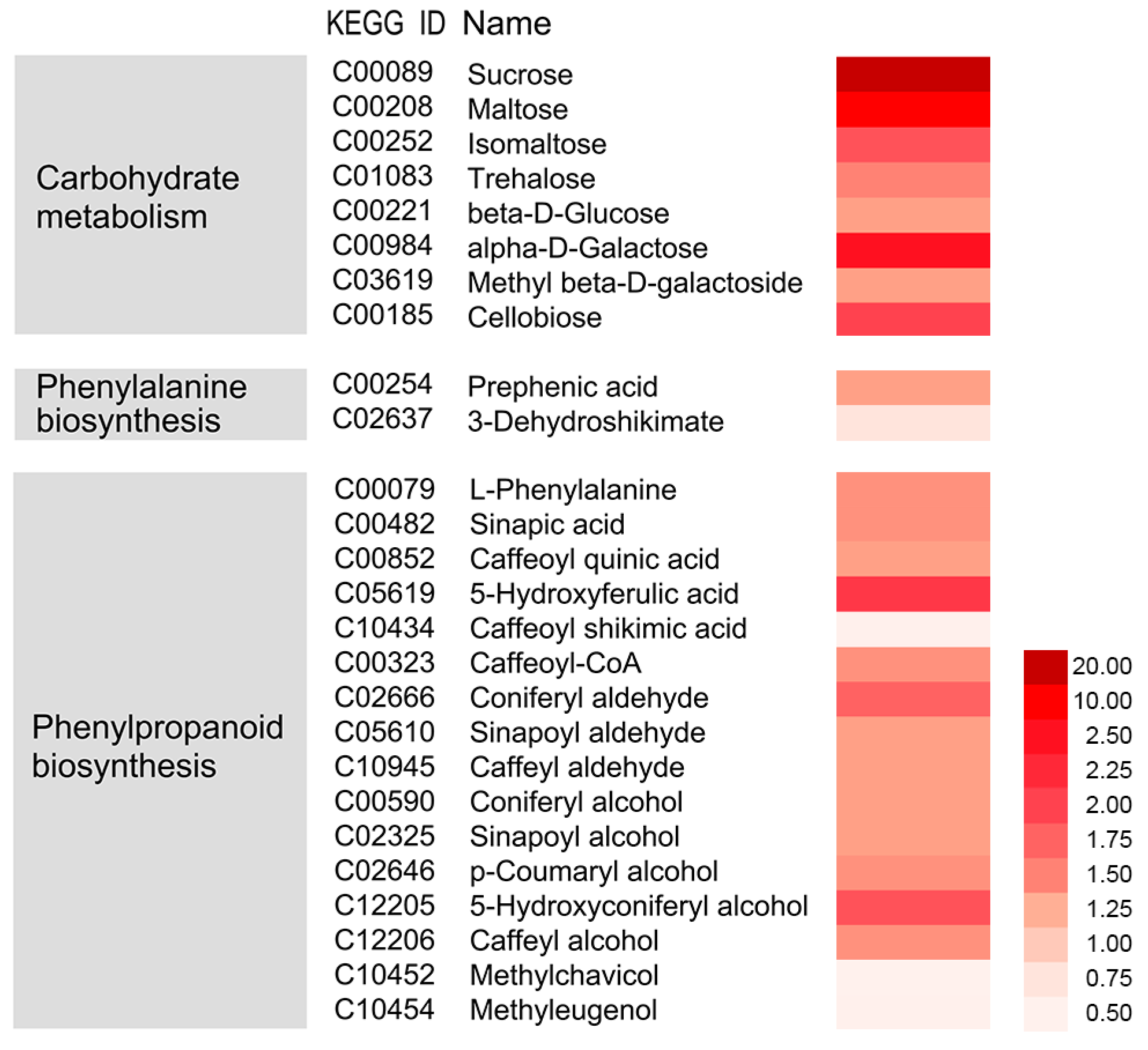
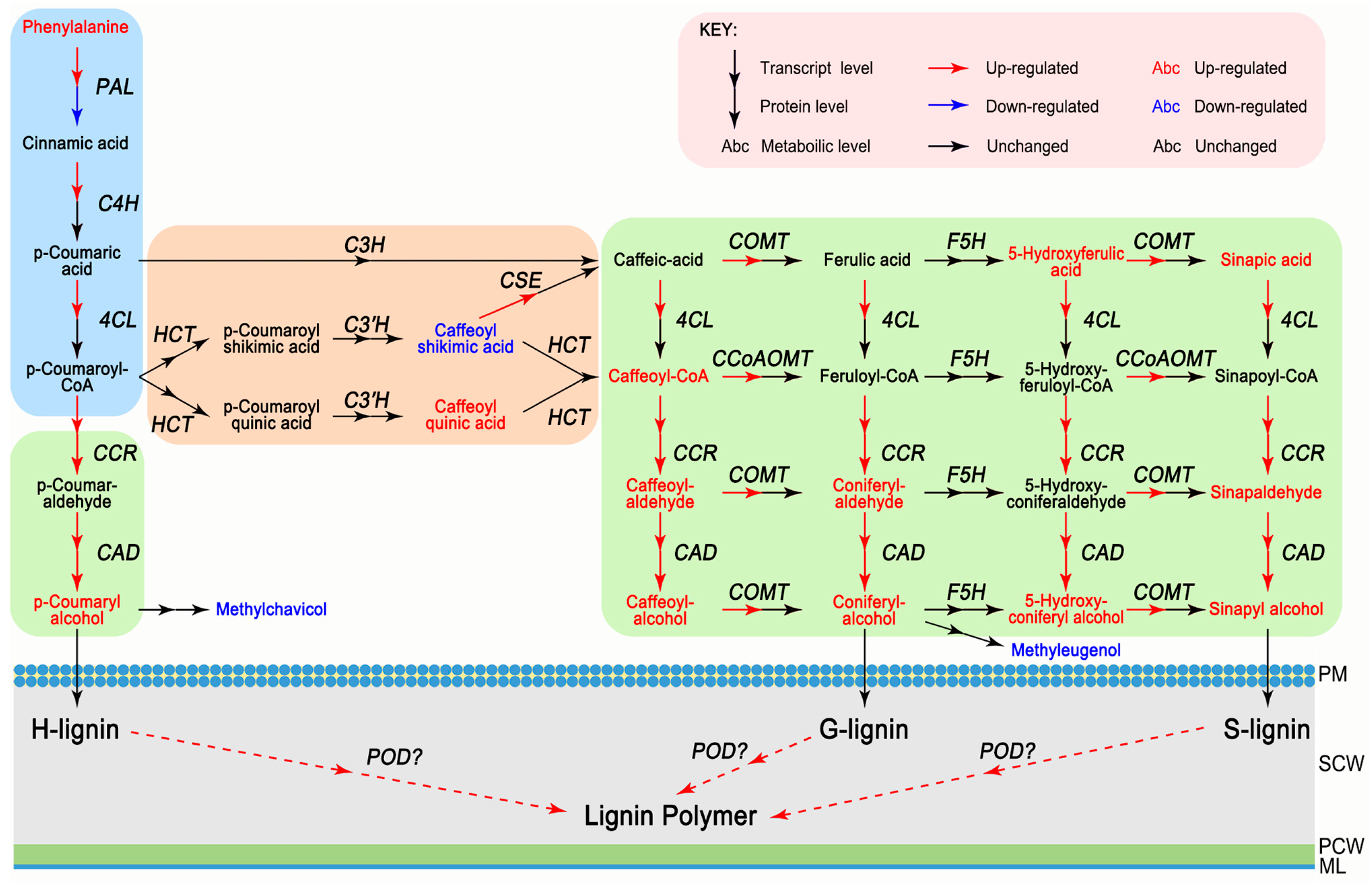
3.4. Integrative Analysis of Metabolome, Proteome, and Transcriptome
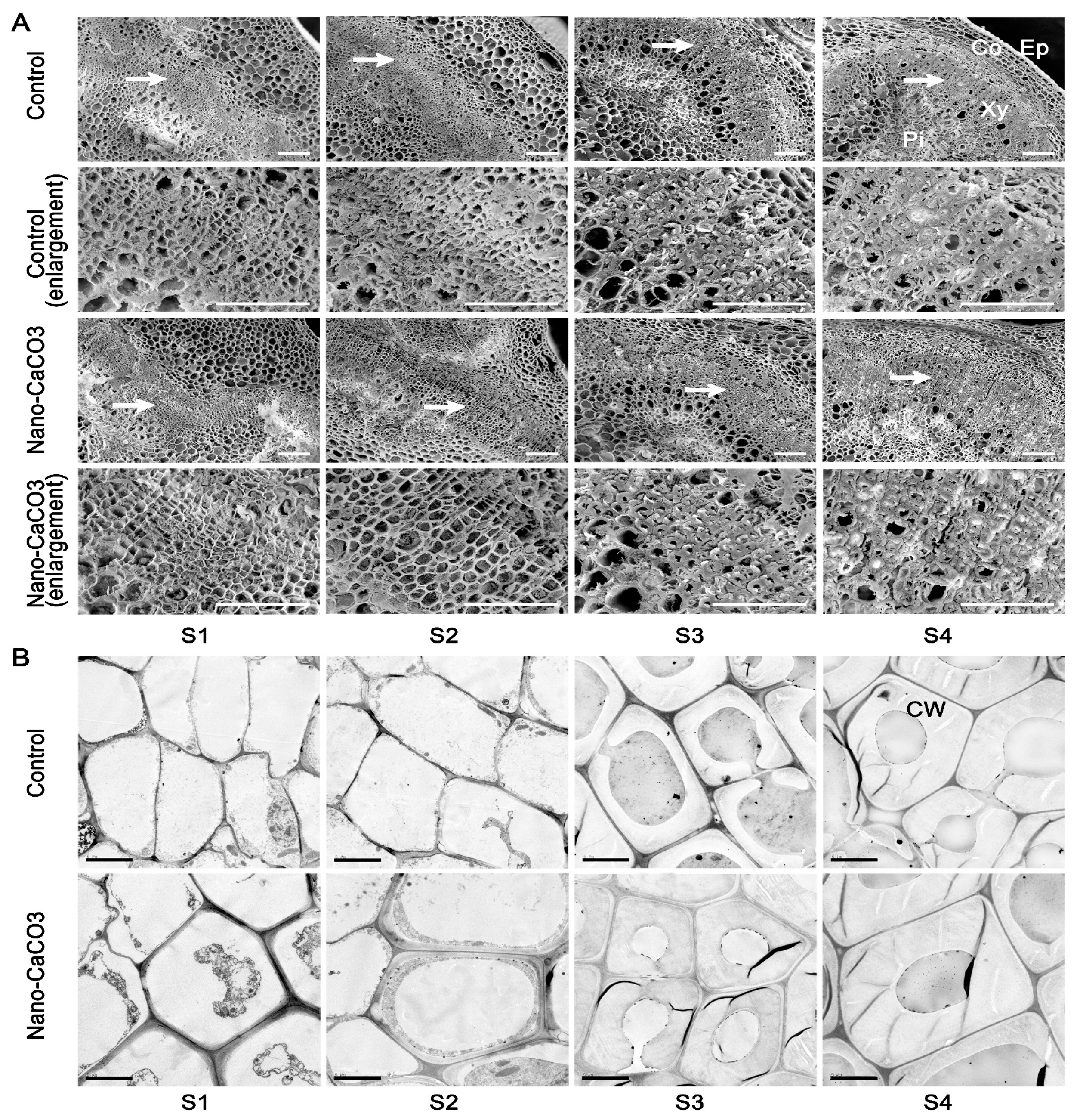
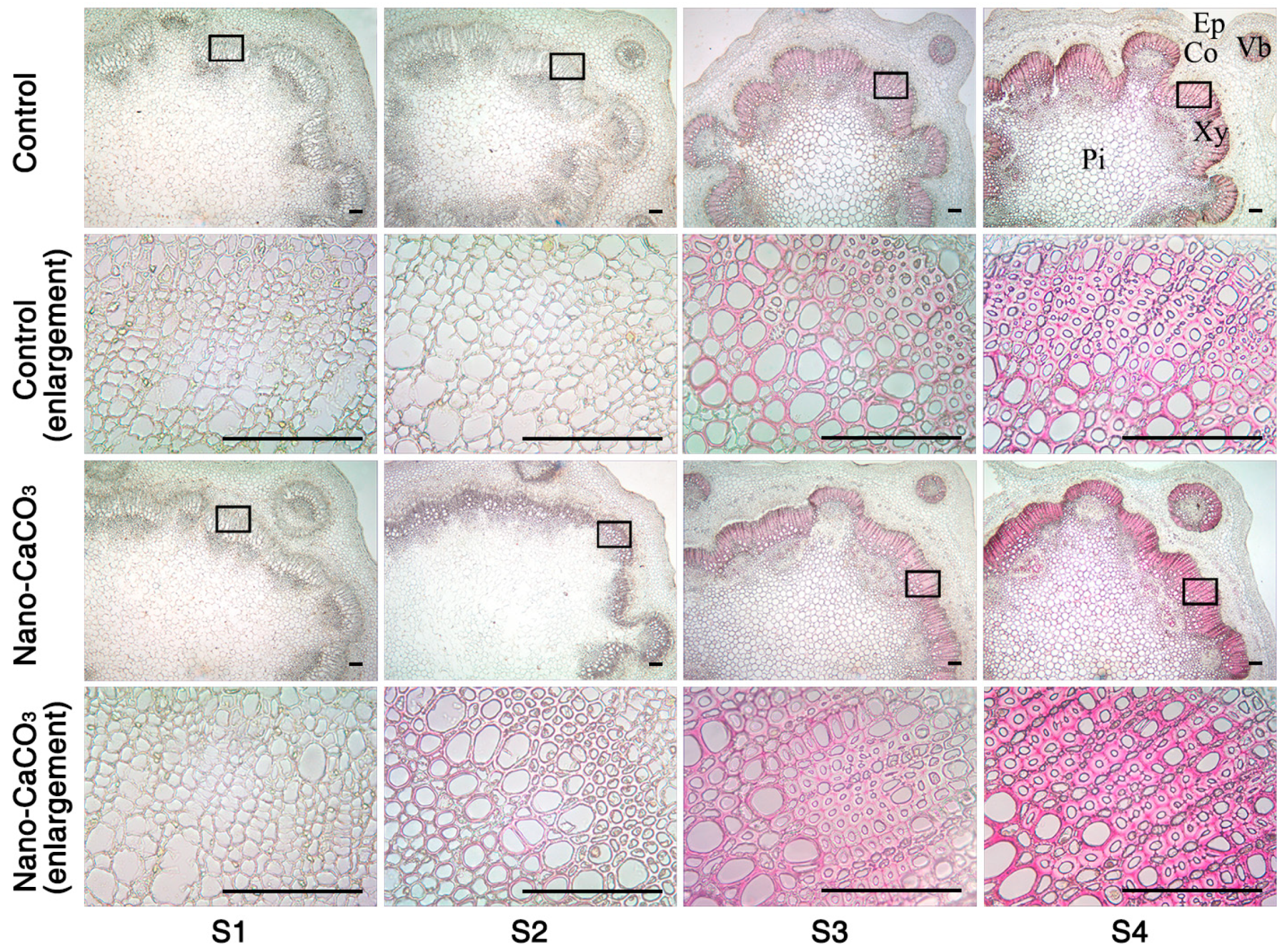
3.5. Microstructures
3.6. Cell Wall Compositions
3.7. Histochemical Staining
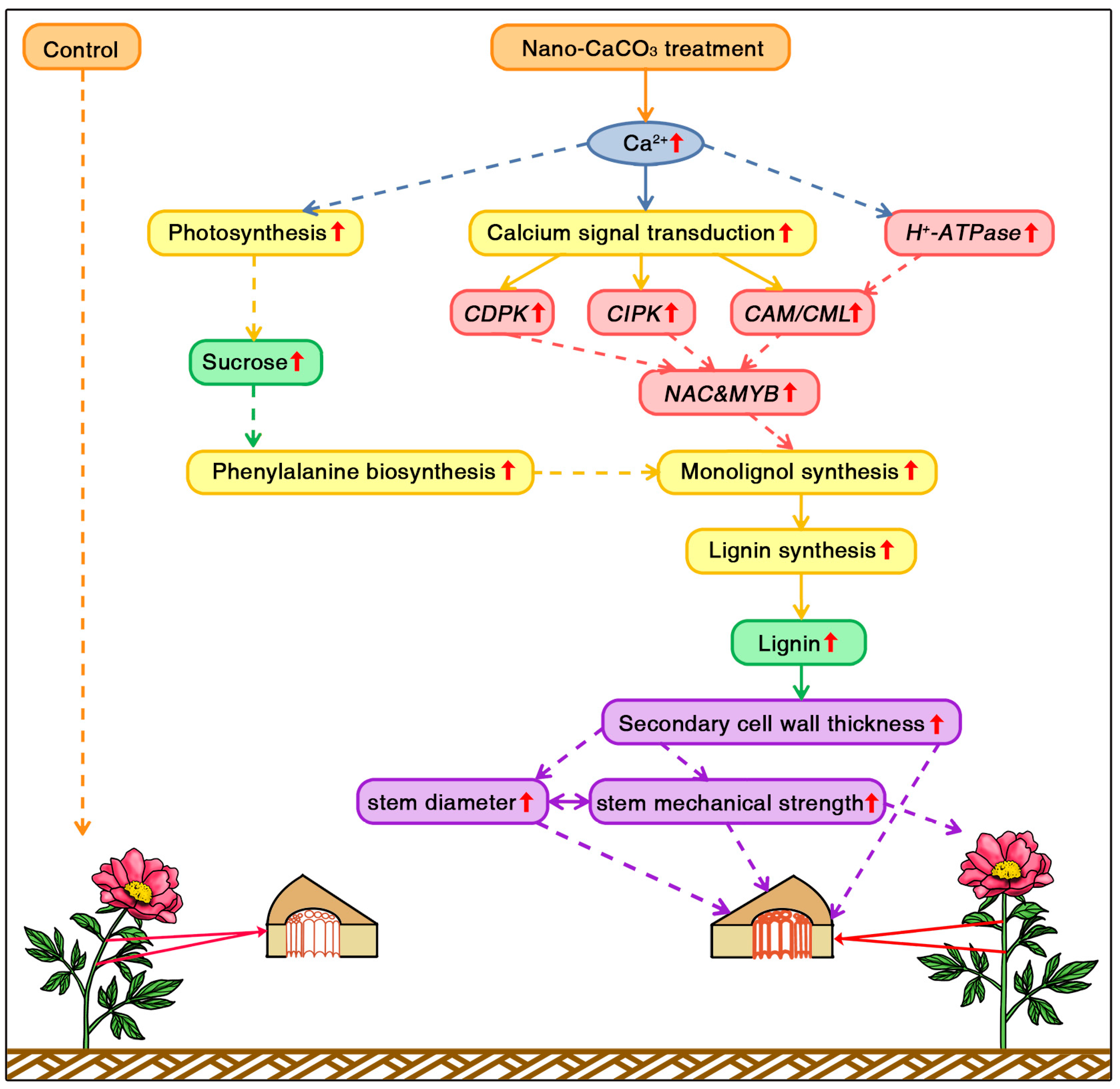
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wang, L.; Zheng, L.; Yu, X. Studies on the application and flora market trend of cut peonies in China and overseas. Heilongjiang Agric. Sci. 2011, 2, 147–149. (In Chinese) [Google Scholar]
- Li, X.J.; Yang, Y.; Yao, J.L.; Chen, G.X.; Li, X.H.; Zhang, Q.F.; Wu, C.Y. FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol. Biol. 2009, 69, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Hepler, P.K.; Winship, L. Calcium at the cell wall-cytoplast Interface. J. Integr. Plant Biol. 2010, 52, 147–160. [Google Scholar] [CrossRef]
- Hepler, P.K. Calcium: A central regulator of plant growth and development. Plant Cell 2005, 17, 2142–2155. [Google Scholar] [CrossRef] [PubMed]
- Arfaoui, A.; Hadrami, A.E.; Adam, L.R.; Daayf, F. Pre-treatment with calcium enhanced defense-related genes’ expression in the soybean’s isoflavones pathway in response to Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 2016, 93, 12–21. [Google Scholar] [CrossRef]
- Michailidis, M.; Karagiannis, E.; Tanou, G.; Karamanoli, K.; Lazaridou, A.; Matsi, T.; Molassiotis, A. Metabolomic and physico-chemical approach unravel dynamic regulation of calcium in sweet cherry fruit physiology. Plant Physiol. Biochem. 2017, 116, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Shams, M.; Etemadi, N.; Baninasab, B.; Ramin, A.A.; Khoshgoftarmanesh, A.H. Effect of boron and calclum on growth and quality of ‘Easy lover’ cut rose. J. Plant Nutr. 2012, 35, 1303–1313. [Google Scholar] [CrossRef]
