The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers
Abstract
1. Introduction
2. The Biological Function of PARP1 and Its Enzymatic Activity
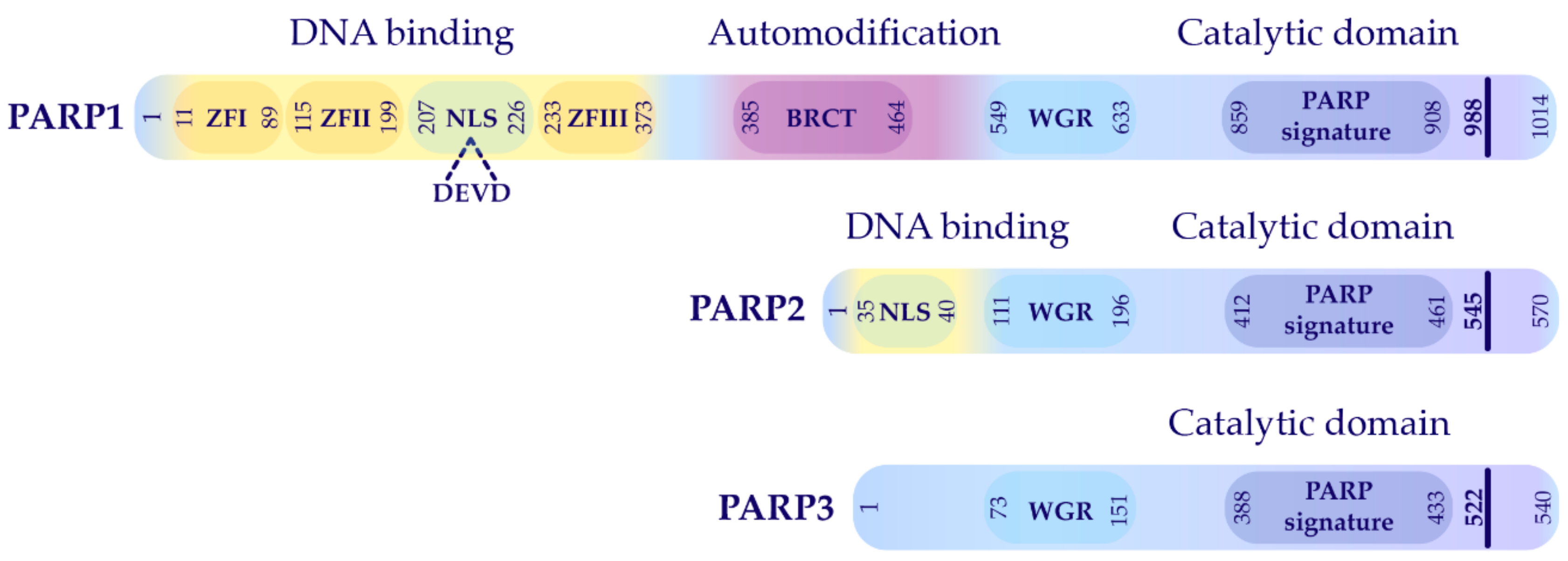
2.1. PARP1 Structure and Functional Domains
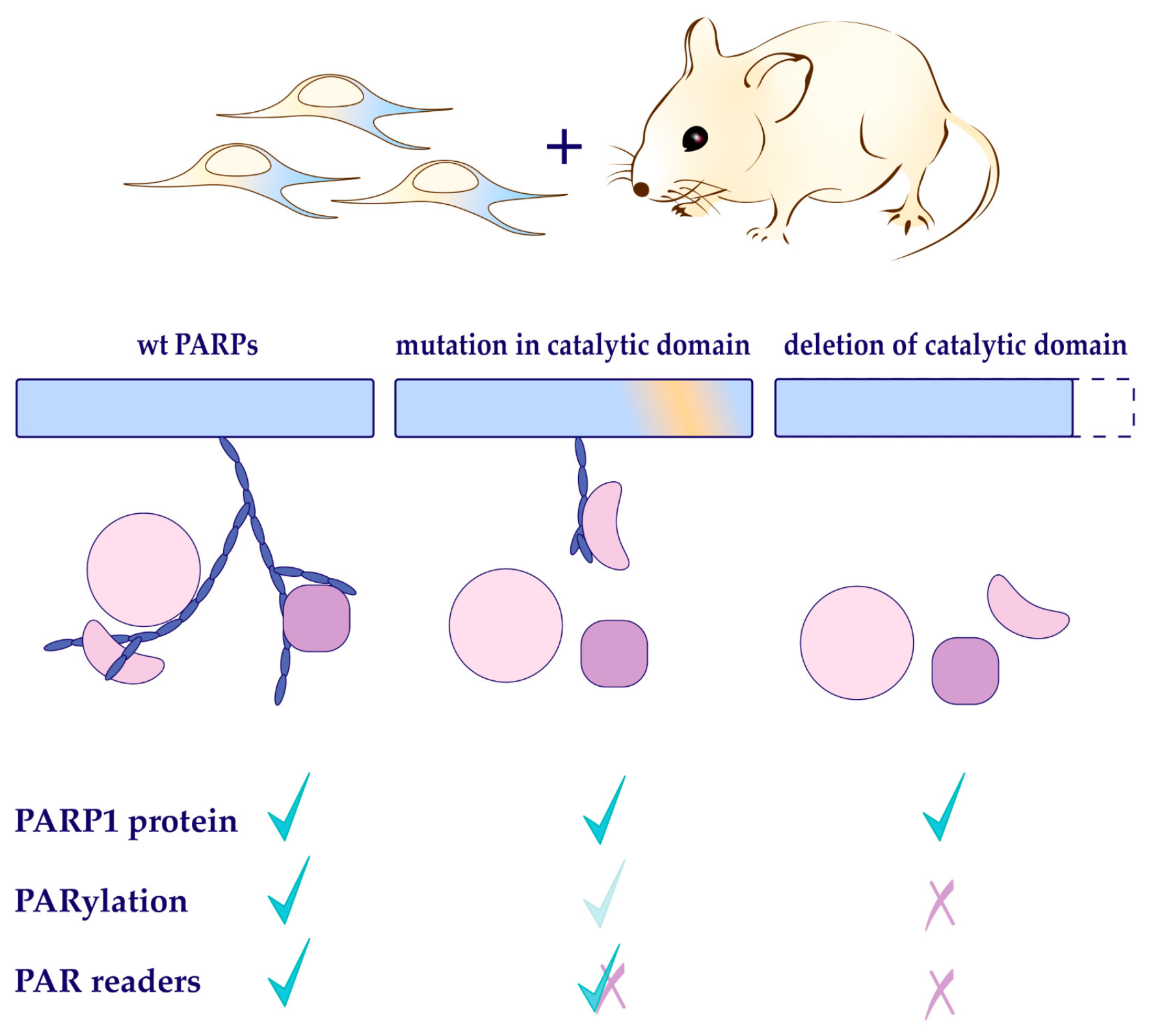
2.2. Functional Domain Mapping of PARP1
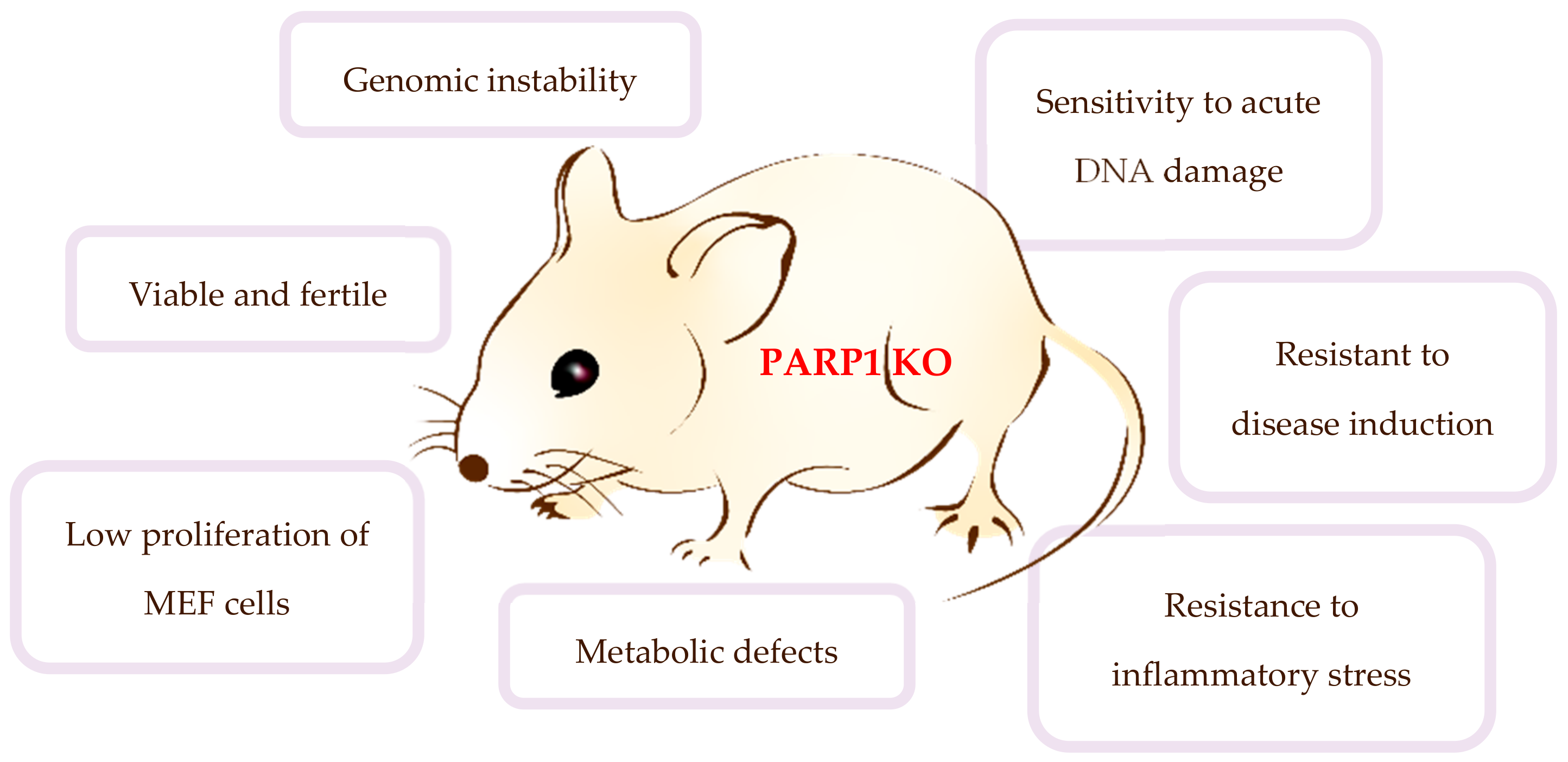
2.3. Biological Functions of PARP1
3. PAR-Binding Motifs and PAR Reader Proteins
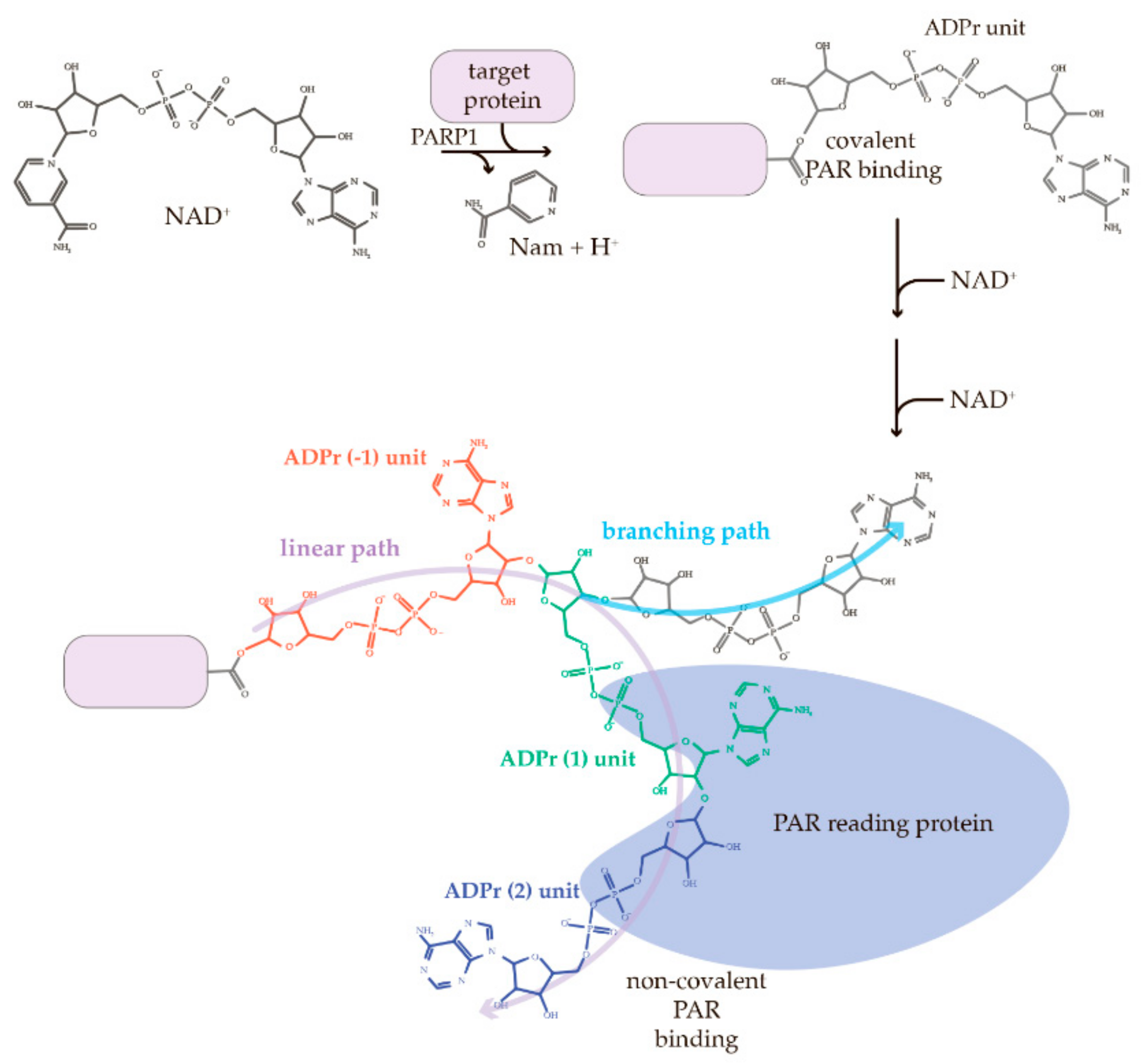
3.1. PARylation and PAR
3.2. PAR-Binding Motifs
3.3. PAR-Binding Proteins and Their Functions
3.4. Length- and Branching-Dependent PAR-Binding
4. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, C.; Vyas, A.; Kassab, M.A.; Singh, A.K.; Yu, X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 2017, 45, 8129–8141. [Google Scholar] [CrossRef] [PubMed]
- Schuhwerk, H.; Atteya, R.; Siniuk, K.; Wang, Z.Q. PARPing for balance in the homeostasis of poly(ADP-ribosyl)ation. Semin Cell Dev. Biol. 2017, 63, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Wang, Z.-Q. Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front. BioSci. 2009, 14, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Slade, D.; Dunstan, M.S.; Barkauskaite, E.; Weston, R.; Lafite, P.; Dixon, N.; Ahel, M.; Leys, D.; Ahel, I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 2011, 477, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly (ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Alemasova, E.E.; Lavrik, O.I. Poly (ADP-ribosyl) ation by PARP1: Reaction mechanism and regulatory proteins. Nucleic Acids Res. 2019, 47, 3811–3827. [Google Scholar] [CrossRef]
- Hottiger, M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 2015, 84, 227–263. [Google Scholar] [CrossRef]
- Beck, C.; Robert, I.; Reina-San-Martin, B.; Schreiber, V.; Dantzer, F. Poly (ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014, 329, 18–25. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610. [Google Scholar] [CrossRef]
- Hou, W.H.; Chen, S.H.; Yu, X. Poly-ADP ribosylation in DNA damage response and cancer therapy. Mutat. Res. 2019, 780, 82–91. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Eisemann, T.; Riccio, A.A.; Pascal, J.M. PARP family enzymes: Regulation and catalysis of the poly (ADP-ribose) posttranslational modification. Curr. Opin. Struct. Biol. 2018, 53, 187–198. [Google Scholar] [CrossRef]
- Jubin, T.; Kadam, A.; Jariwala, M.; Bhatt, S.; Sutariya, S.; Gani, A.; Gautam, S.; Begum, R. The PARP family: Insights into functional aspects of poly (ADP-ribose) polymerase-1 in cell growth and survival. Cell Prolif. 2016, 49, 421–437. [Google Scholar] [CrossRef]
- Keung, M.Y.T.; Wu, Y.; Vadgama, J.V. PARP Inhibitors as a Therapeutic Agent for Homologous Recombination Deficiency in Breast Cancers. J. Clin. Med. 2019, 8, 435. [Google Scholar] [CrossRef]
- Kunze, F.A.; Hottiger, M.O. Regulating Immunity via ADP-Ribosylation: Therapeutic Implications and Beyond. Trends Immunol 2019, 40, 159–173. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Servent, K.M.; Rogers, E.E.; Pascal, J.M. A third zinc-binding domain of human poly (ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J. Biol. Chem. 2008, 283, 4105–4114. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage–dependent poly (ADP-ribosyl) ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef]
- Loeffler, P.A.; Cuneo, M.J.; Mueller, G.A.; DeRose, E.F.; Gabel, S.A.; London, R.E. Structural studies of the PARP-1 BRCT domain. BMC Struct. Biol. 2011, 11, 37. [Google Scholar] [CrossRef]
- Gagné, J.-P.; Rouleau, M.; Poirier, G.G. PARP-1 Activation—Bringing the Pieces Together. Science 2012, 336, 678–679. [Google Scholar] [CrossRef]
- Mansoorabadi, S.O.; Wu, M.; Tao, Z.; Gao, P.; Pingali, S.V.; Guo, L.; Liu, H.-w. Conformational activation of poly (ADP-ribose) polymerase-1 upon DNA binding revealed by small-angle X-ray scattering. Biochemistry 2014, 53, 1779–1788. [Google Scholar] [CrossRef]
- Buelow, B.; Uzunparmak, B.; Paddock, M.; Scharenberg, A.M. Structure/function analysis of PARP-1 in oxidative and nitrosative stress-induced monomeric ADPR formation. PLoS ONE 2009, 4, e6339. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Crystal structures of poly (ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011, 286, 10690–10701. [Google Scholar] [CrossRef]
