Predicting Disease Related microRNA Based on Similarity and Topology
Abstract
1. Introduction
2. Materials and Methods
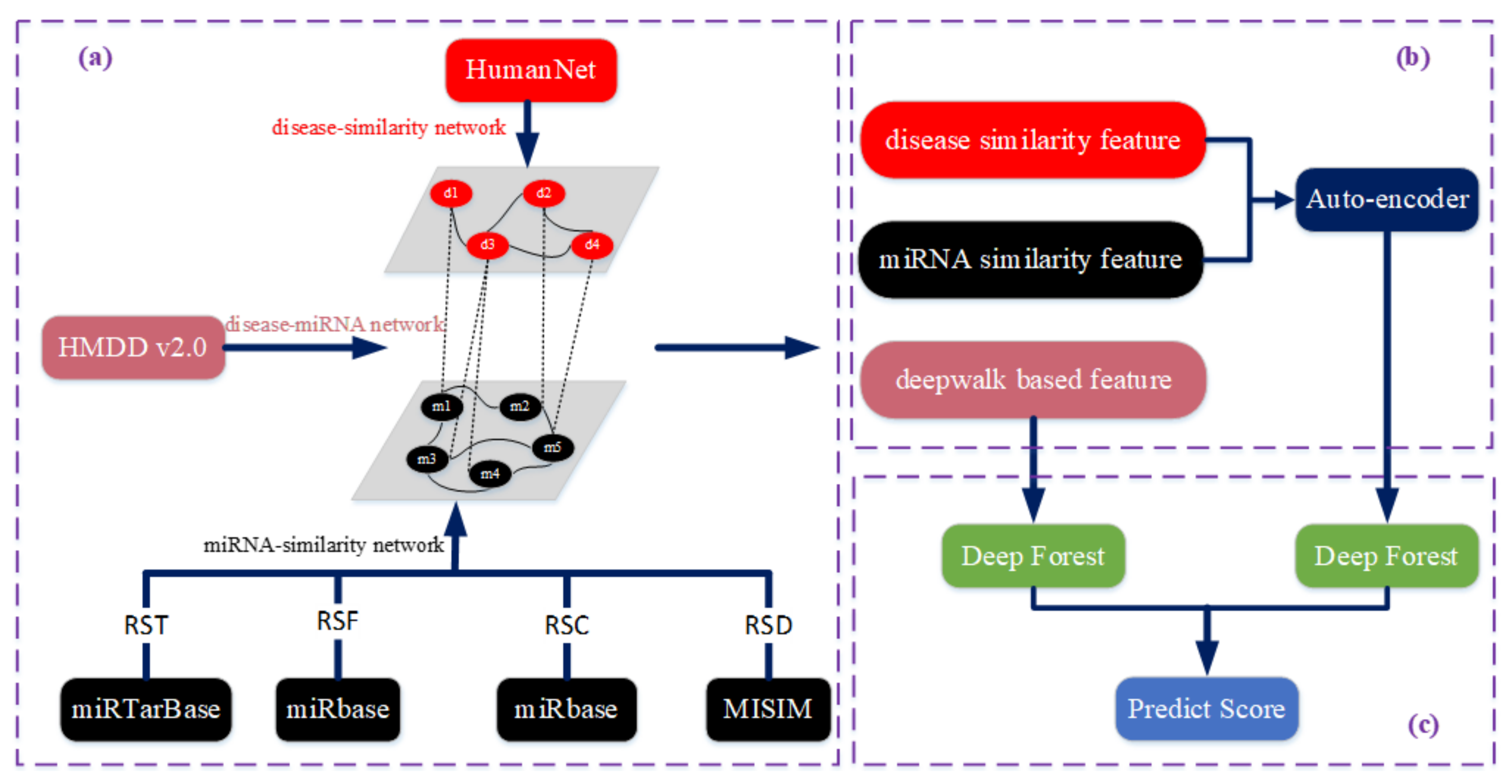
2.1. Constructing the Bilayer Network
2.2. Feature Extraction and Selection
2.2.1. Mining Network Topology Information Based on Deepwalk
2.2.2. Constructing the Sample Space
2.3. Deep Forest-Based Association Prediction
3. Result
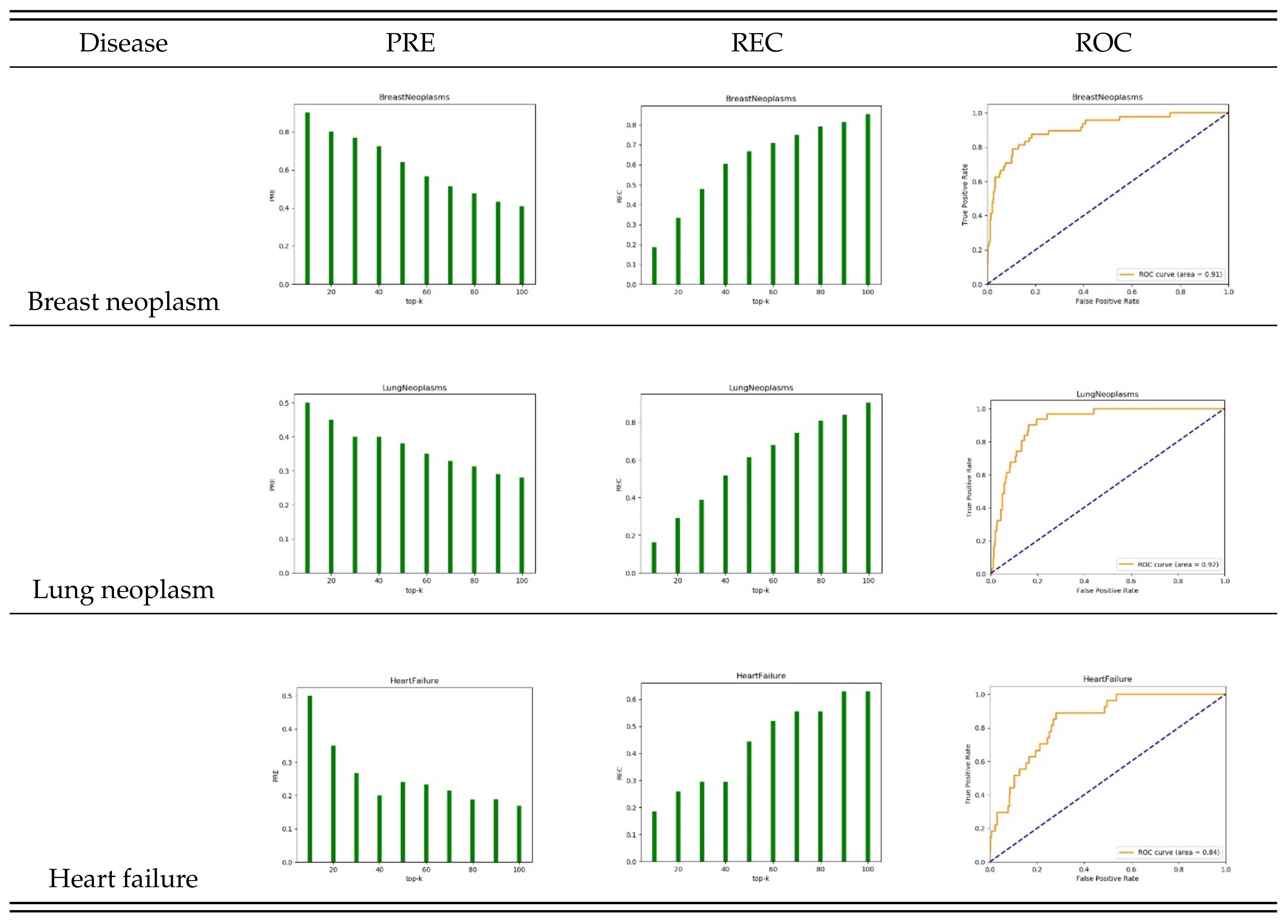
3.1. Evaluation Metrics
3.2. Performance of STIM
3.3. Comparison with Other Methods
3.4. Case Study: Lung Neoplasm
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, B.; Fang, L.; Liu, F.; Wang, X.; Chen, J.; Chou, K.C. Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS ONE 2015, 10, e0121501. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wu, Z.; Kong, W. Improving Clustering of MicroRNA Microarray Data by Incorporating Functional Similarity. Curr. Bioinform. 2018, 13, 34–41. [Google Scholar] [CrossRef]
- Liao, Z.; Li, D.; Wang, X.; Li, L.; Zou, Q. Cancer diagnosis through IsomiR expression with machine learning method. Curr. Bioinform. 2018, 13, 57–63. [Google Scholar] [CrossRef]
- Lü, L.; Zhou, T. Link prediction in complex networks: A survey. Phys. A Stat. Mech. Its Appl. 2011, 390, 1150–1170. [Google Scholar] [CrossRef]
- Jiang, L.; Xiao, Y.; Ding, Y.; Tang, J.; Guo, F. FKL-Spa-LapRLS: An accurate method for identifying human microRNA-disease association. BMC Genom. 2018, 19, 911. [Google Scholar] [CrossRef]
- Jiang, L.; Xiao, Y.; Ding, Y.; Tang, J.; Guo, F. Discovering Cancer Subtypes via an Accurate Fusion Strategy on Multiple Profile Data. Front. Genet. 2019, 10, 20. [Google Scholar] [CrossRef]
- Zou, Q.; Li, J.; Song, L.; Zeng, X.; Wang, G. Similarity computation strategies in the microRNA-disease network: a survey. Brief. Funct. Genom. 2015, 15, 55–64. [Google Scholar] [CrossRef]