- Lee, C.H.; Nam, M.K. Enhancement of stem firmness in standard chrysanthemum ‘Baekma’ by foliar spray of liquid calcium compounds. Korean J. Hortic. Sci. Technol. 2011, 29, 298–305. [Google Scholar]
- Perik, R.R.J.; Razé, D.; Ferrante, A.; Doorn, W.G.V. Stem bending in cut Gerbera jamesonii flowers: Effects of a pulse treatment with sucrose and calcium ions. Postharvest Biol. Technol. 2014, 98, 7–13. [Google Scholar] [CrossRef]
- Li, C.Z.; Tao, J.; Zhao, D.Q.; You, C.; Ge, J.T. Effect of calcium sprays on mechanical strength and cell wall fractions of herbaceous peony (Paeonia lactiflora Pall.) inflorescence stems. Int. J. Mol. Sci. 2012, 13, 4704–4713. [Google Scholar] [CrossRef]
- Zhang, J. Effect of Calcium and Calmodulin on Neck-Bending of Gerbera Cut-Flower. Master’s Thesis, Fujian Agriculture Forest University, Fuzhou, China, 2008. (In Chinese). [Google Scholar]
- Matschi, S.; Werner, S.; Schulze, W.X.; Legen, J.; Hilger, H.H.; Romeis, T. Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 2013, 73, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Pareek, C.S. Transcriptome analysis on RNA-seq data. J. Next Gener. Seq. Appl. 2015, S1, 1–2. [Google Scholar]
- Yeung, E.S. Genome-wide correlation between mRNA and protein in a single cell. Angew. Chem. Int. Ed. 2011, 50, 583–585. [Google Scholar] [CrossRef] [PubMed]