- Smulson, M.; Istock, N.; Ding, R.; Cherney, B. Deletion Mutants of Poly (ADP-Ribose) Polymerase A Support a Model of Cyclic Association and Dissociation of Enzyme from DNA Ends During DNA Repair. Biochemistry 1994, 33, 6186–6191. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Ruhl, D.D.; Planck, J.L.; Kraus, W.L.; Pascal, J.M. The Zn3 domain of human poly (ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly (ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010, 285, 18877–18887. [Google Scholar] [CrossRef]
- Ali, A.A.; Timinszky, G.; Arribas-Bosacoma, R.; Kozlowski, M.; Hassa, P.O.; Hassler, M.; Ladurner, A.G.; Pearl, L.H.; Oliver, A.W. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012, 19, 685. [Google Scholar] [CrossRef]
- Altmeyer, M.; Messner, S.; Hassa, P.O.; Fey, M.; Hottiger, M.O. Molecular mechanism of poly (ADP-ribosyl) ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009, 37, 3723–3738. [Google Scholar] [CrossRef]
- Bonfiglio, J.J.; Fontana, P.; Zhang, Q.; Colby, T.; Gibbs-Seymour, I.; Atanassov, I.; Bartlett, E.; Zaja, R.; Ahel, I.; Matic, I. Serine ADP-ribosylation depends on HPF1. Mol. Cell 2017, 65, 932–940.e936. [Google Scholar] [CrossRef]
- Simonin, F.; Menissier-de Murcia, J.; Poch, O.; Muller, S.; Gradwohl, G.; Molinete, M.; Penning, C.; Keith, G.; de Murcia, G. Expression and site-directed mutagenesis of the catalytic domain of human poly (ADP-ribose) polymerase in Escherichia coli. Lysine 893 is critical for activity. J. Biol. Chem. 1990, 265, 19249–19256. [Google Scholar]
- Simonin, F.; Poch, O.; Delarue, M.; De Murcia, G. Identification of potential active-site residues in the human poly (ADP-ribose) polymerase. J. Biol. Chem. 1993, 268, 8529–8535. [Google Scholar]
- Marsischky, G.T.; Wilson, B.A.; Collier, R.J. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation Evidence for active site similarities to the ADP-ribosylating toxins. J. Biol. Chem. 1995, 270, 3247–3254. [Google Scholar] [CrossRef]
- Miranda, E.A.; Dantzer, F.; Ofarrell, M.; Demurcia, G.; Demurcia, J.M. Characterization of a gain-of-function mutant of poly (ADP-ribose) polymerase. Biochem. Biophys. Res. Commun. 1995, 212, 317–325. [Google Scholar] [CrossRef]
- Rolli, V.; O’Farrell, M.; Ménissier-de Murcia, J.; de Murcia, G. Random mutagenesis of the poly (ADP-ribose) polymerase catalytic domain reveals amino acids involved in polymer branching. Biochemistry 1997, 36, 12147–12154. [Google Scholar] [CrossRef]
- Chen, S.-H.; Yu, X. Targeting dePARylation selectively suppresses DNA repair–defective and PARP inhibitor–resistant malignancies. Sci. Adv. 2019, 5, eaav4340. [Google Scholar] [CrossRef]
- Feng, F.Y.; De Bono, J.S.; Rubin, M.A.; Knudsen, K.E. Chromatin to clinic: The molecular rationale for PARP1 inhibitor function. Mol. Cell 2015, 58, 925–934. [Google Scholar] [CrossRef]
- Rajawat, J.; Shukla, N.; Mishra, D.P. Therapeutic Targeting of Poly(ADP-Ribose) Polymerase-1 (PARP1) in Cancer: Current Developments, Therapeutic Strategies, and Future Opportunities. Med. Res. Rev. 2017, 37, 1461–1491. [Google Scholar] [CrossRef]
- Jeggo, P.A. DNA repair: PARP - another guardian angel? Curr Biol 1998, 8, R49–R51. [Google Scholar] [CrossRef]
- Wang, Z.-Q.; Auer, B.; Stingl, L.; Berghammer, H.; Haidacher, D.; Schweiger, M.; Wagner, E.F. Mice lacking ADPRT and poly (ADP-ribosyl) ation develop normally but are susceptible to skin disease. Genes Dev. 1995, 9, 509–520. [Google Scholar] [CrossRef]
- Satoh, M.S.; Lindahl, T. Role of poly (ADP-ribose) formation in DNA repair. Nature 1992, 356, 356. [Google Scholar] [CrossRef]
- Lindahl, T.; Satoh, M.S.; Poirier, G.G.; Klungland, A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem. Sci. 1995, 20, 405–411. [Google Scholar] [CrossRef]
- De Murcia, J.M.; Niedergang, C.; Trucco, C.; Ricoul, M.; Dutrillaux, B.; Mark, M.; Oliver, F.J.; Masson, M.; Dierich, A.; LeMeur, M. Requirement of poly (ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA 1997, 94, 7303–7307. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Stingl, L.; Morrison, C.; Jantsch, M.; Los, M.; Schulze-Osthoff, K.; Wagner, E.F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997, 11, 2347–2358. [Google Scholar] [CrossRef] [PubMed]
