Autophagy and Nutrients Management in Plants
Abstract
1. Introduction
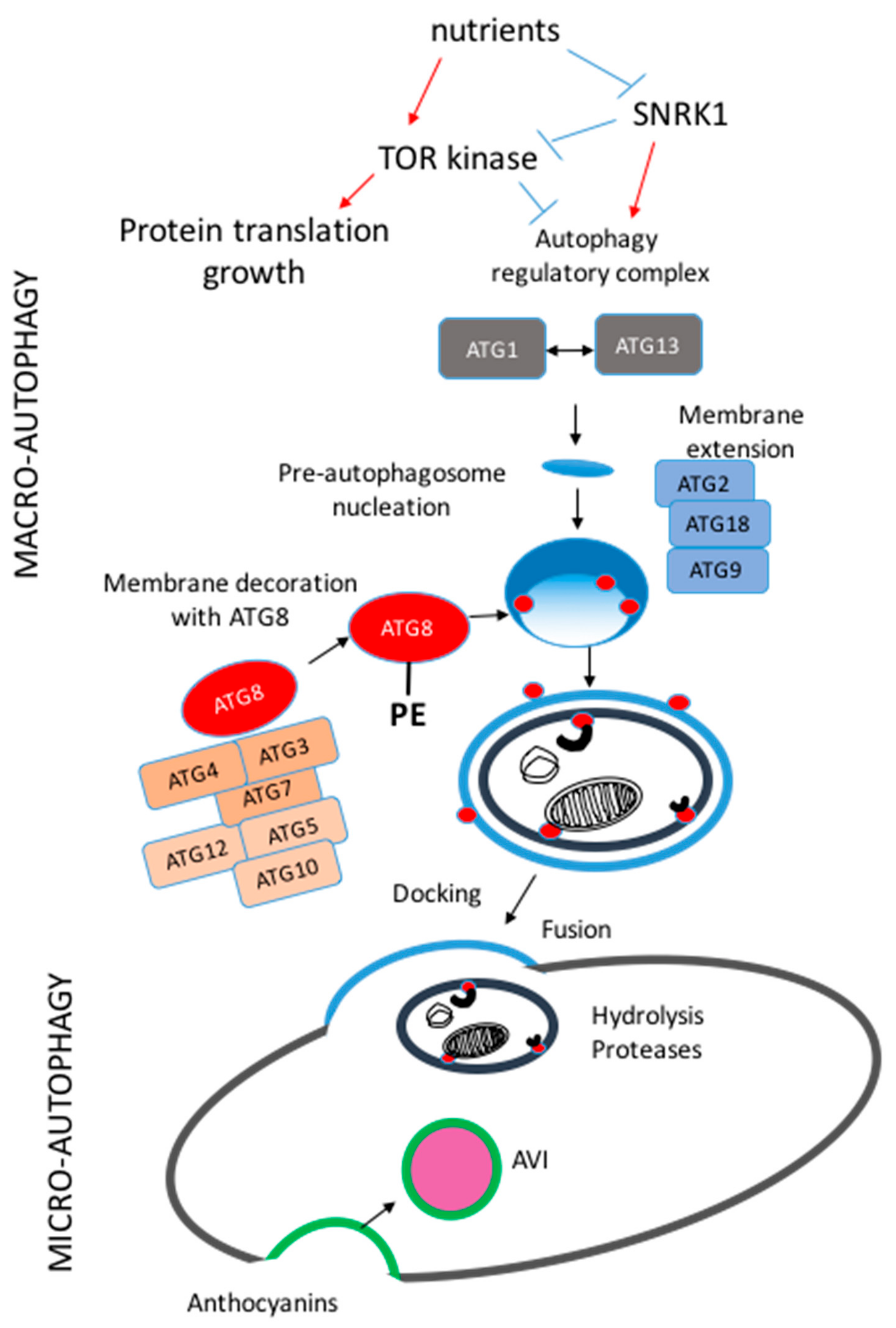
2. Molecular Machinery of Macro-Autophagy in Plants
3. Selective Macro-Autophagy
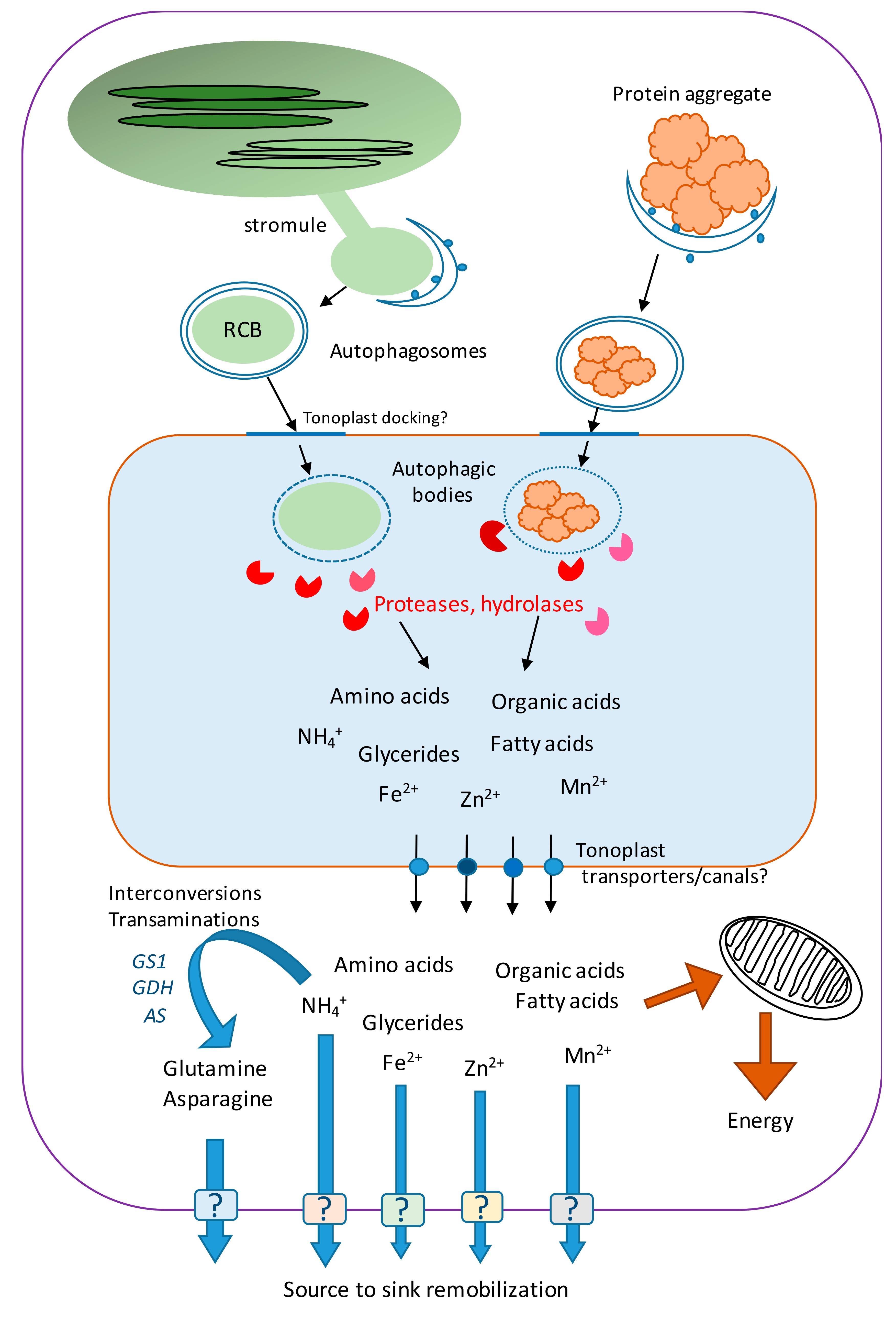
4. Nutrient Remobilization after Organelle and Protein Degradation in Senescing Leaves
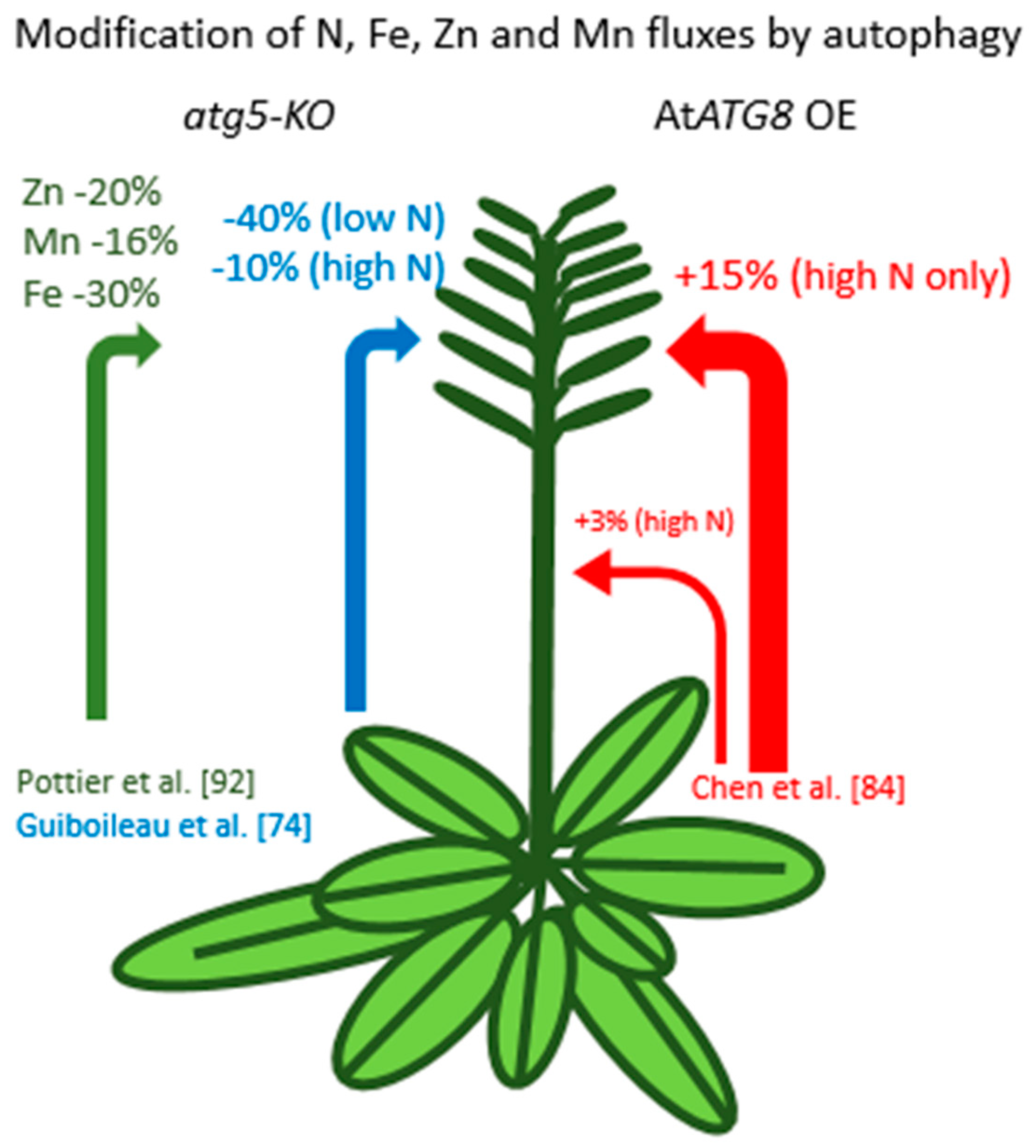
5. Role of Autophagy in Nitrogen Recycling
6. Cross-Talk between Autophagy and Senescence-Related Cysteine Proteases
7. Autophagy and Other Nutrients
8. Conclusions
Funding
Conflicts of Interest
References
- Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Chen, Q.; Havé, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant. Biol. 2017, 39, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant. Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Irani, N.G.; Lu, Y.; Riedl, K.; Schwartz, S.; Grotewold, E. The Formation of anthocyanic vacuolar inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant. 2010, 3, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Chanoca, A.; Kovinich, N.; Burkel, B.; Stecha, S.; Bohorquez-Restrepo, A.; Ueda, T.; Eliceiri, K.W.; Grotewold, E.; Otegui, M.S. Anthocyanin vacuolar inclusions form by a microautophagy mechanism. Plant. Cell 2015, 27, 2545–2559. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Clément, G.; Anne, P.; Routaboul, J.; Guiboileau, A.; Soulay, F.; Shirasu, K.; Yoshimoto, K. Stitching together the multiple dimensions of autophagy using metabolomic and transcriptomic analyses reveals new impacts of autophagy defects on metabolism, development and plant response to environment. Plant. Cell 2014, 26, 1857–1877. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Vierstra, R.D. Autophagic recycling: Lessons from yeast help define the process in plants. Curr Opin. Plant. Biol. 2005, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G.; Papini, A. Ultrastructure of autophagy in plant cells: A review. Autophagy 2013, 9, 1922–1936. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.I.; Cho, H.J.; Jung, J.H.; Yoshimoto, K.; Shirasu, K.; Park, O.K. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010, 64, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Galili, G. Degradation of organelles or specific organelle components via selective autophagy in plant cells. Int. J. Mol. Sci. 2014, 15, 7624–7638. [Google Scholar] [CrossRef] [PubMed]
- Avila-Ospina, L.; Marmagne, A.; Soulay, F.; Masclaux-Daubresse, C. Identification of Barley (Hordeum vulgare L.) Autophagy genes and their expression levels during leaf senescence, chronic nitrogen limitation and in response to dark exposure. Agron-Basel 2016, 6, 15. [Google Scholar] [CrossRef]