- Langley, S.R.; Dwyer, J.; Drozdov, I.; Yin, X.; Mayr, M. Proteomics: From single molecules to biological pathways. Cardiovasc. Res. 2013, 97, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.Q.; Han, C.X.; Tao, J.; Wang, J.; Hao, Z.J.; Geng, Q.P.; Du, B. Effects of inflorescence stem structure and cell wall components on the mechanical strength of inflorescence stem in herbaceous peony. Int. J. Mol. Sci. 2012, 13, 4993–5009. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaefer, L.; Wold, B. Lorian schaeffer and barbara wold mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Gong, S.J.; Hao, Z.J.; Meng, J.S.; Tao, J. Quantitative proteomics analysis of herbaceous peony in response to paclobutrazol inhibition of lateral branching. Int. J. Mol. Sci. 2015, 16, 24332–24352. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Tao, J.; Han, C.X.; Ge, J.T. Actin as an alternative internal control gene for gene expression analysis in herbaceous peony (Paeonia lactiflora Pall.). Afr. J. Agric. Res. 2012, 7, 2153–2159. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zhao, D.Q.; Wang, R.; Meng, J.S.; Li, Z.Y.; Wu, Y.Q.; Tao, J. Ameliorative effects of melatonin on dark-induced leaf senescence in gardenia (Gardenia jasminoides Ellis): Leaf morphology, anatomy, physiology and transcriptome. Sci. Rep. 2017, 7, 10423. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Z.; Tao, J.; Sun, Y.; Kong, F.; Geng, Q.P.; Du, B. Effects of spraying calcium on the inflorescence stem quality and leaf photosynthesis of herbaceous peony (Paeonia lactiflora Pall.). Chin. J. Ecol. 2012, 31, 2817–2822. (In Chinese) [Google Scholar]
- Kontturi, E.J. Surface Chemistry of Cellulose: From Natural Fibres to Model Surfaces. Ph.D. Thesis, Eindhoven University Technology, Eindhoven, Germany, 2005. [Google Scholar]