- Oliver, F.J.; de la Rubia, G.; Rolli, V.; Ruiz-Ruiz, M.C.; de Murcia, G.; Ménissier-de Murcia, J. Importance of poly (ADP-ribose) polymerase and its cleavage in apoptosis Lesson from an uncleavable mutant. J. Biol. Chem. 1998, 273, 33533–33539. [Google Scholar] [CrossRef] [PubMed]
- Trucco, C.; Javier Oliver, F.; de Murcia, G.; Ménissier-de Murcia, J. DNA repair defect in poly (ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998, 26, 2644–2649. [Google Scholar] [CrossRef] [PubMed]
- Cuzzocrea, S.; Wang, Z.-Q. Role of poly (ADP-ribose) glycohydrolase (PARG) in shock, ischemia and reperfusion. Pharmacol. Res. 2005, 52, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.M.; Cortes, U.; Wang, Z.Q. Poly(ADP-ribose) polymerase: A guardian angel protecting the genome and suppressing tumorigenesis. Biochim. Et Biophys. Acta 2001, 1552, 27–37. [Google Scholar] [CrossRef]
- Di Fagagna, F.d.A.; Hande, M.P.; Tong, W.-M.; Lansdorp, P.M.; Wang, Z.-Q.; Jackson, S.P. Functions of poly (ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat. Genet. 1999, 23, 76. [Google Scholar] [CrossRef]
- Menissier de Murcia, J.; Ricoul, M.; Tartier, L.; Niedergang, C.; Huber, A.; Dantzer, F.; Schreiber, V.; Ame, J.C.; Dierich, A.; LeMeur, M.; et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo. J. 2003, 22, 2255–2263. [Google Scholar] [CrossRef]
- Boehler, C.; Gauthier, L.R.; Mortusewicz, O.; Biard, D.S.; Saliou, J.M.; Bresson, A.; Sanglier-Cianferani, S.; Smith, S.; Schreiber, V.; Boussin, F.; et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. USA 2011, 108, 2783–2788. [Google Scholar] [CrossRef]
- Herceg, Z.; Wang, Z.-Q. Failure of poly (ADP-ribose) polymerase cleavage by caspases leads to induction of necrosis and enhanced apoptosis. Mol. Cell. Biol. 1999, 19, 5124–5133. [Google Scholar] [CrossRef]
- Boulares, A.H.; Yakovlev, A.G.; Ivanova, V.; Stoica, B.A.; Wang, G.; Iyer, S.; Smulson, M. Role of poly (ADP-ribose) polymerase (PARP) cleavage in apoptosis Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J. Biol. Chem. 1999, 274, 22932–22940. [Google Scholar] [CrossRef]
- Los, M.; Mozoluk, M.; Ferrari, D.; Stepczynska, A.; Stroh, C.; Renz, A.; Herceg, Z.; Wang, Z.-Q.; Schulze-Osthoff, K. Activation and caspase-mediated inhibition of PARP: A molecular switch between fibroblast necrosis and apoptosis in death receptor signaling. Mol. Biol. Cell 2002, 13, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Pétrilli, V.; Herceg, Z.; Hassa, P.O.; Patel, N.S.; Di Paola, R.; Cortes, U.; Dugo, L.; Filipe, H.-M.; Thiemermann, C.; Hottiger, M.O. Noncleavable poly (ADP-ribose) polymerase-1 regulates the inflammation response in mice. J. Clin. Investig. 2004, 114, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Hassa, P.O.; Covic, M.; Hasan, S.; Imhof, R.; Hottiger, M.O. The enzymatic and DNA binding activity of PARP-1 are not required for NF-κB coactivator function. J. Biol. Chem. 2001, 276, 45588–45597. [Google Scholar] [CrossRef] [PubMed]
- Oliver, F.J.; Ménissier-de Murcia, J.; Nacci, C.; Decker, P.; Andriantsitohaina, R.; Muller, S.; de la Rubia, G.; Stoclet, J.C.; de Murcia, G. Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly (ADP-ribose) polymerase-1 deficient mice. Embo J. 1999, 18, 4446–4454. [Google Scholar] [CrossRef] [PubMed]
- Endres, M.; Wang, Z.-Q.; Namura, S.; Waeber, C.; Moskowitz, M.A. Ischemic brain injury is mediated by the activation of poly (ADP-ribose) polymerase. J. Cereb. Blood Flow Metab. 1997, 17, 1143–1151. [Google Scholar] [CrossRef]
- Eliasson, M.J.; Sampei, K.; Mandir, A.S.; Hurn, P.D.; Traystman, R.J.; Bao, J.; Pieper, A.; Wang, Z.-Q.; Dawson, T.M.; Snyder, S.H. Poly (ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat. Med. 1997, 3, 1089. [Google Scholar] [CrossRef]
- Kauppinen, T.M. Multiple roles for poly (ADP-ribose) polymerase-1 in neurological disease. Neurochem. Int. 2007, 50, 954–958. [Google Scholar] [CrossRef]