- Wang, L.; Ping, P.; Kuang, L.; Ye, S.; Pei, T. A Novel Approach Based on Bipartite Network to Predict Human Microbe-Disease Associations. Curr. Bioinform. 2018, 13, 141–148. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, X.; Zou, Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Brief. Bioinform. 2016, 17, 193–203. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, X.; He, Z.; Zou, Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Wang, W.; Deng, G.; Bing, J.; Zou, Q. Prediction of Potential Disease-Associated MicroRNAs by Using Neural Networks. Mol. Ther. Nucleic Acids 2019, 16, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Liu, L.; Lü, L.; Zou, Q. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics 2018, 34, 2425–2432. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Hao, Y.; Wang, G.; Juan, L.; Zhang, T.; Teng, M.; Liu, Y.; Wang, Y. Prioritization of disease microRNAs through a human phenome-microRNAome network. BMC Syst. Biol. 2010, 4, S2. [Google Scholar] [CrossRef]
- Jiang, Q.; Hao, Y.; Wang, G.; Zhang, T.; Wang, Y. Weighted network-based inference of human microrna-disease associations. In Proceedings of the 2010 Fifth International Conference on Frontier of Computer Science and Technology, Changchun, China, 18–22 August 2010; pp. 431–435. [Google Scholar]
- Pan, L.; Song, B. P systems with rule production and removal. Fund. Inform. 2020, 171, 313–329. [Google Scholar]
- Song, B.; Kong, Y. Solution to PSPACE-complete problem using P systems with active membranes with time-freeness. Math. Probl. Eng. 2019, 2019, 5793234. [Google Scholar] [CrossRef]
- Xuan, P.; Han, K.; Guo, M.; Guo, Y.; Li, J.; Ding, J.; Liu, Y.; Dai, Q.; Li, J.; Teng, Z.; et al. Prediction of microRNAs associated with human diseases based on weighted k most similar neighbors. PLoS ONE 2013, 8, e70204. [Google Scholar] [CrossRef]
- Chen, X.; Liu, M.X.; Yan, G.Y. RWRMDA: predicting novel human microRNA–disease associations. Mol. Biosyst. 2012, 8, 2792–2798. [Google Scholar] [CrossRef]