- Wang, J.; Zhu, J.; Lin, Q.; Li, X.; Teng, N.; Li, Z.; Li, B.; Zhang, A.; Lin, J. Effects of stem structure and cell wall components on bending strength in wheat. Chin. Sci. Bull. 2006, 51, 815–823. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Morrison, W.H.; Himmelsbach, D.S.; Poole, F.L.; Ye, Z. Essential role of caffeoyl coenzyme a O-methyltransferase in lignin biosynthesis in woody poplar plants. Plant Physiol. 2000, 124, 563–577. [Google Scholar] [CrossRef]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Bieniek, A. Mineral composition of fruit of actinidia arguta and actinidia purpurea and some of their hybrid cultivars grown in northeastern Poland. Pol. J. Environ. Stud. 2012, 21, 1543–1550. [Google Scholar]
- Li, Z.; Tan, X.F.; Lu, K.; Liu, Z.M.; Wu, L.L. The effect of CaCl2 on calcium content, photosynthesis, and chlorophyll fluorescence of tung tree seedlings under drought conditions. Photosynthetica 2017, 55, 553–560. [Google Scholar] [CrossRef]
- Islam, M.Z.; Mele, M.A.; Baek, J.P.; Kang, H.M. Cherry tomato qualities affected by foliar spraying with boron and calcium. Hortic. Environ. Biotechnol. 2016, 57, 46–52. [Google Scholar] [CrossRef]
- Kayal, W.E.; Paliyath, G.; Sullivan, J.A.; Subramanian, J. Phospholipase D inhibition by hexanal is associated with calcium signal transduction events in raspberry. Hortic. Res. 2017, 4, 17042. [Google Scholar] [CrossRef]
- Bakeer, S.M. Effect of ammonium nitrate fertilizer and calcium chloride foliar spray on fruit cracking and sunburn of Manfalouty pomegranate trees. Sci. Hortic. 2016, 209, 300–308. [Google Scholar] [CrossRef]
- Bivi, M.S.H.R.; Paiko, A.S.; Khairulmazmi, A.; Akhtar, M.S.; Idris, A.S. Control of basal stem rot disease in oil palm by supplementation of calcium, copper, and salicylic acid. Plant Pathol. J. 2016, 32, 396–406. [Google Scholar] [CrossRef]
