iPSCs-Based Neural 3D Systems: A Multidimensional Approach for Disease Modeling and Drug Discovery
Abstract
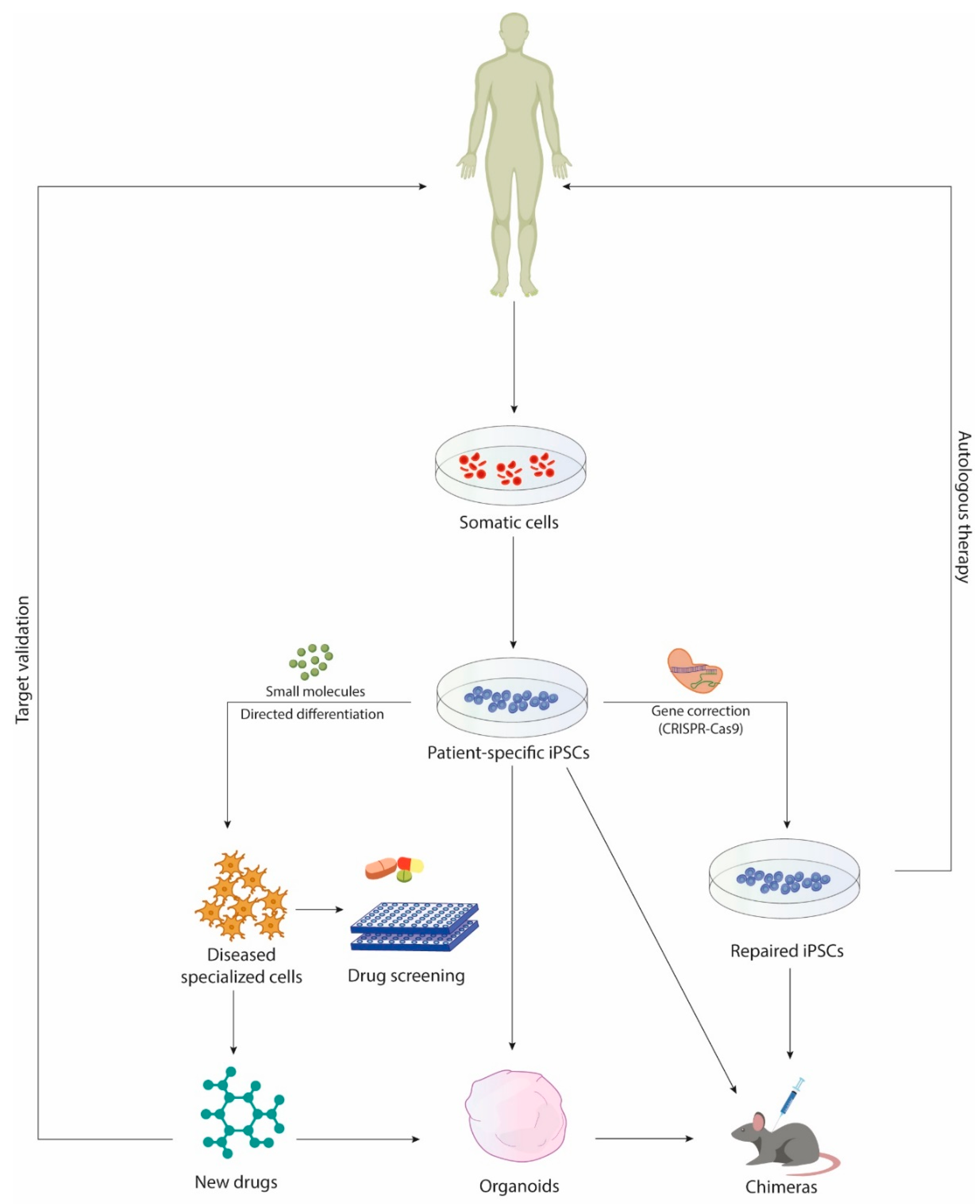
1. Introduction
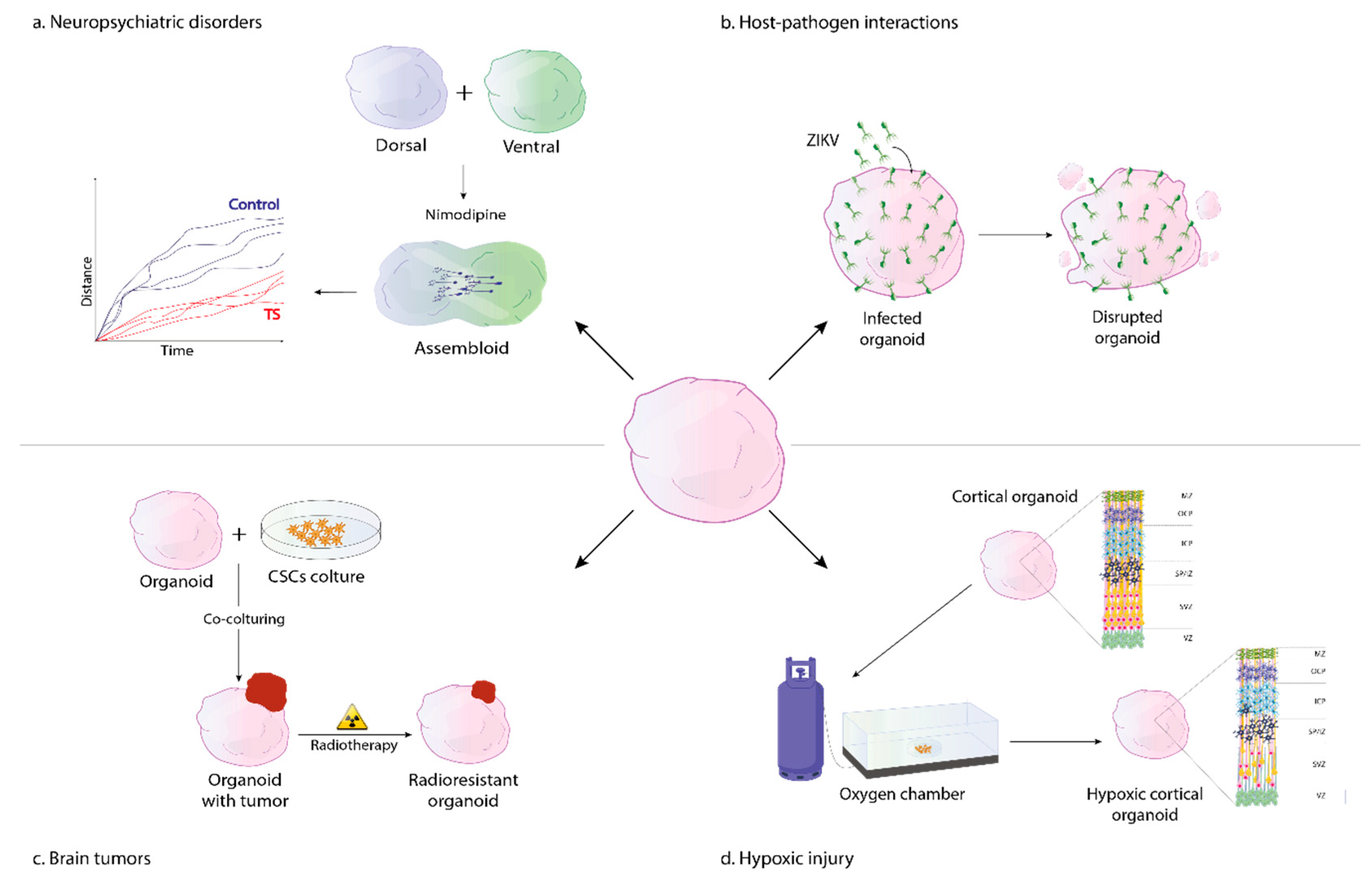
2. Modeling Neuropsychiatric and Neurodevelopmental Disorders
2.1. Timothy Syndrome
2.2. Autism Spectrum Disorders
2.3. Tuberous Sclerosis Complex
3. Organoid Models of Neural Hypoxic Injury
4. 3D Models of Host-Pathogen Interactions
4.1. Zika Virus
4.2. Herpesviruses
5. Models of Brain Cancer
6. New Frontiers in Neural Organoid Research: Human-Animal Chimeras
Neural Chimeras
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Roubal, I.; Park, S.J.; Kim, Y. Derivation of Neural Precursor Cells from Human Embryonic Stem Cells for DNA Methylomic Analysis. Methods Mol. Biol. Clifton NJ 2016, 1341, 345–357. [Google Scholar]
- Guloglu, M.O.; Larsen, A. Dopaminergic Differentiation of Human Embryonic Stem Cells on PA6-Derived Adipocytes. Methods Mol. Biol. Clifton NJ 2016, 1341, 235–244. [Google Scholar]
- Ben-Shushan, E.; Feldman, E.; Reubinoff, B.E. Notch signaling regulates motor neuron differentiation of human embryonic stem cells. Stem Cells Dayt. Ohio 2015, 33, 403–415. [Google Scholar] [CrossRef]
- Krencik, R.; Zhang, S.-C. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat. Protoc. 2011, 6, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Manley, N.C.; Priest, C.A.; Denham, J.; Wirth, E.D.; Lebkowski, J.S. Human Embryonic Stem Cell-Derived Oligodendrocyte Progenitor Cells: Preclinical Efficacy and Safety in Cervical Spinal Cord Injury. Stem Cells Transl. Med. 2017, 6, 1917–1929. [Google Scholar] [CrossRef]
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.-H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016, 22, 1358–1367. [Google Scholar] [CrossRef]
- Urbach, A.; Schuldiner, M.; Benvenisty, N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells Dayt. Ohio 2004, 22, 635–641. [Google Scholar] [CrossRef]
- Mandai, M.; Watanabe, A.; Kurimoto, Y.; Hirami, Y.; Morinaga, C.; Daimon, T.; Fujihara, M.; Akimaru, H.; Sakai, N.; Shibata, Y.; et al. Autologous Induced Stem-Cell–Derived Retinal Cells for Macular Degeneration. N. Engl. J. Med. 2017, 376, 1038–1046. [Google Scholar] [CrossRef]
- Takahashi, J. Preparing for first human trial of induced pluripotent stem cell-derived cells for Parkinson’s disease: An interview with Jun Takahashi. Regen. Med. 2019, 14, 93–95. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. “Reprogrammed” stem cells approved to mend human hearts for the first time. Nature 2018, 557, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Park, I.-H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-specific induced pluripotent stem cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.D.; Yu, J.; Rose, F.F.; Mattis, V.B.; Lorson, C.L.; Thomson, J.A.; Svendsen, C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009, 457, 277–280. [Google Scholar] [CrossRef]
- Yoshida, M.; Kitaoka, S.; Egawa, N.; Yamane, M.; Ikeda, R.; Tsukita, K.; Amano, N.; Watanabe, A.; Morimoto, M.; Takahashi, J.; et al. Modeling the early phenotype at the neuromuscular junction of spinal muscular atrophy using patient-derived iPSCs. Stem Cell Rep. 2015, 4, 561–568. [Google Scholar] [CrossRef]
- Camnasio, S.; Delli Carri, A.; Lombardo, A.; Grad, I.; Mariotti, C.; Castucci, A.; Rozell, B.; Lo Riso, P.; Castiglioni, V.; Zuccato, C.; et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol. Dis. 2012, 46, 41–51. [Google Scholar] [CrossRef]
- HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell 2012, 11, 264–278. [CrossRef]
- Juopperi, T.A.; Kim, W.R.; Chiang, C.-H.; Yu, H.; Margolis, R.L.; Ross, C.A.; Ming, G.; Song, H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol. Brain 2012, 5, 17. [Google Scholar] [CrossRef]