- Doelling, J.H.; Walker, J.M.; Friedman, E.M.; Thompson, A.R.; Vierstra, R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 33105–33114. [Google Scholar] [CrossRef] [PubMed]
- Hanaoka, H.; Noda, T.; Shirano, Y.; Kato, T.; Hayashi, H.; Shibata, D.; Tabata, S.; Ohsumi, Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant. Physiol. 2002, 129, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.R.; Doelling, J.H.; Suttangkakul, A.; Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant. Physiol. 2005, 138, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant. J. 2005, 42, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in Rice (Oryza sativa L.). Dna. Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Pei, D.; Zhang, W.; Sun, H.; Wei, X.; Yue, J.; Wang, H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant. Cell Rep. 2014, 33, 1697–1710. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.J.; Suttangkakul, A.; Vierstra, R.D. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant. Physiol. 2009, 149, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.-m.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.-h.; Peng, X.-b.; Sun, M.-x. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. Dna. Res. 2015, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Chen, M.; Wang, E.H.; Hu, L.Q.; Hawkesford, M.J.; Zhong, L.; Chen, Z.; Xu, Z.S.; Li, L.C.; Zhou, Y.B.; et al. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. Bmc. Genom. 2016, 17, 797. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, X.; Jia, X.; Wang, N.; Gong, X.; Ma, F. characterization of an autophagy-related gene MdATG8i from apple. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vierstra, R.D. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 2014, 10, 1466–1467. [Google Scholar] [CrossRef] [PubMed]
- Le Bars, R.; Marion, J.; Le Borgne, R.; Satiat-Jeunemaitre, B.; Bianchi, M.W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Chung, K.P.; Cui, Y.; Lin, W.; Gao, C.; Kang, B.-H.; Jiang, L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 4, E426–E435. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Chung, K.P.; Luo, M.; Jiang, L. Autophagosome biogenesis and the endoplasmic reticulum: A plant perspective. Trends Plant Sci. 2018, 23, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Marshall, R.S.; Li, F. Understanding and exploiting the roles of autophagy in plants through multi-omics approaches. Plant. Sci. 2018, 274, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant. Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.; Park, E.; Dinesh-Kumar, S.P. Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc. Natl. Acad. Sci. USA 2014, 111, 863–868. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.R.; Suttangkakul, A.; Vierstra, R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008, 178, 1339–1353. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant. J. 2010, 62, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Dobrenel, T.; Marchive, C.; Sormani, R.; Moreau, M.; Mozzo, M.; Montane, M.H.; Menand, B.; Robaglia, C.; Meyer, C. Regulation of plant growth and metabolism by the TOR kinase. Biochem. Soc. Trans. 2011, 39, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.B.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181. [Google Scholar] [CrossRef] [PubMed]
- Soto-Burgos, J.; Bassham, D.C. SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Bassham, D.C. Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Floyd, B.E.; Morriss, S.C.; MacIntosh, G.C.; Bassham, D.C. What to eat: Evidence for selective autophagy in plants. J. Integr. Plant. Biol. 2012, 54, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vierstra, R.D. Regulator and substrate Dual roles for the ATG1-ATG13 kinase complex during autophagic recycling in Arabidopsis. Autophagy 2012, 8. [Google Scholar] [CrossRef][Green Version]
- Farre, J.C.; Subramani, S. Mechanistic insights into selective autophagy pathways: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2016, 17, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Ohsumi, Y.; Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010, 584, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Hua, Z.H.; Mali, S.; McLoughlin, F.; Vierstra, R.D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 2019, 177, 766. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.P.; Nair, U.; Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Dynamic regulation of the 26s proteasome: From synthesis to degradation. frontiers in molecular biosciences. Front. Mol. Biosci. 2019, 6. [Google Scholar] [CrossRef]
- Yoshimoto, K.; Ohsumi, Y. Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant. Cell Physiol. 2018, 59, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzynska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.-J.; Fan, B.; Yu, J.-Q.; Chen, Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Lee, H.; Marshall, R.S.; Lomax, A.; Yoon, M.J.; Kim, J.; Kim, J.H.; Vierstra, R.D.; Chung, T. NBR1 mediates selective autophagy of defective proteins in Arabidopsis. J. Exp. Bot. 2019. [Google Scholar] [CrossRef] [PubMed]
