Identification of a Diagnostic Set of Endomyocardial Biopsy microRNAs for Acute Cellular Rejection Diagnostics in Patients after Heart Transplantation Using Next-Generation Sequencing
Abstract
:1. Introduction
2. Materials and Methods
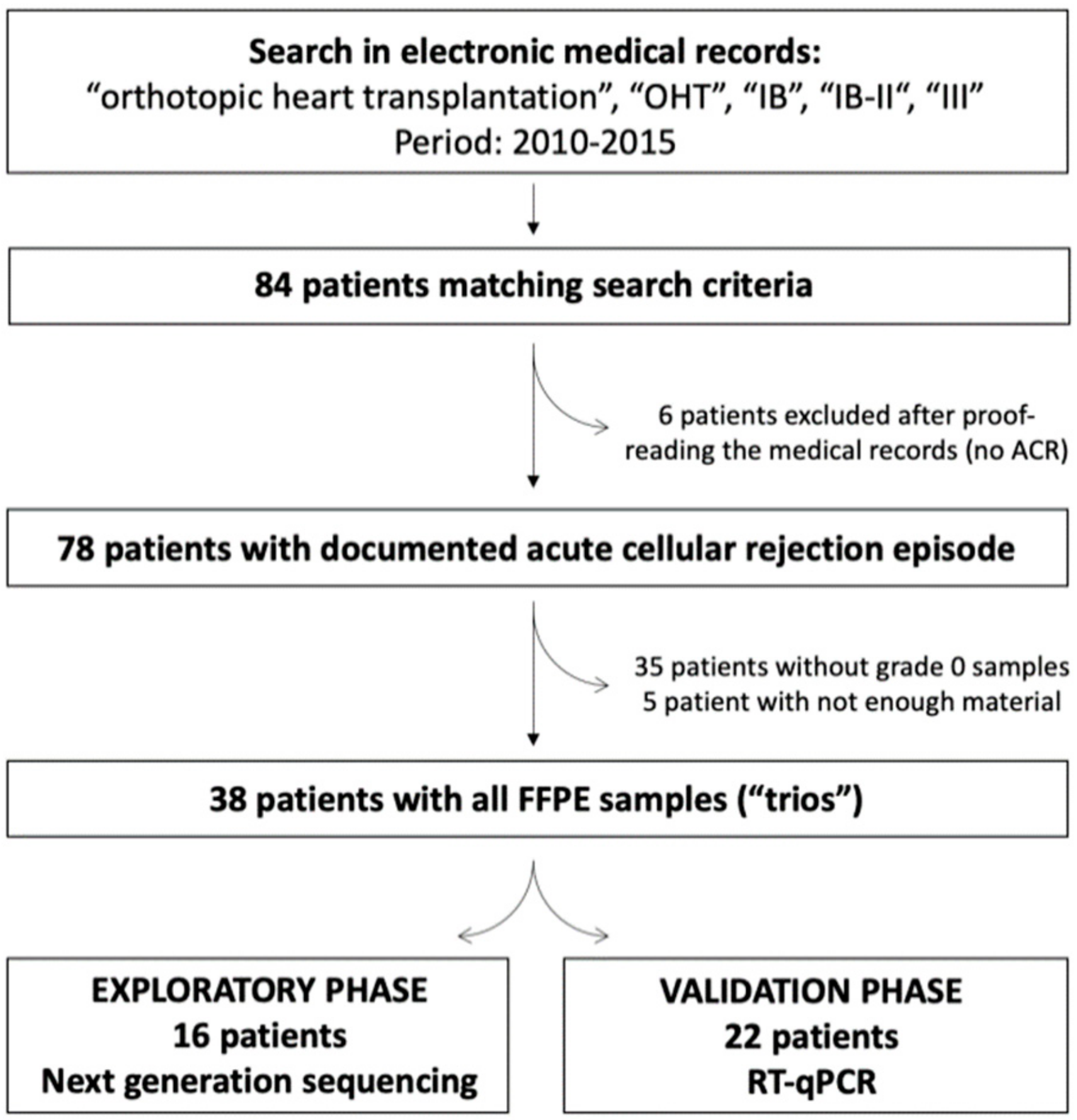
2.1. Study Population
2.2. RNA Isolation
2.3. Exploratory Phase: Next-Generation Sequencing (NGS)
2.4. Validation Phase: Reverse Transcription and Quantitative PCR (RT-qPCR)
2.5. Statistics
3. Results
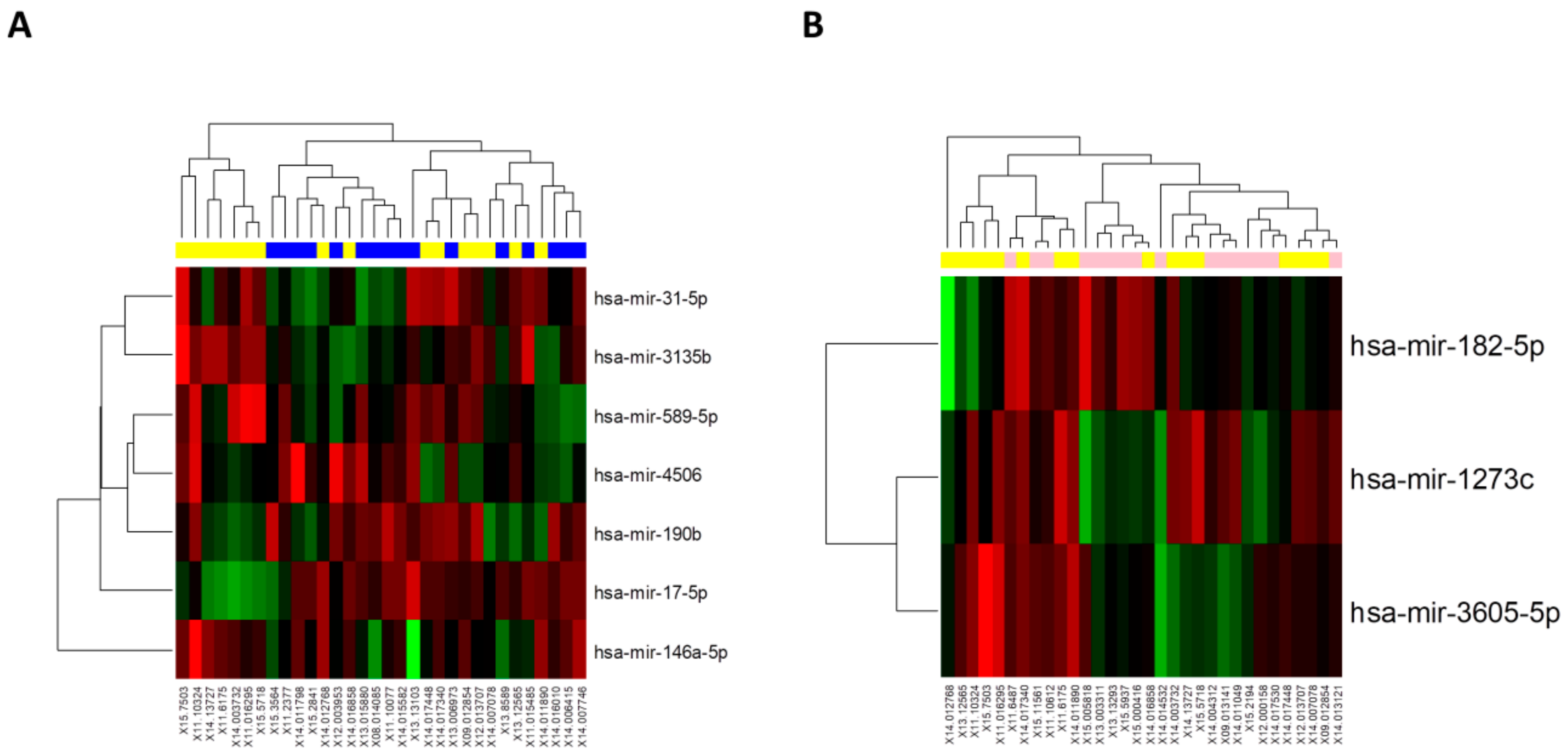
3.1. Small RNA Sequencing Reveals Several Sets of microRNAs Altered during ACR
3.2. Validation of Identified microRNAs Altered during ACR on the Validation Cohort
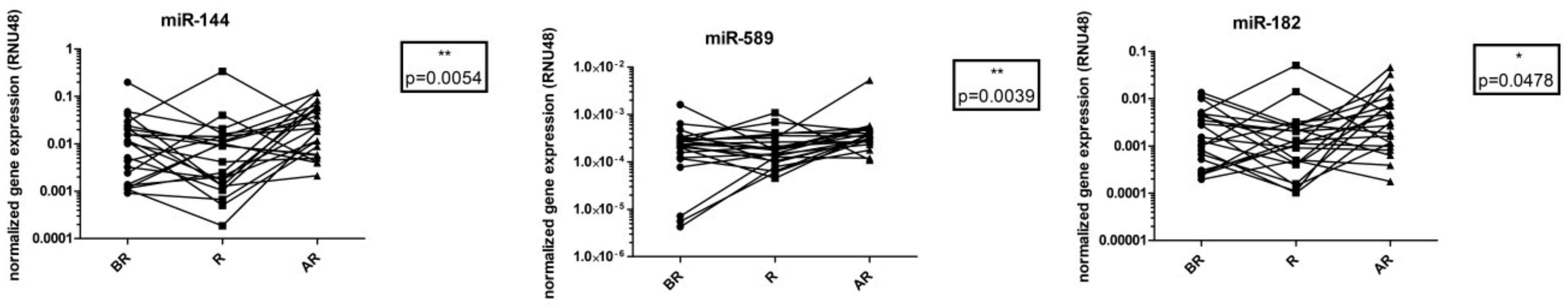
3.3. Dependence of miRNA Levels on Time
3.4. Correlation of Individual microRNAs Expression at Different Time Points
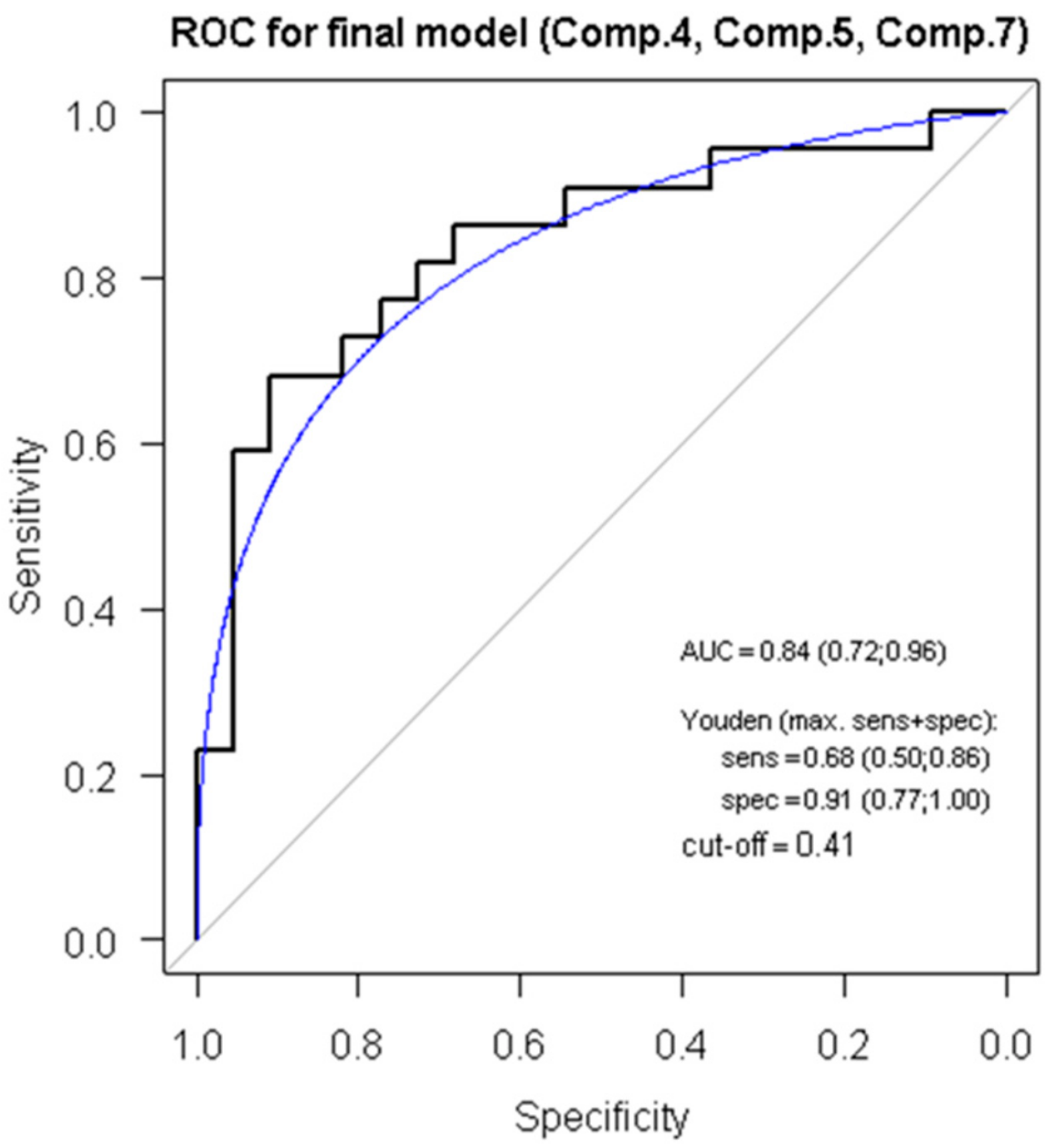
3.5. ROC Analysis, Regression Modelling and Principal Component Analysis Reveals a Score for ACR Diagnostics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.F.; Coats, A.J.S.; Falk, V.; González-Juanatey, J.R.; Harjola, V.-P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail. 2016, 18, 891–975. [Google Scholar] [PubMed]
- Bleumink, G.S.; Knetsch, A.M.; Sturkenboom, M.C.J.M.; Straus, S.M.J.M.; Hofman, A.; Deckers, J.W.; Witteman, J.C.M.; Stricker, B.H.C. Quantifying the heart failure epidemic: Prevalence, incidence rate, lifetime risk and prognosis of heart failure The Rotterdam Study. Eur. Heart J. 2004, 25, 1614–1619. [Google Scholar] [CrossRef] [PubMed]
- Kannel, W.B. Incidence and epidemiology of heart failure. Heart Fail. Rev. 2000, 5, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Lund, L.H.; Edwards, L.B.; Kucheryavaya, A.Y.; Benden, C.; Christie, J.D.; Dipchand, A.I.; Dobbels, F.; Goldfarb, S.B.; Levvey, B.J.; Meiser, B.; et al. The registry of the International Society for Heart and Lung Transplantation: Thirty-first official adult heart transplant report—2014; focus theme: Retransplantation. J. Heart Lung Transpl. 