- Shi, H.; Xu, J.; Zhang, G.; Xu, L.; Li, C.; Wang, L.; Zhao, Z.; Jiang, W.; Guo, Z.; Li, X. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst. Biol. 2013, 7, 101. [Google Scholar] [CrossRef]
- Xuan, P.; Han, K.; Guo, Y.; Li, J.; Li, X.; Zhong, Y.; Zhang, Z.; Ding, J. Prediction of potential disease-associated microRNAs based on random walk. Bioinformatics 2015, 31, 1805–1815. [Google Scholar] [CrossRef]
- Zou, Q.; Li, X.; Jiang, Y.; Zhao, Y.; Wang, G. BinMemPredict: A web server and software for predicting membrane protein types. Curr. Proteom. 2013, 10, 2–9. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, G.; Jin, S.; Li, Y.; Wang, Y. Predicting human microRNA-disease associations based on support vector machine. Int. J. Data Min. Bioinform. 2013, 8, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, C.X.; Lv, J.Y.; Li, Y.S.; Xiao, Y.; Shao, T.T.; Huo, X.; Li, X.; Zou, Y.; Han, Q.L.; et al. Prioritizing candidate disease miRNAs by topological features in the miRNA target–dysregulated network: Case study of prostate cancer. Mol. Cancer Ther. 2011, 10, 1857–1866. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, G.Y. Semi-supervised learning for potential human microRNA-disease associations inference. Sci. Rep. 2014, 4, 5501. [Google Scholar] [CrossRef]
- Liu, B.; Wang, X.; Zou, Q.; Dong, Q.; Chen, Q. Protein remote homology detection by combining Chou’s pseudo amino acid composition and profile-based protein representation. Mol. Inform. 2013, 32, 775–782. [Google Scholar] [CrossRef]
- Chen, B.; Ding, Y.; Wild, D.J. Assessing drug target association using semantic linked data. PLoS Comput. Biol. 2012, 8, e1002574. [Google Scholar] [CrossRef]
- Chris Nicholson, A.G. Deeplearning4j: Open-source Distributed Deep Learning for the JVM. 2017. Available online: https://Deeplearning4j.org.