- Hafren, A.; Macia, J.L.; Love, A.J.; Milner, J.J.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, E2026–E2035. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Belhaj, K.; Maqbool, A.; Chaparro-Garcia, A.; Pandey, P.; Petre, B.; Tabassum, N.; Cruz-Mireles, N.; Hughes, R.K.; Sklenar, J.; et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Pandey, P.; Tumtas, Y.; Sanguankiattichai, N.; Belhaj, K.; Duggan, C.; Leary, A.Y.; Segretin, M.E.; Contreras, M.P.; Savage, Z.; et al. Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Honig, A.; Avin-Wittenberg, T.; Galili, G. Selective autophagy in the aid of plant germination and response to nutrient starvation. Autophagy 2012, 8, 838–839. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Avin-Wittenberg, T.; Galili, G. Involvement of autophagy in the direct ER to vacuole protein trafficking route in plants. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Li, F.Q.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26S proteasome Is mediated by the dual ATG8/Ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Vanhee, C.; Batoko, H. Autophagy involvement in responses to abscisic acid by plant cells. Autophagy 2011, 7, 655–656. [Google Scholar] [CrossRef] [PubMed]
- Hachez, C.; Veljanovski, V.; Reinhardt, H.; Guillaumot, D.; Vanhee, C.; Chaumont, F.; Batoko, H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant. Cell 2014, 26, 4974–4990. [Google Scholar] [CrossRef] [PubMed]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef] [PubMed]
- Kichey, T.; Hirel, B.; Heumez, E.; Dubois, F.; Le Gouis, J. In winter wheat (Triticum aestivum L.), post-anthesis nitrogen uptake and remobilisation to the grain correlates with agronomic traits and nitrogen physiological markers. Field Crop Res 2007, 102, 22–32. [Google Scholar] [CrossRef]
- Hirel, B.; Le Gouis, J.; Ney, B.; Gallais, A. The challenge of improving nitrogen use efficiency in crop plants: Towards a more central role for genetic variability and quantitative genetics within integrated approaches. J. Exp. Bot. 2007, 58, 2369–2387. [Google Scholar] [CrossRef] [PubMed]
- Hortensteiner, S.; Feller, U. Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 2002, 53, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Have, M.; Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar] [CrossRef] [PubMed]
- Lohman, K.N.; Gan, S.; John, M.C.; Amasino, R.M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 1994, 92, 322–328. [Google Scholar] [CrossRef]