- Paşca, S.P.; Portmann, T.; Voineagu, I.; Yazawa, M.; Shcheglovitov, A.; Paşca, A.M.; Cord, B.; Palmer, T.D.; Chikahisa, S.; Nishino, S.; et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat. Med. 2011, 17, 1657–1662. [Google Scholar] [CrossRef]
- Madison, J.M.; Zhou, F.; Nigam, A.; Hussain, A.; Barker, D.D.; Nehme, R.; van der Ven, K.; Hsu, J.; Wolf, P.; Fleishman, M.; et al. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol. Psychiatry 2015, 20, 703–717. [Google Scholar] [CrossRef]
- Mertens, J.; Wang, Q.-W.; Kim, Y.; Yu, D.X.; Pham, S.; Yang, B.; Zheng, Y.; Diffenderfer, K.E.; Zhang, J.; Soltani, S.; et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015, 527, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Mariani, J.; Simonini, M.V.; Palejev, D.; Tomasini, L.; Coppola, G.; Szekely, A.M.; Horvath, T.L.; Vaccarino, F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 12770–12775. [Google Scholar] [CrossRef] [PubMed]
- McCauley, H.A.; Wells, J.M. Pluripotent stem cell-derived organoids: Using principles of developmental biology to grow human tissues in a dish. Dev. Camb. Engl. 2017, 144, 958–962. [Google Scholar] [CrossRef]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef]
- Velasco, S.; Kedaigle, A.J.; Simmons, S.K.; Nash, A.; Rocha, M.; Quadrato, G.; Paulsen, B.; Nguyen, L.; Adiconis, X.; Regev, A.; et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 2019, 570, 523–527. [Google Scholar] [CrossRef]
- Xiang, Y.; Tanaka, Y.; Patterson, B.; Kang, Y.-J.; Govindaiah, G.; Roselaar, N.; Cakir, B.; Kim, K.-Y.; Lombroso, A.P.; Hwang, S.-M.; et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017, 21, 383–398.e7. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Kadoshima, T.; Soen, M.; Narii, N.; Ishida, Y.; Ohgushi, M.; Takahashi, J.; Eiraku, M.; Sasai, Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015, 6, 8896. [Google Scholar] [CrossRef]
- Krefft, O.; Jabali, A.; Iefremova, V.; Koch, P.; Ladewig, J. Generation of Standardized and Reproducible Forebrain-type Cerebral Organoids from Human Induced Pluripotent Stem Cells. J. Vis. Exp. JoVE 2018, 131, e56768. [Google Scholar] [CrossRef] [PubMed]
- Hor, J.H.; Soh, E.S.-Y.; Tan, L.Y.; Lim, V.J.W.; Santosa, M.M.; Winanto, N.; Ho, B.X.; Fan, Y.; Soh, B.-S.; Ng, S.-Y. Cell cycle inhibitors protect motor neurons in an organoid model of Spinal Muscular Atrophy. Cell Death Dis. 2018, 9, 1100. [Google Scholar] [CrossRef] [PubMed]
- Di Lullo, E.; Kriegstein, A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017, 18, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Kim, Y.H.; Hebisch, M.; Sliwinski, C.; Lee, S.; D’Avanzo, C.; Chen, H.; Hooli, B.; Asselin, C.; Muffat, J.; et al. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature 2014, 515, 274–278. [Google Scholar] [CrossRef]
- Fan, W.; Sun, Y.; Shi, Z.; Wang, H.; Deng, J. Mouse induced pluripotent stem cells-derived Alzheimer’s disease cerebral organoid culture and neural differentiation disorders. Neurosci. Lett. 2019, 711, 134433. [Google Scholar] [CrossRef]
- Raja, W.K.; Mungenast, A.E.; Lin, Y.-T.; Ko, T.; Abdurrob, F.; Seo, J.; Tsai, L.-H. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS ONE 2016, 11, e0161969. [Google Scholar] [CrossRef]
- Amin, N.; Tan, X.; Ren, Q.; Zhu, N.; Botchway, B.O.A.; Hu, Z.; Fang, M. Recent advances of induced pluripotent stem cells application in neurodegenerative diseases. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 95, 109674. [Google Scholar] [CrossRef]
- Barker, R.A.; Parmar, M.; Studer, L.; Takahashi, J. Human Trials of Stem Cell-Derived Dopamine Neurons for Parkinson’s Disease: Dawn of a New Era. Cell Stem Cell 2017, 21, 569–573. [Google Scholar] [CrossRef]