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2. 0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2013, 42, D1070–D1074. [Google Scholar] [CrossRef]
- Lee, I.; Blom, U.M.; Wang, P.I.; Shim, J.E.; Marcotte, E.M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011, 21, 1109–1121. [Google Scholar] [CrossRef]
- Liu, B. BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief. Bioinform. 2017, 20, 1280–1294. [Google Scholar] [CrossRef]
- Chen, W.; Lv, H.; Nie, F.; Lin, H. i6mA-Pred: Identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics 2019, 35, 2796–2800. [Google Scholar] [CrossRef]
- Zhu, X.J.; Feng, C.Q.; Lai, H.Y.; Chen, W.; Hao, L. Predicting protein structural classes for low-similarity sequences by evaluating different features. Knowl. Based Syst. 2019, 163, 787–793. [Google Scholar] [CrossRef]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference On Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]
- Mnih, A.; Hinton, G.E. A scalable hierarchical distributed language model. In Proceedings of the NIPS’08 21st International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–10 December 2008; pp. 1081–1088. [Google Scholar]
- Chen, W.; Yang, H.; Feng, P.; Ding, H.; Lin, H. iDNA4mC: Identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics 2017, 33, 3518–3523. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.Q.; Zhang, Z.Y.; Zhu, X.J.; Lin, Y.; Chen, W.; Tang, H.; Lin, H. iTerm-PseKNC: A sequence-based tool for predicting bacterial transcriptional terminators. Bioinformatics 2018, 35, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Zhang, Z.; Eyben, F.; Schuller, B. Autoencoder-based unsupervised domain adaptation for speech emotion recognition. IEEE Signal Process. Lett. 2014, 21, 1068–1072. [Google Scholar]
- Yang, F.; Xu, Q.; Li, B.; Ji, Y. Ship detection from thermal remote sensing imagery through region-based deep forest. IEEE Geosci. Remote Sens. Lett. 2018, 15, 449–453. [Google Scholar] [CrossRef]
- Su, R.; Liu, X.; Wei, L.; Zou, Q. Deep-Resp-Forest: A deep forest model to predict anti-cancer drug response. Methods 2019, 166, 91–102. [Google Scholar] [CrossRef]
- Dao, F.Y.; Lv, H.; Wang, F.; Feng, C.Q.; Ding, H.; Chen, W.; Lin, H. Identify origin of replication in Saccharomyces cerevisiae using two-step feature selection technique. Bioinformatics 2018, 35, 2075–2083. [Google Scholar] [CrossRef]
- Wei, L.; Xing, P.; Zeng, J.; Chen, J.; Su, R.; Guo, F. Improved prediction of protein–protein interactions using novel negative samples, features, and an ensemble classifier. Artif. Intell. Med. 2017, 83, 67–74. [Google Scholar] [CrossRef]
- Wei, L.; Wan, S.; Guo, J.; Wong, K.K. A novel hierarchical selective ensemble classifier with bioinformatics application. Artif. Intell. Med. 2017, 83, 82–90. [Google Scholar] [CrossRef]
- Tan, J.X.; Li, S.H.; Zhang, Z.M.; Chen, C.X.; Chen, W.; Tang, H.; Lin, H. Identification of hormone binding proteins based on machine learning methods. Math. Biosci. Eng. 2019, 16, 2466–2480. [Google Scholar] [CrossRef]