- Pružinská, A.; Shindo, T.; Niessen, S.; Kaschani, F.; Tóth, R.; Millar, A.H.; van der Hoorn, R.A.L. Major Cys protease activities are not essential for senescence in individually darkened Arabidopsis leaves. Bmc. Plant. Biol. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Chiba, A.; Ishida, H.; Nishizawa, N.K.; Makino, A.; Mae, T. Exclusion of ribulose-1,5-biphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant. Cell Physiol. 2003, 44, 914–921. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Izumi, M.; Wada, S.; Makino, A. Roles of autophagy in chloroplast recycling. Biochim. Et Biophys. ACTA-Bioenerg. 2014, 1837, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Yoshimoto, K.; Izumi, M.; Reisen, D.; Yano, Y.; Makino, A.; Ohsumi, Y.; Hanson, M.R.; Mae, T. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant. Physiol. 2008, 148, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Wada, S.; Makino, A.; Ishida, H. The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiology 2010, 154, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Ishida, H.; Nakamura, S.; Hidema, J. Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant. Cell 2017, 29, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Hidema, J.; Wada, S.; Kondo, E.; Kurusu, T.; Kuchitsu, K.; Makino, A.; Ishida, H. Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant. Physiol. 2015, 167, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Nakamura, S.; Li, N. Autophagic Turnover of chloroplasts: Its roles and regulatory mechanisms in response to sugar starvation. Front. Plant. Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Hidema, J.; Sakamoto, W.; Ishida, H.; Izumi, M. selective elimination of membrane-damaged chloroplasts via microautophagy. Plant. Physiol. 2018, 177, 1007–1026. [Google Scholar] [CrossRef] [PubMed]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataillé, M.; Avice, J.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef] [PubMed]
- Guiboileau, A.; Avila-Ospina, L.; Yoshimoto, K.; Soulay, F.; Azzopardi, M.; Marmagne, A.; Lothier, J.; Masclaux-Daubresse, C. Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 2013, 199, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic Recycling plays a central role in maize nitrogen remobilization. Plant. Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Hayashida, Y.; Izumi, M.; Kurusu, T.; Hanamata, S.; Kanno, K.; Kojima, S.; Yamaya, T.; Kuchitsu, K.; Makino, A.; et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant. Physiol. 2015, 168, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.S.; Elander, P.H.; Dalman, K.; Beganovic, M.; Yilmaz, J.L.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef] [PubMed]