- Espuny-Camacho, I.; Michelsen, K.A.; Gall, D.; Linaro, D.; Hasche, A.; Bonnefont, J.; Bali, C.; Orduz, D.; Bilheu, A.; Herpoel, A.; et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron 2013, 77, 440–456. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Mariani, J.; Coppola, G.; Zhang, P.; Abyzov, A.; Provini, L.; Tomasini, L.; Amenduni, M.; Szekely, A.; Palejev, D.; Wilson, M.; et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 2015, 162, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Iefremova, V.; Manikakis, G.; Krefft, O.; Jabali, A.; Weynans, K.; Wilkens, R.; Marsoner, F.; Brändl, B.; Müller, F.-J.; Koch, P.; et al. An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Rep. 2017, 19, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell–derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–20289. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef] [PubMed]
- Pașca, A.M.; Park, J.-Y.; Shin, H.-W.; Qi, Q.; Revah, O.; Krasnoff, R.; O’Hara, R.; Willsey, A.J.; Palmer, T.D.; Pașca, S.P. Human 3D cellular model of hypoxic brain injury of prematurity. Nat. Med. 2019, 25, 784. [Google Scholar] [CrossRef]
- Brown, R.M.; Rana, P.S.J.B.; Jaeger, H.K.; O’Dowd, J.M.; Balemba, O.B.; Fortunato, E.A. Human Cytomegalovirus Compromises Development of Cerebral Organoids. J. Virol. 2019. [Google Scholar] [CrossRef]
- Groveman, B.R.; Foliaki, S.T.; Orru, C.D.; Zanusso, G.; Carroll, J.A.; Race, B.; Haigh, C.L. Sporadic Creutzfeldt-Jakob disease prion infection of human cerebral organoids. Acta Neuropathol. Commun. 2019, 7, 90. [Google Scholar] [CrossRef]
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically engineered cerebral organoids model brain tumour formation. Nat. Methods 2018, 15, 631–639. [Google Scholar] [CrossRef]
- Insel, T.R.; Landis, S.C. Twenty-Five Years of Progress: The View from NIMH and NINDS. Neuron 2013, 80, 561–567. [Google Scholar] [CrossRef]
- Demjaha, A.; MacCabe, J.H.; Murray, R.M. How Genes and Environmental Factors Determine the Different Neurodevelopmental Trajectories of Schizophrenia and Bipolar Disorder. Schizophr. Bull. 2012, 38, 209–214. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smoller, J.W.; Andreassen, O.A.; Edenberg, H.J.; Faraone, S.V.; Glatt, S.J.; Kendler, K.S. Psychiatric genetics and the structure of psychopathology. Mol. Psychiatry 2019, 24, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, T.; Feng, G. Modeling psychiatric disorders for developing effective treatments. Nat. Med. 2015, 21, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Geschwind, D.H.; Flint, J. Genetics and genomics of psychiatric disease. Science 2015, 349, 1489–1494. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, M.; Zhang, F. Applications of CRISPR-Cas systems in neuroscience. Nat. Rev. Neurosci. 2016, 17, 36–44. [Google Scholar] [CrossRef]
- Hoffmann, A.; Ziller, M.; Spengler, D. Progress in iPSC-Based Modeling of Psychiatric Disorders. Int. J. Mol. Sci. 2019, 20, 4896. [Google Scholar] [CrossRef]
- Stachowiak, E.K.; Benson, C.A.; Narla, S.T.; Dimitri, A.; Chuye, L.E.B.; Dhiman, S.; Harikrishnan, K.; Elahi, S.; Freedman, D.; Brennand, K.J.; et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia-computational anatomy and genomics, role of FGFR1. Transl. Psychiatry 2017, 7, 6. [Google Scholar] [CrossRef]
- Kepecs, A.; Fishell, G. Interneuron cell types are fit to function. Nature 2014, 505, 318–326. [Google Scholar] [CrossRef]
- Gelman, D.M.; Marín, O.; Rubenstein, J.L.R. The Generation of Cortical Interneurons. In Jasper’s Basic Mechanisms of the Epilepsies, 4th ed.; Noebels, J.L., Avoli, M., Rogawski, M.A., Olsen, R.W., Delgado-Escueta, A.V., Eds.; National Center for Biotechnology Information: Bethesda, MD, USA, 2012. [Google Scholar]