2014, 33, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Stoica, S.C.; Cafferty, F.; Pauriah, M.; Taylor, C.J.; Sharples, L.D.; Wallwork, J.; Large, S.R.; Parameshwar, J. The cumulative effect of acute rejection on development of cardiac allograft vasculopathy. J. Heart Lung Transpl. 2006, 25, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.H.; Revelo, M.P.; Miller, D.V.; Snow, G.L.; Budge, D.; Stehlik, J.; Molina, K.M.; Selzman, C.H.; Drakos, S.G.; Rami, A.; et al. ISHLT pathology antibody mediated rejection score correlates with increased risk of cardiovascular mortality: A retrospective validation analysis. J. Heart Lung Transpl. 2016, 35, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Caves, P.K.; Stinson, E.B.; Billingham, M.E.; Shumway, N.E. Serial transvenous biopsy of the transplanted human heart. Improved management of acute rejection episodes. Lancet 1974, 1, 821–826. [Google Scholar] [CrossRef]
- Stewart, S.; Winters, G.L.; Fishbein, M.C.; Tazelaar, H.D.; Kobashigawa, J.; Abrams, J.; Andersen, C.B.; Angelini, A.; Berry, G.J.; Burke, M.M.; et al. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J. Heart Lung Transpl. 2005, 24, 1710–1720. [Google Scholar] [CrossRef]
- Crespo-Leiro, M.G.; Zuckermann, A.; Bara, C.; Mohacsi, P.; Schulz, U.; Boyle, A.; Ross, H.J.; Parameshwar, J.; Zakliczyński, M.; Fiocchi, R.; et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II). Transplantation 2012, 94, 1172–1177. [Google Scholar] [CrossRef]
- Tang, Z.; Kobashigawa, J.; Rafiei, M.; Stern, L.K.; Hamilton, M. The natural history of biopsy-negative rejection after heart transplantation. J. Transpl. 2013, 2013, 236720. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Falzone, L.; Lupo, G.; La Rosa, G.R.M.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Rapisarda, E.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers 2019, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Scola, L.; Zanghì, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging 2018, 10, 1000–1014. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Stiegelbauer, V.; Vychytilova-Faltejskova, P.; Ivan, C.; Ling, H.; Winter, E.; Zhang, X.; Goblirsch, M.; Wulf-Goldenberg, A.; Ohtsuka, M.; et al. Genome-Wide miRNA Analysis Identifies miR-188-3p as a Novel Prognostic Marker and Molecular Factor Involved in Colorectal Carcinogenesis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 1323–1333. [Google Scholar] [CrossRef]
- Bekris, L.M.; Leverenz, J.B. The biomarker and therapeutic potential of miRNA in Alzheimer’s disease. Neurodegener. Dis. Manag. 2015, 5, 61–74. [Google Scholar] [CrossRef]
- Candido, S.; Lupo, G.; Pennisi, M.; Basile, M.S.; Anfuso, C.D.; Petralia, M.C.; Gattuso, G.; Vivarelli, S.; Spandidos, D.A.; Libra, M.; et al. The analysis of miRNA expression profiling datasets reveals inverse microRNA patterns in glioblastoma and Alzheimer’s disease. Oncol. Rep. 2019, 42, 911–922. [Google Scholar] [CrossRef]
- Evangelatos, G.; Fragoulis, G.E.; Koulouri, V.; Lambrou, G.I. MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact. Autoimmun. Rev. 2019, 2019, 102391. [Google Scholar] [CrossRef]
- Yang, L.; Bian, Y.; Li, Z.; Yan, Y.; Li, J.; Li, W.; Zeng, L. Identification of potential biomarkers and pathways in ulcerative colitis with combined public mRNA and miRNA expression microarray data analysis. J. Gastrointest. Oncol. 2019, 10, 847–858. [Google Scholar] [CrossRef]
- Mirzaei, H.; Ferns, G.A.; Avan, A.; Mobarhan, M.G. Cytokines and MicroRNA in Coronary Artery Disease. Adv. Clin. Chem. 2017, 82, 47–70. [Google Scholar]
- Wong, L.L.; Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.-T. MicroRNA and Heart Failure. Int. J. Mol. Sci. 2016, 17, 502. [Google Scholar] [CrossRef]
- Briasoulis, A.; Sharma, S.; Telila, T.; Mallikethi-Reddy, S.; Papageorgiou, N.; Oikonomou, E.; Tousoulis, D. MicroRNAs in Atrial Fibrillation. Curr. Med. Chem. 2019, 26, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Fedrigo, M.; Santovito, D.; Natarelli, L.; Castellani, C.; De Pascale, F.; Toscano, G.; Fraiese, A.; Feltrin, G.; Benazzi, E.; et al. MicroRNA signatures in cardiac biopsies and detection of allograft rejection. J. Heart Lung Transpl. 