- Xia, T.; Xiao, D.; Liu, D.; Chai, W.; Gong, Q.; Wang, N.N. Heterologous expression of ATG8c from Soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Chen, M.; Zhong, L.; Liu, J.M.; Xu, Z.S.; Li, L.C.; Zhou, Y.B.; Guo, C.H.; Ma, Y.Z. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 2015, 468, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.M.; Zhang, P.P.; Zhu, R.H.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.Y.; Wang, D.; Gong, Q.Q. autophagy is rapidly induced by salt stress and is required for salt tolerance in Arab. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Sun, X.; Jia, X.; Huo, L.Q.; Che, R.M.; Gong, X.Q.; Wang, P.; Ma, F.W. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant. Cell Env. 2018, 41, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, P.; Jia, X.; Huo, L.Q.; Che, R.M.; Ma, F.W. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant. Biotechnol. J. 2018, 16, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Soulay, F.; Saudemont, J.; Elmayan, T.; Marmagne, A.; Masclaux-Daubresse, C. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant. Cell Physiol. 2019, 60, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhen, X.; Li, X.; Li, N.; Xu, F. Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- James, M.; Masclaux-Daubresse, C.; Marmagne, A.; Azzopardi, M.; Laine, P.; Goux, D.; Etienne, P.; Trouverie, J. A new role for SAG12 cysteine protease in roots of Arabidopsis thaliana. Front. Plant. Sci. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- James, M.; Poret, M.; Masclaux-Daubresse, C.; Marmagne, A.; Coquet, L.; Jouenne, T.; Chan, P.; Trouverie, J.; Etienne, P. SAG12, a major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant. Cell Physiol. 2018, 59, 2052–2063. [Google Scholar] [CrossRef] [PubMed]
- Havé, M.; Balliau, T.; Cottyn-Boitte, B.; Derond, E.; Cueff, G.; Soulay, F.; Lornac, A.; Reichman, P.; Dissmeyer, N.; Avice, J.-C.; et al. Increase of proteasome and papain-like cysteine protease activities in autophagy mutants: backup compensatory effect or pro cell-death effect? J. Exp. Bot. 2018, 69, 1369–1385. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Murakami, S.; Yamamoto, Y.; Chatani, H.; Kondo, Y.; Nakano, T.; Yokota, A.; Sato, F. The DNA-binding protease, CND41, and the degradation of ribulose-1,5-bisphosphate carboxylase/oxygenase in senescent leaves of tobacco. Planta 2004, 220, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Mancias, J.D.; Wang, X.X.; Gygi, S.P.; Harper, J.W.; Kimmelman, A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 509, 105. [Google Scholar] [CrossRef] [PubMed]
- Santana-Codina, N.; Mancias, J.D. The Role of NCOA4-Mediated Ferritinophagy in Health and Disease. Pharmaceuticals 2018, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Pottier, M.; Dumont, J.; Masclaux-Daubresse, C.; Thomine, S. Autophagy is essential for optimal translocation of iron to seeds in Arabidopsis. J. Exp. Bot. 2019, 70, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, M.; Kimura, K.; Makino, A.; Ishida, H. Autophagy is induced under Zn limitation and contributes to Zn-limited stress tolerance in Arabidopsis (Arabidopsis thaliana). Soil Sci. Plant Nutr. 2017, 63, 342–350. [Google Scholar] [CrossRef]