- Bartolini, G.; Ciceri, G.; Marín, O. Integration of GABAergic interneurons into cortical cell assemblies: Lessons from embryos and adults. Neuron 2013, 79, 849–864. [Google Scholar] [CrossRef]
- Marín, O.; Valiente, M.; Ge, X.; Tsai, L.-H. Guiding Neuronal Cell Migrations. Cold Spring Harb. Perspect. Biol. Feb. 2010. [Google Scholar] [CrossRef]
- Watanabe, M.; Buth, J.E.; Vishlaghi, N.; de la Torre-Ubieta, L.; Taxidis, J.; Khakh, B.S.; Coppola, G.; Pearson, C.A.; Yamauchi, K.; Gong, D.; et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 2017, 21, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.-D.; Göke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.-P.; Lokman, H.; et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 2016, 19, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef]
- Betancur, C. Etiological heterogeneity in autism spectrum disorders: More than 100 genetic and genomic disorders and still counting. Brain Res. 2011, 1380, 42–77. [Google Scholar] [CrossRef]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Wang, P.; Mokhtari, R.; Pedrosa, E.; Kirschenbaum, M.; Bayrak, C.; Zheng, D.; Lachman, H.M. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Curatolo, P.; Moavero, R.; de Vries, P.J. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol. 2015, 14, 733–745. [Google Scholar] [CrossRef]
- Sabatini, D.M. Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. USA 2017, 114, 11818–11825. [Google Scholar] [CrossRef]
- Mizuguchi, M.; Takashima, S. Neuropathology of tuberous sclerosis. Brain Dev. 2001, 23, 508–515. [Google Scholar] [CrossRef]
- Blair, J.D.; Hockemeyer, D.; Bateup, H.S. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat. Med. 2018, 24, 1568–1578. [Google Scholar] [CrossRef]
- Blair, J.D.; Bateup, H.S. New frontiers in modeling tuberous sclerosis with human stem cell-derived neurons and brain organoids. Dev. Dyn 2019. [Google Scholar] [CrossRef] [PubMed]
- Crino, P.B. The mTOR signalling cascade: Paving new roads to cure neurological disease. Nat. Rev. Neurol. 2016, 12, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, N.; Jablonska, B.; Scafidi, J.; Vaccarino, F.M.; Gallo, V. Neurobiology of premature brain injury. Nat. Neurosci. 2014, 17, 341–346. [Google Scholar] [CrossRef]
- Fels, D.R.; Koumenis, C. The PERK/eIF2α/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol. Ther. 2006, 5, 723–728. [Google Scholar] [CrossRef]
- Mestas, J.; Hughes, C.C.W. Of Mice and Not Men: Differences between Mouse and Human Immunology. J. Immunol. 2004, 172, 2731–2738. [Google Scholar] [CrossRef]
- Takao, K.; Miyakawa, T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA 2015, 112, 1167–1172. [Google Scholar] [CrossRef]
- Aragao, M.D.F.V.; van der Linden, V.; Brainer-Lima, A.M.; Coeli, R.R.; Rocha, M.A.; da Silva, P.S.; de Carvalho, M.D.C.G.; van der Linden, A.; de Holanda, A.C.; Valenca, M.M. Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: Retrospective case series study. BMJ 2016, 353, i1901. [Google Scholar] [CrossRef]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika Virus and Birth Defects-Reviewing the Evidence for Causality. Available online: https://www.nejm.org/doi/10.1056/NEJMsr1604338 (accessed on 26 July 2019).
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.M.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef]
- Garcez, P.P.; Loiola, E.C.; da Costa, R.M.; Higa, L.M.; Trindade, P.; Delvecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef]