- Yu, X.N.; Lu, G.P.; Cheng, F.Y.; Zheng, L.W. Effect of calcium on the stem quality of cut herbaceous peony. J. Hunan Agric. Univ. 2010, 36, 531–535. (In Chinese) [Google Scholar] [CrossRef]
- Chen, G.H.; Deng, H.B.; Zhang, G.L.; Tang, W.B.; Huang, H. The correlation of stem characters and lodging resistance and combining ability analysis in rice. Sci. Agric. Sin. 2016, 49, 407–417. [Google Scholar]
- Sreeja, R.; Balaji, S.; Arul, L.; Kumari, A.N.; Bapu, J.R.K.; Subramanian, A. Association of lignin and Flexible Culm 1 (FC1) ortholog in imparting culm strength and lodging resistance in kodo millet (Paspalum scrobiculatum L.). Mol. Breed. 2016, 36, 149. [Google Scholar] [CrossRef]
- Speck, T.; Burgert, I. Plant stems: Functional design and mechanics. Annu. Rev. Mat. Res. 2011, 41, 169–193. [Google Scholar] [CrossRef]
- Rojas, V.; Ulacio, D.; Elena Sanabria, M.; Auxiliadora, J.M. Effect of calcium, trichoderma and broccoli on cell wall thickness and cellular area of garlic, to control white rot. Bol. Centro Investig. Biol. 2009, 43, 183–195. [Google Scholar]
- Aohara, T.; Kotake, T.; Kaneko, Y.; Takatsuji, H.; Tsumuraya, Y.; Kawasaki, S. Rice BRITTLE CULM 5 (BRITTLE NODE) is involved in secondary cell wall formation in the sclerenchyma tissue of nodes. Plant Cell Physiol. 2009, 50, 1886–1897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, B.C.; Qian, Q.; Yu, Y.C.; Li, R.; Zhang, J.W.; Liu, X.L.; Zeng, D.L.; Li, J.Y.; Zhou, Y.H. Brittle Culm 12, a dual–targeting kinesin–4 protein, controls cell-cycle progression and wall properties in rice. Plant J. 2010, 63, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Faraji, M.; Fonseca, L.L.; Escamilla-Treviño, L.; Barros-Rios, J.; Engle, N.; Yang, Z.K.; Tschaplinski, T.J.; Dixon, R.A.; Voit, E.O. Mathematical