- Naumann, C.; Mueller, J.; Sakhonwasee, S.; Wieghaus, A.; Hause, G.; Heisters, M.; Buerstenbinder, K.; Abel, S. The local phosphate deficiency response activates endoplasmic reticulum stress-dependent autophagy. Plant. Physiol. 2019, 179, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Avin-Wittenberg, T.; Bajdzienko, K.; Wittenberg, G.; Alseekh, S.; Tohge, T.; Bock, R.; Giavalisco, P.; Fernie, A.R. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in arabidopsis seedlings under carbon starvation. Plant Cell 2015, 27, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.A.S.; Cavalcanti, J.H.F.; Medeiros, D.B.; Nunes-Nesi, A.; Avin-Wittenberg, T.; Fernie, A.R.; Araújo, W.L. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant. Physiol. 2017, 175, 62–76. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yu, L.; Xu, C.G. Dual role of autophagy in lipid metabolism in Arabidopsis. Plant Cell 2019, 31, 1598–1613. [Google Scholar] [CrossRef] [PubMed]
- Havé, M.; Luo, J.; Tellier, F.; Balliau, T.; Cueff, G.; Chardon, F.; Zivy, M.; Rajjou, L.; Cacas, J.-L.; Masclaux-Daubresse, C. Proteomic and lipidomic analyses of the Arabidopsis atg5 autophagy mutant reveal major changes in ER and peroxisome metabolisms and in lipid composition. New Phytol. 2019, 223, 1461–1477. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Shinozaki, D.; Luo, J.; Pottier, M.; Havé, M.; Marmagne, A.; Reisdorf-Cren, M.; Chardon, F.; Thomine, S.; Yoshimoto, K.; et al. Autophagy and Nutrients Management in Plants. Cells 2019, 8, 1426. https://doi.org/10.3390/cells8111426
Chen Q, Shinozaki D, Luo J, Pottier M, Havé M, Marmagne A, Reisdorf-Cren M, Chardon F, Thomine S, Yoshimoto K, et al. Autophagy and Nutrients Management in Plants. Cells. 2019; 8(11):1426. https://doi.org/10.3390/cells8111426
Chicago/Turabian StyleChen, Qinwu, Daiki Shinozaki, Jie Luo, Mathieu Pottier, Marien Havé, Anne Marmagne, Michèle Reisdorf-Cren, Fabien Chardon, Sébastien Thomine, Kohki Yoshimoto, and et al. 2019. "Autophagy and Nutrients Management in Plants" Cells 8, no. 11: 1426. https://doi.org/10.3390/cells8111426
APA StyleChen, Q., Shinozaki, D., Luo, J., Pottier, M., Havé, M., Marmagne, A., Reisdorf-Cren, M., Chardon, F., Thomine, S., Yoshimoto, K., & Masclaux-Daubresse, C. (2019). Autophagy and Nutrients Management in Plants. Cells, 8(11), 1426. https://doi.org/10.3390/cells8111426