2018, 37, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, M.; Qu, X.; Mah, A.; Xiong, X.; Harris, A.G.C.; Phillips, L.K.; Martinez, O.M.; Krams, S.M. Differential expression of microRNAs during allograft rejection. Am. J. Transpl. 2012, 12, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Kaul, V.; Qu, X.; Xiong, X.; Lau, A.H.; Iwai, N.; Martinez, O.M.; Krams, S.M. Absence of mir-182 Augments Cardiac Allograft Survival. Transplantation 2017, 101, 524–530. [Google Scholar] [CrossRef]
- Dambal, S.; Shah, M.; Mihelich, B.; Nonn, L. The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Res. 2015, 43, 7173–7188. [Google Scholar] [CrossRef]
- Xu, Z.; Ramachandran, S.; Gunasekaran, M.; Zhou, F.; Trulock, E.; Kreisel, D.; Hachem, R.; Mohanakumar, T. MicroRNA-144 dysregulates the transforming growth factor-β signalling cascade and contributes to the development of bronchiolitis obliterans syndrome after human lung transplantation. J. Heart Lung Transpl. 2015, 34, 1154–1162. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Hu, Y.-R.; Zhao, J.-Y.; Li, S.-F.; Ma, X.; Wu, S.-G.; Lu, J.-B.; Qiu, Y.-R.; Sha, Y.-H.; Wang, Y.-C.; et al. An agomir of miR-144-3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS ONE 2014, 9, e94997. [Google Scholar] [CrossRef]
- Halushka, M.K. MicroRNA-144 is unlikely to play a role in bronchiolitis obliterans syndrome. J. Heart Lung Transpl. 2016, 35, 543. [Google Scholar] [CrossRef]
- Li, C.; Liu, T.; Qi, F.; Li, F.; Zhu, L.; Wang, P.; Wang, H. Analysis of intragraft microRNA expression in a mouse-to-rat cardiac xenotransplantation model. Microsurgery 2014, 34, 44–50. [Google Scholar] [CrossRef]