- Li, Y.; Muffat, J.; Omer, A.; Bosch, I.; Lancaster, M.A.; Sur, M.; Gehrke, L.; Knoblich, J.A.; Jaenisch, R. Induction of expansion and folding in human cerebral organoids. Cell Stem Cell 2017, 20, 385–396.e3. [Google Scholar] [CrossRef] [PubMed]
- Kalay, E.; Yigit, G.; Aslan, Y.; Brown, K.E.; Pohl, E.; Bicknell, L.S.; Kayserili, H.; Li, Y.; Tüysüz, B.; Nürnberg, G.; et al. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat. Genet. 2011, 43, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, E.; Wason, A.; Ramani, A.; Gooi, L.M.; Keller, P.; Pozniakovsky, A.; Poser, I.; Noack, F.; Telugu, N.S.; Calegari, F.; et al. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J. 2016, 35, 803–819. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, E.; Ramani, A.; Karow, U.; Gottardo, M.; Natarajan, K.; Gooi, L.M.; Goranci-Buzhala, G.; Krut, O.; Peters, F.; Nikolic, M.; et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell 2017, 20, 397–406.e5. [Google Scholar] [CrossRef]
- Nowakowski, T.J.; Pollen, A.A.; Di Lullo, E.; Sandoval-Espinosa, C.; Bershteyn, M.; Kriegstein, A.R. Expression Analysis Highlights AXL as a Candidate Zika Virus Entry Receptor in Neural Stem Cells. Cell Stem Cell 2016, 18, 591–596. [Google Scholar] [CrossRef]
- Wells, M.F.; Salick, M.R.; Wiskow, O.; Ho, D.J.; Worringer, K.A.; Ihry, R.J.; Kommineni, S.; Bilican, B.; Klim, J.R.; Hill, E.J.; et al. Genetic Ablation of AXL Does Not Protect Human Neural Progenitor Cells and Cerebral Organoids from Zika Virus Infection. Cell Stem Cell 2016, 19, 703–708. [Google Scholar] [CrossRef]
- Xu, M.; Lee, E.M.; Wen, Z.; Cheng, Y.; Huang, W.-K.; Qian, X.; Tcw, J.; Kouznetsova, J.; Ogden, S.C.; Hammack, C.; et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016, 22, 1101–1107. [Google Scholar] [CrossRef]
- Zhou, T.; Tan, L.; Cederquist, G.Y.; Fan, Y.; Hartley, B.J.; Mukherjee, S.; Tomishima, M.; Brennand, K.J.; Zhang, Q.; Schwartz, R.E.; et al. High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 2017, 21, 274–283.e5. [Google Scholar] [CrossRef]
- Xu, Y.-P.; Qiu, Y.; Zhang, B.; Chen, G.; Chen, Q.; Wang, M.; Mo, F.; Xu, J.; Wu, J.; Zhang, R.-R.; et al. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 2019, 29, 265. [Google Scholar] [CrossRef]
- Neu, N.; Duchon, J.; Zachariah, P. TORCH infections. Clin. Perinatol. 2015, 42, 77–103. [Google Scholar] [CrossRef]
- Kimberlin, D.W. Neonatal herpes simplex infection. Clin. Microbiol. Rev. 2004, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Muller, W.J.; Jones, C.A.; Koelle, D.M. Immunobiology of herpes simplex virus and cytomegalovirus infections of the fetus and newborn. Curr. Immunol. Rev. 2010, 6, 38–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knipe, D.M.; Raja, P.; Lee, J.S. Clues to mechanisms of herpesviral latent infection and potential cures. Proc. Natl. Acad. Sci. USA 2015, 112, 11993–11994. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, L.; Bonasoni, M.P.; Santini, D.; Piccirilli, G.; Chiereghin, A.; Petrisli, E.; Dolcetti, R.; Guerra, B.; Piccioli, M.; Lanari, M.; et al. Congenital cytomegalovirus infection: Patterns of fetal brain damage. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, E419–E427. [Google Scholar]
- Gabrielli, L.; Bonasoni, M.P.; Lazzarotto, T.; Lega, S.; Santini, D.; Foschini, M.P.; Guerra, B.; Baccolini, F.; Piccirilli, G.; Chiereghin, A.; et al. Histological findings in foetuses congenitally infected by cytomegalovirus. J. Clin. Virol. 2009, 46, S16–S21. [Google Scholar] [CrossRef]
- D’Aiuto, L.; Bloom, D.C.; Naciri, J.N.; Smith, A.; Edwards, T.G.; McClain, L.; Callio, J.A.; Jessup, M.; Wood, J.; Chowdari, K.; et al. Modeling Herpes Simplex Virus 1 Infections in Human Central Nervous System Neuronal Cells Using Two- and Three-Dimensional Cultures Derived from Induced Pluripotent Stem Cells. J. Virol. 2019, 93, e00111–e00119. [Google Scholar] [CrossRef]