models of lignin biosynthesis. Biotechnol. Biofuels 2018, 11, 34. [Google Scholar] [CrossRef]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Blee, K.A.; Choi, J.W.; O’Connell, A.P.; Schuch, W.; Lewis, N.G.; Bolwell, G.P. A lignin-specific peroxidase in tobacco whose antisense suppression leads to vascular tissue modification. Phytochemistry 2003, 64, 163–176. [Google Scholar] [CrossRef]
- Lv, G.S.; Tang, D.J.; Chen, F.D.; Sun, Y.; Fang, W.M.; Guan, Z.Y.; Liu, Z.L.; Chen, S.M. The anatomy and physiology of spray cut chrysanthemum pedicels, and expression of a caffeic acid 3–O–methyltransferase homologue. Postharvest Biol. Technol. 2011, 60, 244–250. [Google Scholar] [CrossRef]
- Li, Y.; Liu, G.B.; Li, J.; You, Y.L.; Zhao, H.M.; Liang, H.; Mao, P.S. Acid detergent lignin, lodging resistance index, and expression of the caffeic acid O-methyltransferase gene in brown midrib-12 sudangrass. Jpn. J. Breed. 2015, 65, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Choi, H.; An, G. Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 2015, 57, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Ma, Q.J.; Sun, C.H.; Sun, M.H.; You, C.X.; Hao, Y.J. Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 2016, 156, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Lee, C.H.; Ye, Z.H. Functional characterization of poplar wood–associated NAC domain transcription factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Yamaguchiz, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.J.; Campbell, M.M. Expansion and diversifcation of the populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Mele, G.; Ori, N.; Sato, Y.; Hake, S. The knotted1–like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 2003, 17, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.L.; Lee, C.H.; Zhong, R.Q.; Ye, Z.H. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 2009, 21, 248–266. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Goué, N.; Igarashi, H.; Ohtani, M.; Nakano, Y.; Mortimer, J.C.; Nishikubo, N.; Kubo, M.; Katayama, Y.; Kakegawa, K.; et al. Vascular-related NAC-domain6 and vascular-related NAC-domain7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010, 153, 906–914. [Google Scholar] [CrossRef]
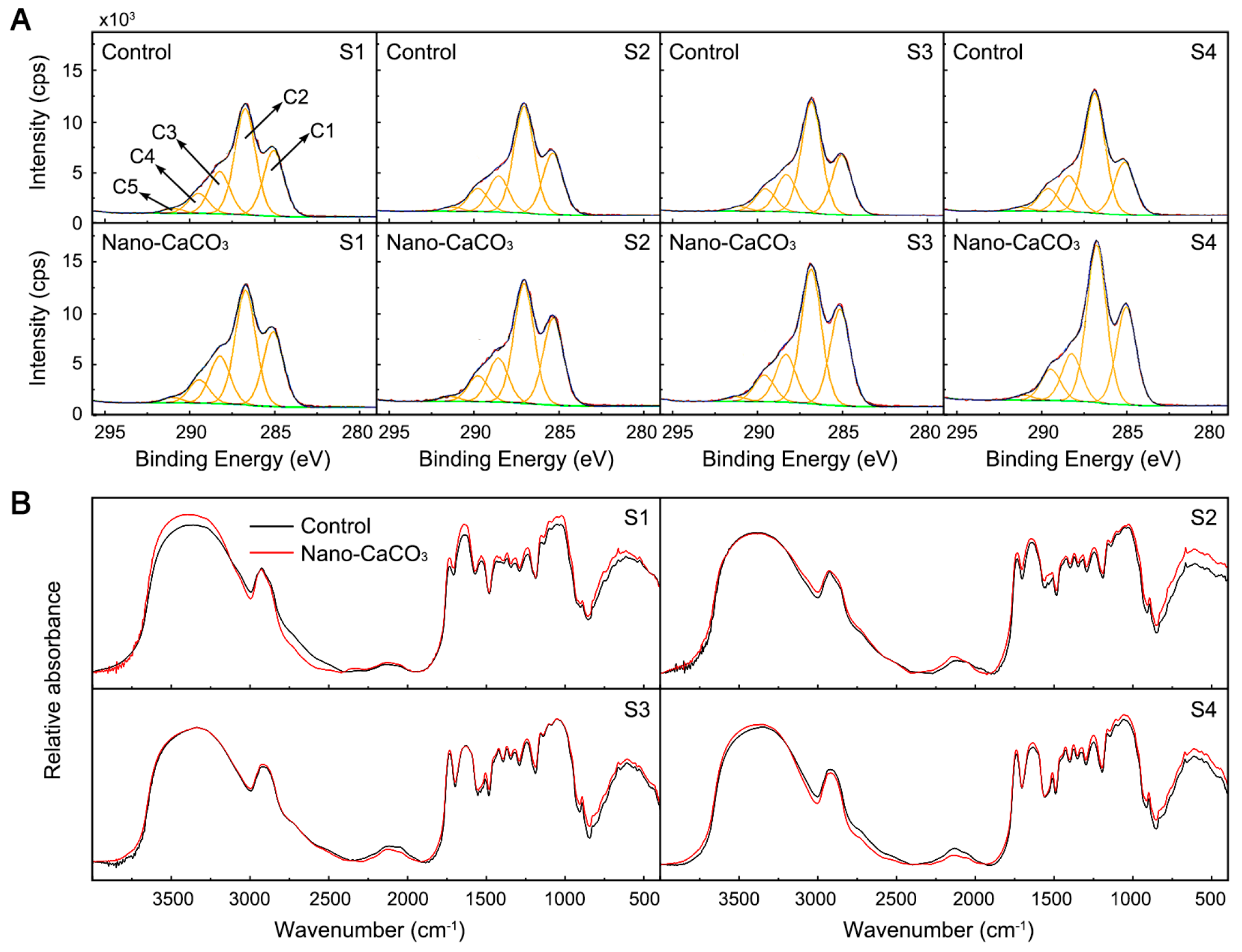
| Stage | Treatment | Element (%) | |||
|---|---|---|---|---|---|
| C | O | N | Ca | ||
| S1 | Control | 60.73 ± 0.38 a | 33.01 ± 0.21 b | 5.90 ± 0.11 a | 0.36 ± 0.05 e |
| Nano-CaCO3 | 59.72 ± 0.76 a | 34.22 ± 0.67 ab | 5.69 ± 0.11 a | 0.38 ± 0.01 e | |
| S2 | Control | 61.09 ± 1.13 a | 34.31 ± 1.07 ab | 4.18 ± 0.35 b | 0.42 ± 0.01 d |
| Nano-CaCO3 | 61.03 ± 0.76 a | 34.25 ± 0.84 ab | 4.29 ± 0.11 b | 0.43 ± 0.02 d | |
| S3 | Control | 61.67 ± 1.54 a | 34.46 ± 1.26 ab | 3.36 ± 0.30 d | 0.52 ± 0.03 c |
| Nano-CaCO3 | 61.78 ± 0.54 a | 33.97 ± 0.55 ab | 3.65 ± 0.09 c | 0.60 ± 0.04 b | |
| S4 | Control | 61.83 ± 2.23 a | 35.00 ± 1.94 a | 2.62 ± 0.26 e | 0.56 ± 0.06 a |
| Nano-CaCO3 | 61.89 ± 0.54 a | 34.37 ± 0.55 ab | 3.11 ± 0.04 d | 0.63 ± 0.06 a | |
| Stage | Treatment | Element (%) | ||||
|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | ||
| S1 | Control | 28.19 ± 0.71 a | 43.83 ± 0.57 a | 17.71 ± 0.15 a | 8.31 ± 0.43 d | 1.96 ± 0.13 ab |
| Nano-CaCO3 | 28.70 ± 3.44 a | 44.05 ± 2.47 a | 15.70 ± 0.46 b | 9.67 ± 0.32 a | 1.87 ± 0.09 abc | |
| S2 | Control | 29.29 ± 3.02 a | 44.24 ± 0.38 a | 15.22 ± 0.62 bc | 9.00 ± 0.56 abc | 1.78 ± 0.22 bcd |