- Van Aelst, L.N.L.; Summer, G.; Li, S.; Gupta, S.K.; Heggermont, W.; De Vusser, K.; Carai, P.; Naesens, M.; Van Cleemput, J.; Van de Werf, F.; et al. RNA Profiling in Human and Murine Transplanted Hearts: Identification and Validation of Therapeutic Targets for Acute Cardiac and Renal Allograft Rejection. Am. J. Transpl. 2016, 16, 99–110. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, K.; Zhou, C.; Gan, Z.; Ma, D.; Ye, P.; Sun, Y.; Wu, J.; Huang, X.; Ren, L.; et al. Knockout of microRNA-155 ameliorates the Th1/Th17 immune response and tissue injury in chronic rejection. J. Heart Lung Transpl. 2017, 36, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Ellims, A.H.; Moore, X.; White, D.A.; Taylor, A.J.; Chin-Dusting, J.; Dart, A.M. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J. Transl. Med. 2015, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Bonauer, A.; Dimmeler, S. The microRNA-17-92 cluster: Still a miRacle? Cell Cycle 2009, 8, 3866–3873. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Pan, Z.; Chen, X.; Wang, L.; Zhang, Y.; Li, S.; Liang, H.; Xu, C.; Zhang, Y.; Wu, Y.; et al. By targeting Stat3 microRNA-17-5p promotes cardiomyocyte apoptosis in response to ischemia followed by reperfusion. Cell. Physiol. Biochem. 2014, 34, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fan, T.; Hu, Q.; Xu, W.; Yang, J.; Xu, C.; Zhang, B.; Chen, J.; Jiang, H. Downregulation of microRNA-17-5p improves cardiac function after myocardial infarction via attenuation of apoptosis in endothelial cells. Mol. Genet. Genom. MGG 2018, 293, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Charrier, H.; Cuvelliez, M.; Dubois-Deruy, E.; Mulder, P.; Richard, V.; Bauters, C.; Pinet, F. Integrative System Biology Analyses Identify Seven MicroRNAs to Predict Heart Failure. Non Coding RNA 2019, 5, 22. [Google Scholar] [CrossRef]
- Infante, T.; Forte, E.; Punzo, B.; Cademartiri, F.; Cavaliere, C.; Soricelli, A.; Salvatore, M.; Napoli, C. Correlation of Circulating miR-765, miR-93-5p, and miR-433-3p to Obstructive Coronary Heart Disease Evaluated by Cardiac Computed Tomography. Am. J. Cardiol. 2019, 124, 176–182. [Google Scholar] [CrossRef]