- Masutani, M.; Suzuki, H.; Kamada, N.; Watanabe, M.; Ueda, O.; Nozaki, T.; Jishage, K.-i.; Watanabe, T.; Sugimoto, T.; Nakagama, H. Poly (ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 2301–2304. [Google Scholar] [CrossRef]
- Burkart, V.; Wang, Z.Q.; Radons, J.; Heller, B.; Herceg, Z.; Stingl, L.; Wagner, E.F.; Kolb, H. Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nat. Med. 1999, 5, 314–319. [Google Scholar] [CrossRef]
- Selvaraj, V.; Soundarapandian, M.M.; Chechneva, O.; Williams, A.J.; Sidorov, M.K.; Soulika, A.M.; Pleasure, D.E.; Deng, W. PARP-1 deficiency increases the severity of disease in a mouse model of multiple sclerosis. J. Biol. Chem. 2009, 284, 26070–26084. [Google Scholar] [CrossRef]
- Bai, P.; Cantó, C.; Oudart, H.; Brunyánszki, A.; Cen, Y.; Thomas, C.; Yamamoto, H.; Huber, A.; Kiss, B.; Houtkooper, R.H. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011, 13, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Devalaraja-Narashimha, K.; Padanilam, B.J. PARP1 deficiency exacerbates diet-induced obesity in mice. J. Endocrinol. 2010, 205, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Rank, L.; Veith, S.; Gwosch, E.C.; Demgenski, J.; Ganz, M.; Jongmans, M.C.; Vogel, C.; Fischbach, A.; Buerger, S.; Fischer, J.M. Analyzing structure–function relationships of artificial and cancer-associated PARP1 variants by reconstituting TALEN-generated HeLa PARP1 knock-out cells. Nucleic Acids Res. 2016, 44, 10386–10405. [Google Scholar] [PubMed]
- Schuhwerk, H.; Bruhn, C.; Siniuk, K.; Min, W.; Erener, S.; Grigaravicius, P.; Krüger, A.; Ferrari, E.; Zubel, T.; Lazaro, D. Kinetics of poly (ADP-ribosyl) ation, but not PARP1 itself, determines the cell fate in response to DNA damage in vitro and in vivo. Nucleic Acids Res. 2017, 45, 11174–11192. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K. Poly (ADP-ribose): An organizer of cellular architecture. J. Cell Biol. 2014, 205, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yu, X. Functions of PARylation in DNA damage repair pathways. Genom. Proteom. Bioinform. 2016, 14, 131–139. [Google Scholar] [CrossRef]
- Erijman, A.; Rosenthal, E.; Shifman, J.M. How structure defines affinity in protein-protein interactions. PLoS ONE 2014, 9, e110085. [Google Scholar] [CrossRef] [PubMed]
- Vivelo, C.A.; Wat, R.; Agrawal, C.; Tee, H.Y.; Leung, A.K. ADPriboDB: The database of ADP-ribosylated proteins. Nucleic Acids Res. 2016, 45, D204–D209. [Google Scholar] [CrossRef]
- Teloni, F.; Altmeyer, M. Readers of poly (ADP-ribose): Designed to be fit for purpose. Nucleic Acids Res. 2015, 44, 993–1006. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, Y.; Li, M.; Yu, X. The oligonucleotide/oligosaccharide-binding fold motif is a poly (ADP-ribose)-binding domain that mediates DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 7278–7283. [Google Scholar] [CrossRef]
- Zhang, F.; Shi, J.; Bian, C.; Yu, X. Poly (ADP-ribose) mediates the BRCA2-dependent early DNA damage response. Cell Rep. 2015, 13, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Isabelle, M.; Gagné, J.-P.; Gallouzi, I.-E.; Poirier, G.G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly (ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012, 125, 4555–4566. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.; Berger, N.D.; Luijsterburg, M.S.; Piett, C.G.; Stanley, F.K.; Schräder, C.U.; Fang, S.; Chan, J.A.; Schriemer, D.C.; Nagel, Z.D. The CHD6 chromatin remodeler is an oxidative DNA damage response factor. Nat. Commun. 2019, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Iles, N.; Rulten, S.; El-Khamisy, S.F.; Caldecott, K.W. APLF (C2orf13) Is a Novel Human Protein Involved in the Cellular Response to Chromosomal DNA Strand Breaks. Mol. Cell. Biol. 2007, 27, 3793–3803. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Michaud, G.A.; Cheng, Z.; Zhang, Y.; Hinds, T.R.; Fan, E.; Cong, F.; Xu, W. Recognition of the iso-ADP-ribose moiety in poly (ADP-ribose) by WWE domains suggests a general mechanism for poly (ADP-ribosyl) ation-dependent ubiquitination. Genes Dev. 2012, 26, 235–240. [Google Scholar] [CrossRef]
- Li, G.Y.; McCulloch, R.D.; Fenton, A.L.; Cheung, M.; Meng, L.; Ikura, M.; Koch, C.A. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl. Acad. Sci. USA 2010, 107, 9129–9134. [Google Scholar] [CrossRef]