| Nano-CaCO3 | 30.78 ± 5.76 a | 43.93 ± 3.28 a | 14.53 ± 0.67 cd | 9.14 ± 0.53 ab | 1.61 ± 0.13 de | |
| S3 | Control | 26.99 ± 2.81 a | 44.02 ± 2.27 a | 17.92 ± 0.11 a | 8.97 ± 0.38 bcd | 2.09 ± 0.05 a |
| Nano-CaCO3 | 28.94 ± 1.66 a | 43.79 ± 1.31 a | 15.96 ± 0.55 b | 9.45 ± 0.26 ab | 1.86 ± 0.01 abc | |
| S4 | Control | 29.52 ± 1.46 a | 44.54 ± 1.08 a | 15.28 ± 0.75 bc | 8.92 ± 0.43 bcd | 1.71 ± 0.14 cd |
| Nano-CaCO3 | 31.80 ± 1.49 a | 45.12 ± 1.07 a | 13.64 ± 0.69 c | 8.44 ± 0.36 cd | 1.43 ± 0.11 e | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.; Tang, Y.; Xia, X.; Sun, J.; Meng, J.; Shang, J.; Tao, J. Integration of Transcriptome, Proteome, and Metabolome Provides Insights into How Calcium Enhances the Mechanical Strength of Herbaceous Peony Inflorescence Stems. Cells 2019, 8, 102. https://doi.org/10.3390/cells8020102
Zhao D, Tang Y, Xia X, Sun J, Meng J, Shang J, Tao J. Integration of Transcriptome, Proteome, and Metabolome Provides Insights into How Calcium Enhances the Mechanical Strength of Herbaceous Peony Inflorescence Stems. Cells. 2019; 8(2):102. https://doi.org/10.3390/cells8020102
Chicago/Turabian StyleZhao, Daqiu, Yuhan Tang, Xing Xia, Jing Sun, Jiasong Meng, Jiali Shang, and Jun Tao. 2019. "Integration of Transcriptome, Proteome, and Metabolome Provides Insights into How Calcium Enhances the Mechanical Strength of Herbaceous Peony Inflorescence Stems" Cells 8, no. 2: 102. https://doi.org/10.3390/cells8020102
APA StyleZhao, D., Tang, Y., Xia, X., Sun, J., Meng, J., Shang, J., & Tao, J. (2019). Integration of Transcriptome, Proteome, and Metabolome Provides Insights into How Calcium Enhances the Mechanical Strength of Herbaceous Peony Inflorescence Stems. Cells, 8(2), 102. https://doi.org/10.3390/cells8020102