- Wang, H.; Chen, F.; Tong, J.; Li, Y.; Cai, J.; Wang, Y.; Li, P.; Hao, Y.; Tian, W.; Lv, Y.; et al. Circulating microRNAs as novel biomarkers for dilated cardiomyopathy. Cardiol. J. 2017, 24, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Yang, J.; Li, Y.; Wang, H. Circulating microRNAs as novel biomarkers for heart failure. Hell. J. Cardiol. HJC 2018, 59, 209–214. [Google Scholar] [CrossRef]
- Long, J.; Jiang, C.; Liu, B.; Dai, Q.; Hua, R.; Chen, C.; Zhang, B.; Li, H. Maintenance of stemness by miR-589-5p in hepatocellular carcinoma cells promotes chemoresistance via STAT3 signalling. Cancer Lett. 2018, 423, 113–126. [Google Scholar] [CrossRef]
- Zhang, F.; Li, K.; Pan, M.; Li, W.; Wu, J.; Li, M.; Zhao, L.; Wang, H. miR-589 promotes gastric cancer aggressiveness by a LIFR-PI3K/AKT-c-Jun regulatory feedback loop. J. Exp. Clin. Cancer Res. CR 2018, 37, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Han, Z.; Xu, Z.; An, C.; Xu, L.; Xin, H. RNA sequencing uncovers the key long non-coding RNAs and potential molecular mechanism contributing to XAV939-mediated inhibition of non-small cell lung cancer. Oncol. Lett. 2019, 17, 4994–5004. [Google Scholar] [CrossRef] [PubMed]
- Novak, J.; Sana, J.; Stracina, T.; Novakova, M.; Slaby, O. Doxorubicin and Liposomal Doxorubicin Differentially Affect Expression of miR-208a and let-7g in Rat Ventricles and Atria. Cardiovasc. Toxicol. 2017, 17, 355–359. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Mean | SD | Median | Min | Max |
|---|---|---|---|---|---|
| Age at OHT (years) | 53 | 10.6 | 58 | 21 | 65 |
| Weight (kg) | 86.2 | 17.2 | 84 | 56 | 124 |
| Height (cm) | 177.5 | 8.66 | 179 | 162 | 195 |
| BMI (kg/m2) | 27.19 | 4.01 | 26.84 | 17.28 | 37.12 |
| Males/females | 35/3 | ||||
| Diagnosis |
| ||||
| Immunosuppression |
| ||||
| Basic laboratory results (at the time of rejection) | |||||
| Urea (mmol/L) | 9.532 | 4.00 | 8.65 | 3.6 | 24.9 |
| Creatinine (μmol/L) | 117.79 | 38.63 | 113.5 | 66 | 221 |
| GFR (mL/min/m2) | 1.03 | 0.34 | 0.97 | 0.50 | 1.76 |
| AST (μkat/L) | 0.48 | 0.21 | 0.45 | 0.16 | 1.34 |
| ALT (μkat/L) | 0.65 | 0.60 | 0.49 | 0.17 | 3.84 |