- Min, W.; Bruhn, C.; Grigaravicius, P.; Zhou, Z.-W.; Li, F.; Krüger, A.; Siddeek, B.; Greulich, K.-O.; Popp, O.; Meisezahl, C.; et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013, 4, 2993. [Google Scholar] [CrossRef]
- Zhou, Z.D.; Chan, C.H.; Xiao, Z.C.; Tan, E.K. Ring finger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adhes. Migr. 2011, 5, 463–471. [Google Scholar] [CrossRef]
- Rack, J.G.; Perina, D.; Ahel, I. Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu. Rev. Biochem. 2016, 85, 431–454. [Google Scholar] [CrossRef]
- Zweifel, M.E.; Leahy, D.J.; Barrick, D. Structure and Notch receptor binding of the tandem WWE domain of Deltex. Structure 2005, 13, 1599–1611. [Google Scholar] [CrossRef]
- Senissar, M.; Manav, M.C.; Brodersen, D.E. Structural conservation of the PIN domain active site across all domains of life. Protein Sci. A Publ. Protein Soc. 2017, 26, 1474–1492. [Google Scholar] [CrossRef]
- Mahajan, A.; Yuan, C.; Lee, H.; Chen, E.S.; Wu, P.Y.; Tsai, M.D. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 2008, 1, re12. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.N.; Williams, R.S.; Lee, M.S. Interactions between BRCT repeats and phosphoproteins: Tangled up in two. Trends Biochem. Sci. 2004, 29, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. Embo J. 1993, 12, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Malanga, M.; Czubaty, A.; Girstun, A.; Staron, K.; Althaus, F.R. Poly (ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008, 283, 19991–19998. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Akitsu, M.; Amano, Y.; Yamashita, K.; Ide, M.; Shimada, K.; Yamashita, A.; Hirano, H.; Arakawa, N.; Maki, T.; et al. The novel PAR-1-binding protein MTCL1 has crucial roles in organizing microtubules in polarizing epithelial cells. J. Cell Sci. 2013, 126, 4671–4683. [Google Scholar] [CrossRef]
- Rulten, S.L.; Rotheray, A.; Green, R.L.; Grundy, G.J.; Moore, D.A.; Gomez-Herreros, F.; Hafezparast, M.; Caldecott, K.W. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014, 42, 307–314. [Google Scholar] [CrossRef]
- Pomeranz-Krummel, D.; Nagai, K. RNA-Binding Domains in Proteins. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; pp. 1733–1735. [Google Scholar]
- Rulten, S.L.; Cortes-Ledesma, F.; Guo, L.; Iles, N.J.; Caldecott, K.W. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 2008, 28, 4620–4628. [Google Scholar] [CrossRef]
- Kim, I.K.; Stegeman, R.A.; Brosey, C.A.; Ellenberger, T. A quantitative assay reveals ligand specificity of the DNA scaffold repair protein XRCC1 and efficient disassembly of complexes of XRCC1 and the poly(ADP-ribose) polymerase 1 by poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2015, 290, 3775–3783. [Google Scholar] [CrossRef]
- El-Khamisy, S.F.; Masutani, M.; Suzuki, H.; Caldecott, K.W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003, 31, 5526–5533. [Google Scholar] [CrossRef]
- Keil, C.; Grobe, T.; Oei, S.L. MNNG-induced cell death is controlled by interactions between PARP-1, poly(ADP-ribose) glycohydrolase, and XRCC1. J. Biol. Chem. 2006, 281, 34394–34405. [Google Scholar] [CrossRef]
- Polo, L.M.; Xu, Y.; Hornyak, P.; Garces, F.; Zeng, Z.; Hailstone, R.; Matthews, S.J.; Caldecott, K.W.; Oliver, A.W.; Pearl, L.H. Efficient Single-Strand Break Repair Requires Binding to Both Poly(ADP-Ribose) and DNA by the Central BRCT Domain of XRCC1. Cell Rep. 2019, 26, 573–581.e575. [Google Scholar] [CrossRef] [PubMed]
- Ahel, I.; Ahel, D.; Matsusaka, T.; Clark, A.J.; Pines, J.; Boulton, S.J.; West, S.C. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 2008, 451, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Kashima, L.; Idogawa, M.; Mita, H.; Shitashige, M.; Yamada, T.; Ogi, K.; Suzuki, H.; Toyota, M.; Ariga, H.; Sasaki, Y.; et al. CHFR protein regulates mitotic checkpoint by targeting PARP-1 protein for ubiquitination and degradation. J. Biol. Chem. 2012, 287, 12975–12984. [Google Scholar] [CrossRef]
- Liu, C.; Wu, J.; Paudyal, S.C.; You, Z.; Yu, X. CHFR is important for the first wave of ubiquitination at DNA damage sites. Nucleic Acids Res. 2013, 41, 1698–1710. [Google Scholar] [CrossRef] [PubMed]