- Rock, D.L.; Fraser, N.W. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature 1983, 302, 523. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. The Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.Ø.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In vivo models of primary brain tumors: Pitfalls and perspectives. Neuro-Oncol. 2012, 14, 979–993. [Google Scholar] [CrossRef]
- Ben-David, U.; Ha, G.; Tseng, Y.-Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, F.; Morishita, W.; Zuniga, E.; Nguyen, J.; Blank, M.; Malenka, R.C.; Garner, C.C. Pharmacotherapy for cognitive impairment in a mouse model of Down syndrome. Nat. Neurosci. 2007, 10, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep 2019, 26, 3203–3211.e5. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Cho, D.-Y.; Lin, S.-Z.; Yang, W.-K.; Lee, H.-C.; Hsu, D.-M.; Lin, H.-L.; Chen, C.-C.; Liu, C.-L.; Lee, W.-Y.; Ho, L.-H. Targeting cancer stem cells for treatment of glioblastoma multiforme. Cell Transplant. 2013, 22, 731–739. [Google Scholar] [CrossRef]
- Ohgaki, H.; Kleihues, P. Epidemiology and etiology of gliomas. Acta Neuropathol. (Berl.) 2005, 109, 93–108. [Google Scholar] [CrossRef]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef]
- Ellison, D.; Love, S.; Chimelli, L.M.C.; Harding, B.; Lowe, J.S.; Vinters, H.V.; Brandner, S.; Yong, W.H. Neuropathology E-Book: A Reference Text of CNS Pathology; Elsevier Health Sciences, 2012; ISBN 978-0-7234-3746-8. [Google Scholar]
- Liu, C.; Oikonomopoulos, A.; Sayed, N.; Wu, J.C. Modeling human diseases with induced pluripotent stem cells: From 2D to 3D and beyond. Dev. Camb. Engl. 2018. [Google Scholar] [CrossRef]
- Decimo, I.; Bifari, F.; Krampera, M.; Fumagalli, G. Neural Stem Cell Niches in Health and Diseases. Curr. Pharm. Des. 2012, 18, 1755–1783. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.-R.; Ueno, Y.; Zheng, Y.-W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-F.; Su, J.; Kim, H.S.; Chang, B.; Papatsenko, D.; Zhao, R.; Yuan, Y.; Gingold, J.; Xia, W.; Darr, H.; et al. Modeling familial cancer with induced pluripotent stem cells. Cell 2015, 161, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wert, K.J.; Shvartsman, D.; Melton, D.A.; Jaenisch, R. Establishment of human pluripotent stem cell-derived pancreatic β-like cells in the mouse pancreas. Proc. Natl. Acad. Sci. USA. 2018, 115, 3924–3929. [Google Scholar] [CrossRef]
- Dye, B.R.; Dedhia, P.H.; Miller, A.J.; Nagy, M.S.; White, E.S.; Shea, L.D.; Spence, J.R. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 2016. [Google Scholar] [CrossRef]
- Sugimura, R.; Jha, D.K.; Han, A.; Soria-Valles, C.; da Rocha, E.L.; Lu, Y.-F.; Goettel, J.A.; Serrao, E.; Rowe, R.G.; Malleshaiah, M.; et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 2017, 545, 432–438. [Google Scholar] [CrossRef]
- Assawachananont, J.; Mandai, M.; Okamoto, S.; Yamada, C.; Eiraku, M.; Yonemura, S.; Sasai, Y.; Takahashi, M. Transplantation of Embryonic and Induced Pluripotent Stem Cell-Derived 3D Retinal Sheets into Retinal Degenerative Mice. Stem Cell Rep. 2014, 2, 662–674. [Google Scholar] [CrossRef]
- Xu, R.; Brawner, A.T.; Li, S.; Liu, J.-J.; Kim, H.; Xue, H.; Pang, Z.P.; Kim, W.-Y.; Hart, R.P.; Liu, Y.; et al. OLIG2 Drives Abnormal Neurodevelopmental Phenotypes in Human iPSC-Based Organoid and Chimeric Mouse Models of Down Syndrome. Cell Stem Cell 2019, 24, 908–926.e8. [Google Scholar] [CrossRef]
- Aguayo, A.J.; Kasarjian, J.; Skamene, E.; Kongshavn, P.; Bray, G.M. Myelination of mouse axons by Schwann cells transplanted from normal and abnormal human nerves. Nature 1977, 268, 753–755. [Google Scholar] [CrossRef]