| CRP (mg/L) | 8.27 | 12.06 | 2.70 | 0.50 | 48.30 |
| Erythrocytes (×1012/L) | 4.42 | 0.66 | 4.29 | 3.20 | 5.99 |
| Leukocytes (×109/L) | 7.70 | 3.24 | 7.15 | 2.80 | 19.60 |
| Hemoglobin (g/L) | 126.38 | 16.48 | 128.50 | 89.00 | 173.00 |
| Hematocrit(L/L) | 0.38 | 0.05 | 0.39 | 0.27 | 0.50 |
| Thrombocytes (×109/L) | 201.71 | 59.04 | 209.50 | 93.00 | 305.00 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nováková, T.; Macháčková, T.; Novák, J.; Hude, P.; Godava, J.; Žampachová, V.; Oppelt, J.; Zlámal, F.; Němec, P.; Bedáňová, H.; et al. Identification of a Diagnostic Set of Endomyocardial Biopsy microRNAs for Acute Cellular Rejection Diagnostics in Patients after Heart Transplantation Using Next-Generation Sequencing. Cells 2019, 8, 1400. https://doi.org/10.3390/cells8111400
Nováková T, Macháčková T, Novák J, Hude P, Godava J, Žampachová V, Oppelt J, Zlámal F, Němec P, Bedáňová H, et al. Identification of a Diagnostic Set of Endomyocardial Biopsy microRNAs for Acute Cellular Rejection Diagnostics in Patients after Heart Transplantation Using Next-Generation Sequencing. Cells. 2019; 8(11):1400. https://doi.org/10.3390/cells8111400
Chicago/Turabian StyleNováková, Tereza, Táňa Macháčková, Jan Novák, Petr Hude, Július Godava, Víta Žampachová, Jan Oppelt, Filip Zlámal, Petr Němec, Helena Bedáňová, and et al. 2019. "Identification of a Diagnostic Set of Endomyocardial Biopsy microRNAs for Acute Cellular Rejection Diagnostics in Patients after Heart Transplantation Using Next-Generation Sequencing" Cells 8, no. 11: 1400. https://doi.org/10.3390/cells8111400
APA StyleNováková, T., Macháčková, T., Novák, J., Hude, P., Godava, J., Žampachová, V., Oppelt, J., Zlámal, F., Němec, P., Bedáňová, H., Slabý, O., Bienertová-Vašků, J., Špinarová, L., & Krejčí, J. (2019). Identification of a Diagnostic Set of Endomyocardial Biopsy microRNAs for Acute Cellular Rejection Diagnostics in Patients after Heart Transplantation Using Next-Generation Sequencing. Cells, 8(11), 1400. https://doi.org/10.3390/cells8111400