- Caleca, L.; Colombo, M.; van Overeem Hansen, T.; Lázaro, C.; Manoukian, S.; Parsons, M.T.; Spurdle, A.B.; Radice, P. GFP-Fragment Reassembly Screens for the Functional Characterization of Variants of Uncertain Significance in Protein Interaction Domains of the BRCA1 and BRCA2 Genes. Cancers 2019, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Shi, J.; Chen, S.-H.; Bian, C.; Yu, X. The PIN domain of EXO1 recognizes poly (ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015, 43, 10782–10794. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Lin, W.L.; Chen, Z.; Liu, H.W. PARP-1-dependent recruitment of cold-inducible RNA-binding protein promotes double-strand break repair and genome stability. Proc. Natl. Acad. Sci. USA 2018, 115, E1759–e1768. [Google Scholar] [CrossRef]
- Herceg, Z.; Wang, Z.-Q. Functions of poly (ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2001, 477, 97–110. [Google Scholar] [CrossRef]
- Kurokawa, S.; Okuda, A.; Nishizawa, Y.; Furukawa, K.; Sumihiro, A.; Nakaji, Y.; Tanaka, S.; Takehashi, M. Suppression of cell cycle progression by poly (ADP-ribose) polymerase inhibitor PJ34 in neural stem/progenitor cells. Biochem. Biophys. Res. Commun. 2019, 510, 59–64. [Google Scholar] [CrossRef]
- Najafabadi, H.S.; Mnaimneh, S.; Schmitges, F.W.; Garton, M.; Lam, K.N.; Yang, A.; Albu, M.; Weirauch, M.T.; Radovani, E.; Kim, P.M. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 2015, 33, 555. [Google Scholar] [CrossRef] [PubMed]
- Timinszky, G.; Till, S.; Hassa, P.O.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.; Scheffzek, K.; et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar] [CrossRef] [PubMed]
- DaRosa, P.A.; Wang, Z.; Jiang, X.; Pruneda, J.N.; Cong, F.; Klevit, R.E.; Xu, W. Allosteric activation of the RNF146 ubiquitin ligase by a poly (ADP-ribosyl) ation signal. Nature 2015, 517, 223. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Et Biophys. Acta 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, N.S.; Haince, J.-F.; Kang, H.C.; David, K.K.; Andrabi, S.A.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Poly (ADP-ribose)(PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1–dependent cell death (parthanatos). Sci. Signal. 2011, 4, ra20. [Google Scholar] [CrossRef]
- Andrabi, S.A.; Kang, H.C.; Haince, J.F.; Lee, Y.I.; Zhang, J.; Chi, Z.; West, A.B.; Koehler, R.C.; Poirier, G.G.; Dawson, T.M.; et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011, 17, 692–699. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Mickanin, C.; Feng, Y.; Charlat, O.; Michaud, G.A.; Schirle, M.; Shi, X.; Hild, M.; Bauer, A. RNF146 is a poly (ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat. Cell Biol. 2011, 13, 623. [Google Scholar] [CrossRef]
- Mariotti, L.; Pollock, K.; Guettler, S. Regulation of Wnt/beta-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding. Br. J. Pharmacol. 2017, 174, 4611–4636. [Google Scholar] [CrossRef]
- Kang, H.C.; Lee, Y.-I.; Shin, J.-H.; Andrabi, S.A.; Chi, Z.; Gagné, J.-P.; Lee, Y.; Ko, H.S.; Lee, B.D.; Poirier, G.G. Iduna is a poly (ADP-ribose)(PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 14103–14108. [Google Scholar] [CrossRef]
- Liu, Q.; Kistemaker, H.A.; Overkleeft, H.S.; van der Marel, G.A.; Filippov, D.V. Synthesis of ribosyl-ribosyl-adenosine-5′, 5′′, 5′′′(triphosphate)—the naturally occurring branched fragment of poly (ADP ribose). Chem. Commun. 2017, 53, 10255–10258. [Google Scholar] [CrossRef]
- Kistemaker, H.A.; Overkleeft, H.S.; van der Marel, G.A.; Filippov, D.V. Branching of poly (ADP-ribose): Synthesis of the Core Motif. Org. Lett. 2015, 17, 4328–4331. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Kassab, M.A.; Dantzer, F.; Yu, X. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 2018, 9, 3233. [Google Scholar] [CrossRef] [PubMed]
- Pourfarjam, Y.; Ventura, J.; Kurinov, I.; Cho, A.; Moss, J.; Kim, I.-K. Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition. J. Biol. Chem. 2018, 293, 12350–12359. [Google Scholar] [CrossRef]
- Fahrer, J.; Kranaster, R.; Altmeyer, M.; Marx, A.; Bürkle, A. Quantitative analysis of the binding affinity of poly (ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007, 35, e143. [Google Scholar] [CrossRef] [PubMed]
