P65 Targets FGFR1 to Regulate the Survival of Ovarian Granulosa Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Sample Preparation
2.3. Culture of Porcine GCs In Vitro
2.4. Real-Time Quantitative PCR Analysis
2.5. Construction of the FGFR1 5′ Deletion and Luciferase Assay
2.6. Cell Transfection, Proliferation, and the Apoptosis Assay
2.7. Western Blot Analysis
2.8. Chromatin Immunoprecipitation Assay
2.9. Immunohistochemistry Assay
2.10. Statistical Analysis
3. Results
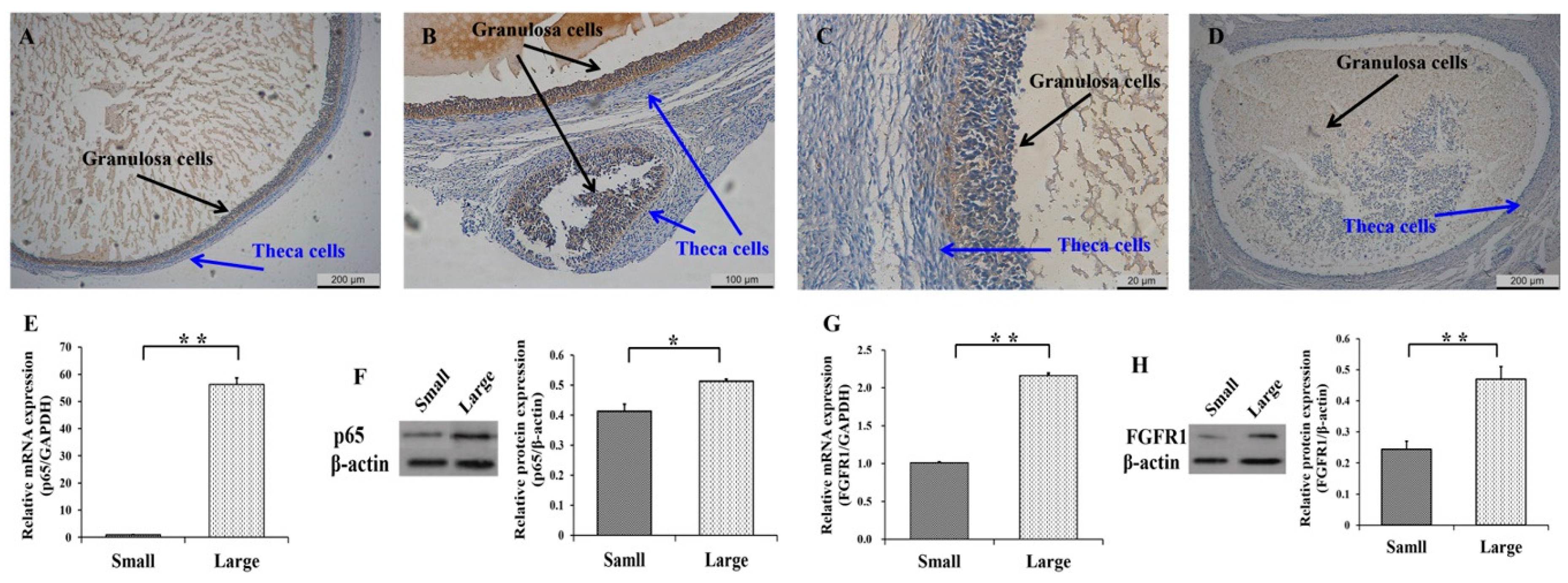
3.1. Expression Patterns of p65 and FGFR1 in Follicles and GCs
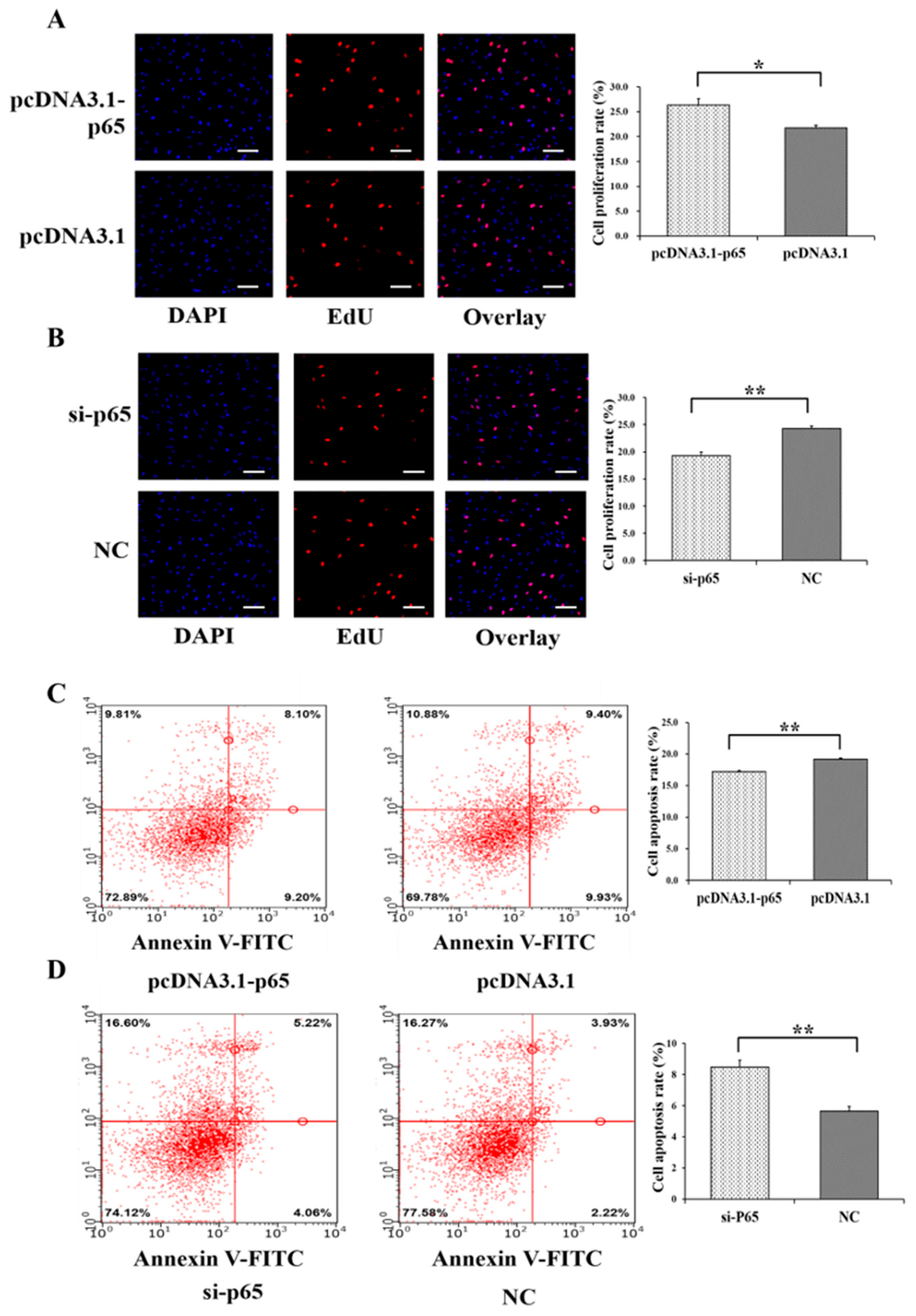