- Reubinoff, B.E.; Itsykson, P.; Turetsky, T.; Pera, M.F.; Reinhartz, E.; Itzik, A.; Ben-Hur, T. Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1134–1140. [Google Scholar] [CrossRef]
- Maria, S.; Helle, B.; Tristan, L.; Gaynor, S.; Arnar, A.; Michele, M.; Teresia, O.; Oliver, C.; Roger, S.; Penelope, H.; et al. Improved cell therapy protocol for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC and non-human primate iPSC-derived DA neurons. Stem Cells Dayt. Ohio 2013, 31, 1548–1562. [Google Scholar]
- Yuan, T.; Liao, W.; Feng, N.-H.; Lou, Y.-L.; Niu, X.; Zhang, A.-J.; Wang, Y.; Deng, Z.-F. Human induced pluripotent stem cell-derived neural stem cells survive, migrate, differentiate, and improve neurologic function in a rat model of middle cerebral artery occlusion. Stem Cell Res. Ther. 2013, 4, 73. [Google Scholar] [CrossRef] [PubMed]
- Basuodan, R.; Basu, A.P.; Clowry, G.J. Human neural stem cells dispersed in artificial ECM form cerebral organoids when grafted in vivo. J. Anat. 2018, 233, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Daviaud, N.; Friedel, R.H.; Zou, H. Vascularization and Engraftment of Transplanted Human Cerebral Organoids in Mouse Cortex. eNeuro 2018. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Kratochvil, M.J.; Seymour, A.J.; Li, T.L.; Paşca, S.P.; Kuo, C.J.; Heilshorn, S.C. Engineered materials for organoid systems. Nat. Rev. Mater. 2019, 4, 606–622. [Google Scholar] [CrossRef]
- Takebe, T.; Enomura, M.; Yoshizawa, E.; Kimura, M.; Koike, H.; Ueno, Y.; Matsuzaki, T.; Yamazaki, T.; Toyohara, T.; Osafune, K.; et al. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell Stem Cell 2015, 16, 556–565. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666. [Google Scholar] [CrossRef]
- Ormel, P.R.; Vieira de Sá, R.; van Bodegraven, E.J.; Karst, H.; Harschnitz, O.; Sneeboer, M.A.M.; Johansen, L.E.; van Dijk, R.E.; Scheefhals, N.; Berdenis van Berlekom, A.; et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018, 9, 4167. [Google Scholar] [CrossRef]
- Marton, R.M.; Miura, Y.; Sloan, S.A.; Li, Q.; Revah, O.; Levy, R.J.; Huguenard, J.R.; Pașca, S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019, 22, 484–491. [Google Scholar] [CrossRef]
- Amin, N.D.; Paşca, S.P. Building Models of Brain Disorders with Three-Dimensional Organoids. Neuron 2018, 100, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Tiriac, H.; Sridharan, B.P.; Scampavia, L.; Madoux, F.; Seldin, J.; Souza, G.R.; Watson, D.; Tuveson, D.; Spicer, T.P. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS Discov. Adv. Life Sci. 2018, 23, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Phan, N.; Hong, J.J.; Tofig, B.; Mapua, M.; Elashoff, D.; Moatamed, N.A.; Huang, J.; Memarzadeh, S.; Damoiseaux, R.; Soragni, A. A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Czerniecki, S.M.; Cruz, N.M.; Harder, J.L.; Menon, R.; Annis, J.; Otto, E.A.; Gulieva, R.E.; Islas, L.V.; Kim, Y.K.; Tran, L.M.; et al. High-throughput automation enhances kidney organoid differentiation from human pluripotent stem cells and enables multidimensional phenotypic screening. Cell Stem Cell 2018, 22, 929–940. [Google Scholar] [CrossRef] [PubMed]
| Disease | Organoid Type | Days of Differentiation | Phenotype and Rescue | Unique Experimental Feature | Protocol |
|---|---|---|---|---|---|
| Timothy Syndrome [41] | Ventral and dorsal forebrain, assembloids | 4 weeks | GABAergic interneuron abnormalities: altered saltation frequency and shorter saltation length; phenotype rescue by pharmacological modulation of l-type calcium channels | Forebrain assembloids with labelling of specific cell type (Dlxi1/2b::eGFP) | [42] |