- Fahrer, J.; Popp, O.; Malanga, M.; Beneke, S.; Markovitz, D.M.; Ferrando-May, E.; Burkle, A.; Kappes, F. High-affinity interaction of poly(ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry 2010, 49, 7119–7130. [Google Scholar] [CrossRef]
- Yu, D.; Liu, R.; Yang, G.; Zhou, Q. The PARP1-Siah1 Axis Controls HIV-1 Transcription and Expression of Siah1 Substrates. Cell Rep. 2018, 23, 3741–3749. [Google Scholar] [CrossRef]
- Li, M.; Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013, 23, 693–704. [Google Scholar] [CrossRef]
- Eckei, L.; Krieg, S.; Butepage, M.; Lehmann, A.; Gross, A.; Lippok, B.; Grimm, A.R.; Kummerer, B.M.; Rossetti, G.; Luscher, B.; et al. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 2017, 7, 41746. [Google Scholar] [CrossRef]
- Liu, C.; Yu, X. ADP-ribosyltransferases and poly ADP-ribosylation. Curr. Protein Pept. Sci. 2015, 16, 491–501. [Google Scholar] [CrossRef]
- Gagne, J.P.; Isabelle, M.; Lo, K.S.; Bourassa, S.; Hendzel, M.J.; Dawson, V.L.; Dawson, T.M.; Poirier, G.G. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008, 36, 6959–6976. [Google Scholar] [CrossRef]
- Beneke, S. Regulation of chromatin structure by poly(ADP-ribosyl)ation. Front. Genet. 2012, 3, 169. [Google Scholar] [CrossRef] [PubMed]
- Renschler, F.A.; Bruekner, S.R.; Salomon, P.L.; Mukherjee, A.; Kullmann, L.; Schutz-Stoffregen, M.C.; Henzler, C.; Pawson, T.; Krahn, M.P.; Wiesner, S. Structural basis for the interaction between the cell polarity proteins Par3 and Par6. Sci. Signal. 2018, 11, eaam9899. [Google Scholar] [CrossRef] [PubMed]
- Fouquerel, E.; Goellner, E.M.; Yu, Z.; Gagne, J.P.; Barbi de Moura, M.; Feinstein, T.; Wheeler, D.; Redpath, P.; Li, J.; Romero, G.; et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014, 8, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.H.; Yoon, H.; Kim, W.J.; Cha, D.; Choi, J.M. Cell-Penetrating Function of the Poly(ADP-Ribose) (PAR)-Binding Motif Derived from the PAR-Dependent E3 Ubiquitin Ligase Iduna. Int. J. Mol. Sci. 2018, 19, 779. [Google Scholar] [CrossRef] [PubMed]
- Todorova, T.; Bock, F.J.; Chang, P. Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer. Trends Mol. Med. 2015, 21, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lu, L.-Y.; Yang, C.-Y.; Wang, S.; Yu, X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013, 27, 1752–1768. [Google Scholar] [CrossRef]
- Murawska, M.; Hassler, M.; Renkawitz-Pohl, R.; Ladurner, A.; Brehm, A. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011, 7, e1002206. [Google Scholar] [CrossRef]
- Altmeyer, M.; Neelsen, K.J.; Teloni, F.; Pozdnyakova, I.; Pellegrino, S.; Grofte, M.; Rask, M.D.; Streicher, W.; Jungmichel, S.; Nielsen, M.L.; et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 2015, 6, 8088. [Google Scholar] [CrossRef]
- Krietsch, J.; Caron, M.C.; Gagne, J.P.; Ethier, C.; Vignard, J.; Vincent, M.; Rouleau, M.; Hendzel, M.J.; Poirier, G.G.; Masson, J.Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012, 40, 10287–10301. [Google Scholar] [CrossRef]
- Diefenbach, J.; Burkle, A. Introduction to poly(ADP-ribose) metabolism. Cell Mol. Life Sci. 2005, 62, 721–730. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kraus, W.L. The PARP side of the nucleus: Molecular actions, physiological outcomes, and clinical targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Ying, W. Roles of NAD (+), PARP-1, and Sirtuins in Cell Death, Ischemic Brain Injury, and Synchrotron Radiation X-Ray-Induced Tissue Injury. Scientifica 2013, 2013, 691251. [Google Scholar] [CrossRef] [PubMed]
| Mutation | Enzymatic Activity | Amino Acid Role | Reference |
|---|---|---|---|
| K893R K893I | ~40% ~0.2% | The initiation of the poly(ADP-ribosy1)ation reaction | [29] |
| D993E D993A | ~15.2% ~0.2% | The initiation of the poly(ADP-ribosy1)ation reaction | [29] |
| K953R K953I | ~2.9% ~9.8% | Indirect involvement in PARP activity | [29] |
| D914E D914A | ~11.5% ~2.5% | Indirect involvement in PARP activity | [29] |
| E988Q E988A E988K | ~2.2% 0.091% 1.25% | Key residues in the synthesis and elongation of PAR | [30,32] |
| L713F | ~879% | Allosteric effect on the catalytic site | [31] |