- Yang, Z.; Ren, F.; Liu, C.; He, S.; Sun, G.; Gao, Q.; Yao, L.; Zhang, Y.; Miao, R.; Cao, Y.; et al. dbDEMC: A database of differentially expressed miRNAs in human cancers. BMC Genom. Biomed Cent. 2010, 11, S5. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. miR2Disease: A manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2008, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mitra, R.; Maulik, U.; Zhang, M.Q. Development of the human cancer microRNA network. Silence 2010, 1, 6. [Google Scholar] [CrossRef]


| Node Type | Number |
|---|---|
| Disease | 336 |
| miRNA | 577 |
| Disease–miRNA associations | 6441 |
| Disease | STIM | RWRMDA | HDMP | RLSMDA | MIDP |
|---|---|---|---|---|---|
| Acute myeloid leukemia | 0.887 | 0.839 | 0.858 | 0.853 | 0.913 |
| Breast neoplasm | 0.894 | 0.785 | 0.801 | 0.832 | 0.838 |
| Colorectal neoplasms | 0.855 | 0.793 | 0.802 | 0.831 | 0.845 |
| Glioblastoma | 0.857 | 0.68 | 0.7 | 0.714 | 0.786 |
| Heart failure | 0.829 | 0.722 | 0.77 | 0.738 | 0.821 |
| Hepatocellular carcinoma | 0.86 | 0.749 | 0.759 | 0.794 | 0.807 |
| Lung neoplasms | 0.913 | 0.827 | 0.835 | 0.855 | 0.876 |
| Melanoma | 0.857 | 0.784 | 0.79 | 0.807 | 0.837 |
| Ovarian neoplasms | 0.89 | 0.882 | 0.884 | 0.909 | 0.923 |
| Pancreatic neoplasms | 0.909 | 0.871 | 0.895 | 0.887 | 0.945 |
| Prostatic neoplasms | 0.872 | 0.823 | 0.854 | 0.841 | 0.882 |
| Renal cell carcinoma | 0.866 | 0.815 | 0.833 | 0.839 | 0.862 |
| Squamous cell carcinoma | 0.876 | 0.819 | 0.82 | 0.849 | 0.87 |
| Stomach neoplasms | 0.875 | 0.779 | 0.787 | 0.797 | 0.821 |
| Urinary bladder neoplasms | 0.868 | 0.821 | 0.85 | 0.845 | 0.897 |
| Average AUC | 0.874 | 0.799 | 0.816 | 0.826 | 0.862 |
| Rank | miRNA | Evidence | Rank | miRNA | Evidence |
|---|---|---|---|---|---|
| 1 | hsa-mir-130a | dbDEMC,miR2disease | 16 | hsa-mir-149 | dbDEMC |
| 2 | hsa-mir-125b-2 | Unconfirm | 17 | hsa-mir-15a | dbDEMC |
| 3 | hsa-mir-195 | dbDEMC,miR2disease | 18 | hsa-mir-302a | Ref.[43] |
| 4 | hsa-mir-451a | dbDEMC,miR2disease | 19 | hsa-mir-99a | dbDEMC,miR2disease |
| 5 | hsa-mir-128-1 | Unconfirm | 20 | hsa-mir-152 | dbDEMC |
| 6 | hsa-mir-23b | dbDEMC | 21 | hsa-mir-708 | Ref.[43] |
| 7 | hsa-mir-151a | Ref. [42] | 22 | hsa-mir-378a | Ref.[42] |
| 8 | hsa-mir-92a-2 | dbDEMC | 23 | hsa-mir-339 | dbDEMC |
| 9 | hsa-mir-302b | dbDEMC | 24 | hsa-mir-106b | dbDEMC |
| 10 | hsa-mir-193b | dbDEMC | 25 | hsa-mir-215 | dbDEMC |
| 11 | hsa-mir-141 | dbDEMC,miR2Disease | 26 | hsa-mir-130b | dbDEMC |
| 12 | hsa-mir-196b | dbDEMC | 27 | hsa-mir-302c | dbDEMC |
| 13 | hsa-mir-10a | dbDEMC | 28 | hsa-mir-296 | dbDEMC |
| 14 | hsa-mir-429 | dbDEMC,miR2disease | 29 | hsa-mir-320a | Ref.[43] |
| 15 | hsa-mir-328 | dbDEMC | 30 | hsa-mir-20b | dbDEMC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Wang, X.; Gao, P.; Liu, H.; Song, B. Predicting Disease Related microRNA Based on Similarity and Topology. Cells 2019, 8, 1405. https://doi.org/10.3390/cells8111405
Chen Z, Wang X, Gao P, Liu H, Song B. Predicting Disease Related microRNA Based on Similarity and Topology. Cells. 2019; 8(11):1405. https://doi.org/10.3390/cells8111405
Chicago/Turabian StyleChen, Zhihua, Xinke Wang, Peng Gao, Hongju Liu, and Bosheng Song. 2019. "Predicting Disease Related microRNA Based on Similarity and Topology" Cells 8, no. 11: 1405. https://doi.org/10.3390/cells8111405
APA StyleChen, Z., Wang, X., Gao, P., Liu, H., & Song, B. (2019). Predicting Disease Related microRNA Based on Similarity and Topology. Cells, 8(11), 1405. https://doi.org/10.3390/cells8111405