| Autism Spectrum Disorder [43] | Dorsal forebrain | 6 weeks | Transcriptome dysregulation: FOXG1 upregulation; increased production of NPCs and GABAergic neurons; rescue by shRNA attenuation of FOXG1 expression | Lentiviral-mediated expression of shRNA-FOXG1 | [23] |
| Miller-Dieker Syndrome [44] | Forebrain | 4 weeks | Smaller organoids with reduced neuroepithelial loops, impaired vRG divisions, disrupted cortical niche; rescue by gene re-expression or β-catenin activation | Doxycycline-inducible overexpression of LIS1 gene | [45,46] |
| Autosomal recessive primary microcephaly [22] | Undirected | ~3 weeks | Altered vRG morphology and orientation, smaller organoids; rescue by shRNA for CDK5RAP2 | Electroporation-mediated overexpression of CDK5RAP2 and shRNA-CDK5RAP2 | [22] |
| Hypoxic injury [47] | Forebrain | ~11 weeks | Disruption of intermediate progenitors in SVZ; impaired UPR pathway activation and premature neuronal differentiation; rescue by stress response inhibitor (ISRIB) | Gas control chamber and needle-type fiber-optic microsensor to reproduce and monitor low oxygen exposure | [42] |
| Zika virus infection (ZIKV) [29] | Forebrain | ~3 months | Smaller organoids with reduced thickness and increased ventricular lumen, ZIKV-induced cell apoptosis | ZIKV strains: MR766 and FSS13025 (99% amino acid sequence homology to Brazilian ZIKV) | [29] |
| Cytomegalovirus infection (CMV) [48] | Undirected | 8 weeks | Reduced cell proliferation, necrosis, vacuolar and cystic degeneration; impaired cortical lamination | Organoid differentiation from CMV-infected hiPSCs | [46] |
| Creutzfeld-jakob disease (CJD) [49] | Undirected | 5 months | Slow metabolism, protease-resistant PrP deposition, acquired prion seeding activity, increased astrocyte activation | Organoids inoculation with human brain homogenates from sporadic CJD subtypes | [46] |
| Brain tumors [50] | Forebrain | ~4 months | Glioma-like with poor glial differentiation, high cell proliferation, disorganized architecture and downregulation of PI3K-AKT, RAS pathways; CNS-PTEN-like with WNT, TGFβ, and TP53 upregulation; tumor invasiveness upon in vivo transplantation; partial rescue in glioma by EGFR-inhibitors | Electroporation-mediated plasmid nucleofection with overexpression of MYC/inhibition of tumor suppressor genes | [22] |
| PROS | CONS | APPROACH |
|---|---|---|
| 3D multicellular architecture with complex cell compartmentalization | Lack of reproducibility |
|
| Patterning into different brain-like subregions | Cost |
|
| Rough organization into cortical layers | Lack of output and input systems |
|
| Long term culturing | Lack of vascular bed |
|
| Generation of patients’specific disease-relevant cell types | Spatial orientation |
|
| Generation of spontaneously active neural networks | Long term maturation |
|
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costamagna, G.; Andreoli, L.; Corti, S.; Faravelli, I. iPSCs-Based Neural 3D Systems: A Multidimensional Approach for Disease Modeling and Drug Discovery. Cells 2019, 8, 1438. https://doi.org/10.3390/cells8111438
Costamagna G, Andreoli L, Corti S, Faravelli I. iPSCs-Based Neural 3D Systems: A Multidimensional Approach for Disease Modeling and Drug Discovery. Cells. 2019; 8(11):1438. https://doi.org/10.3390/cells8111438
Chicago/Turabian StyleCostamagna, Gianluca, Luca Andreoli, Stefania Corti, and Irene Faravelli. 2019. "iPSCs-Based Neural 3D Systems: A Multidimensional Approach for Disease Modeling and Drug Discovery" Cells 8, no. 11: 1438. https://doi.org/10.3390/cells8111438
APA StyleCostamagna, G., Andreoli, L., Corti, S., & Faravelli, I. (2019). iPSCs-Based Neural 3D Systems: A Multidimensional Approach for Disease Modeling and Drug Discovery. Cells, 8(11), 1438. https://doi.org/10.3390/cells8111438