| Y986S | 11% | Enzymatic activity and PAR chain elongation | [32] |
| R847C E923G G972R | 75% 20% 16% | PAR branching | [32] |
| C908R | <0.5% | Enzymatic activity | [32] |
| T316A W318R | ~0.36% ~0.6% | Involvement in the DNA-dependent PARP1 activation | [24] |
| F44A V48A F44A/V48A | Lower auto-modification | DNA-binding affinity, DNA-dependent PARP-1 activation | [22] |
| Q40A D45A | Low auto-modification | Interactions with the domains essential for DNA-dependent activity | [22] |
| V144E/P149D V144E/P149I | ND | Recruitment towards the damage site | [25] |
| S499A/S507A/S519A | Low HPF1-dependent automodification | Automodification site, HPF1-dependent serine modification | [27] |
| PAR-Binding Motif | Motif Structure | Described | |
|---|---|---|---|
| Zinc finger type | PBZ | C2H2 type CX5CX6HX5H | [76] |
| PbR | C2H2 type CX8CX6HX8H | [77] | |
| RING | C3HC4 type CX2CX9-39CX1-3HX2-3CX2CX4-48CX2C | [78] | |
| Macrodomain | globular α/β/α sandwich β-α-β-α-α-β-β-α-β-α-β | [79] | |
| PBM | [HKR]1 × 2 × 3[AIQVY]4[KR]5[KR]6[AILV]7 [FILPV]8 | [78] | |
| WWE | β2-β1-β6-β5-β4-β3 and/or β2-β1-β5-β3-β4 | [80] | |
| PIN-domain | Compact structure β1-α1-β2-α2-β3-α3-β4-α4-β5 | [81] | |
| FHA domain | Two β sheets with Greek key topology β2-β1-β11-β10-β7-β8 and β4-β3-β5-β6-β9 | [82] | |
| BRCT | β-α-β-β-α-β-α | [83] | |
| OB-fold | Antiparallel β-barrel β1-β2-β3-β5-β4-β1 | [84] | |
| KR-rich domains, SR repeats, RG/RGG repeats | KR-, SR- or RG/RGG-rich repeats | [85,86,87] | |
| RRM | [RK]1G2[FY]3[GA]4[FY]5V6 × 7[FY]8– Xn–[LI]1[FY]2[VI]3 × 4[NG]5L6 β-α-β-β-α-β | [88] | |
| PAR-Binding Motif | Example of Readers | Process | Reference |
|---|---|---|---|
| PBZ | APLF, CHFR | DNA damage, chromatin architecture | [74] |
| PbR | Chk1 | DNA damage, cell cycle regulation | [77] |
| RING | RNF146/Iduna, Siah1, BARD1 | DNA damage regulation, protein degradation, transcription. | [110,117,118] |
| Macrodomain | MacroH2A, PARG, TARG1, MacroD1, MacroD2, macroD3, ALC1, ARTD7, ARTD8, ARTD9, PARP9, PARP14, PARP15, GDAP2 | DNA damage, redox defense, chromatin architecture, protein acetylation, viral infection | [79,119,120] |
| PBM | XRCC1, Aurora-A, NF-kappa-B, BID, CENP-A, ERCC-6, HKDC1, MVP, DNA topoisomerase 2-beta, BUB3, DNA ligase III, condensin complex subunit 1, hnRNP A1, hnRNP A2/B1, Ro(SS-A), H2A, H2B, H3, H4, AIF, MRE11, ATM, DNA-PKcs, KU70, MARCKS, MSH6, XPA, p21, DNA polymerase epsilon, NOS2, CAD, TERT, CTCF, DNMT1, Par6, DEK, WRN, HK1 | DNA damage, immune response, cell cycle regulation, chromatin architecture, telomeres length, stress signaling | [116,121,122,123,124] |
| WWE | RNF146/Iduna, PARP11, PARP13, PARP14, Deltex1 (A and B), Deltex2 (A and B), Deltex4 (A and B), ULF, HUWE1, DDHD2 | DNA damage regulation, protein degradation, mRNA stability | [75,110,125,126] |
| PIN-domain | EXO1, GEN1, SMG5 | DNA damage | [98] |
| FHA domain | APLF, PNKP, APTX | DNA damage | [74,127] |
| BRCT | BARD1, APLF, Ligase4, XRCC1, NBS1 | DNA damage | [74,118,127] |
| OB-fold | BRCA2, SSB1, SSB2, CTC1, MEIOB | DNA damage | [70,71] |
| KR-rich domains, SR repeats, RG/RGG repeats | G3BP, ASF/SF2, CHD6, MTCL1, dMi-2, CIRBP, FUS/TLS, TAF15, EWS | DNA damage, chromatin architecture, stress response, transcription and RNA processing | [72,73,85,86,87,99,128,129] |
| RRM | ASF/SF2, CIRBP, FUS/TLS, TAF15, EWS, NONO | DNA damage, RNA processing | [85,87,99,129,130] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamaletdinova, T.; Fanaei-Kahrani, Z.; Wang, Z.-Q. The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers. Cells 2019, 8, 1625. https://doi.org/10.3390/cells8121625
Kamaletdinova T, Fanaei-Kahrani Z, Wang Z-Q. The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers. Cells. 2019; 8(12):1625. https://doi.org/10.3390/cells8121625
Chicago/Turabian StyleKamaletdinova, Tatiana, Zahra Fanaei-Kahrani, and Zhao-Qi Wang. 2019. "The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers" Cells 8, no. 12: 1625. https://doi.org/10.3390/cells8121625
APA StyleKamaletdinova, T., Fanaei-Kahrani, Z., & Wang, Z.-Q. (2019). The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers. Cells, 8(12), 1625. https://doi.org/10.3390/cells8121625