3.2. Pro-Proliferation and Anti-Apoptotic Effects of p65 on GCs
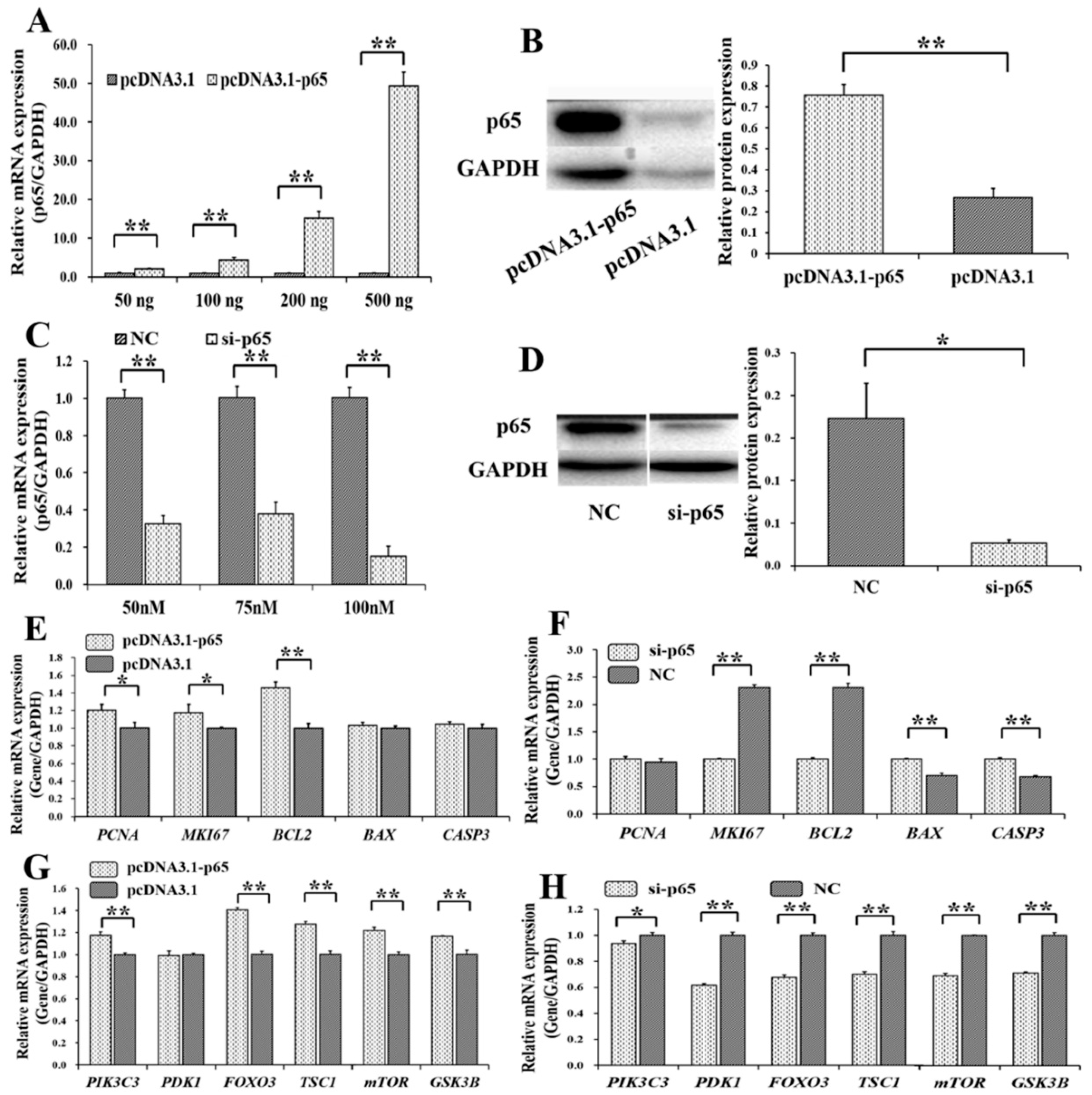
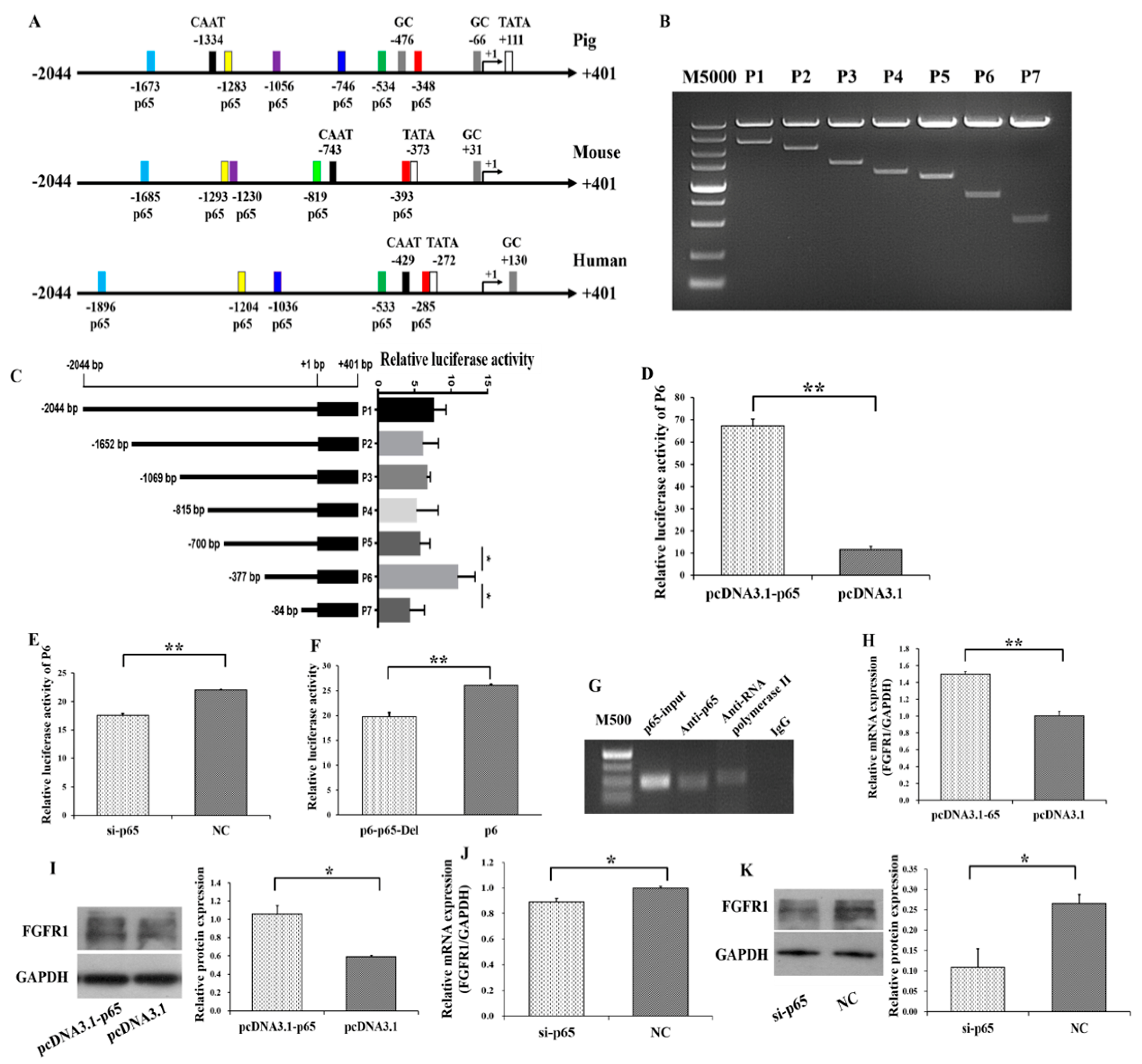
3.3. p65 Targets FGFR1 and Promotes the Expression of FGFR1
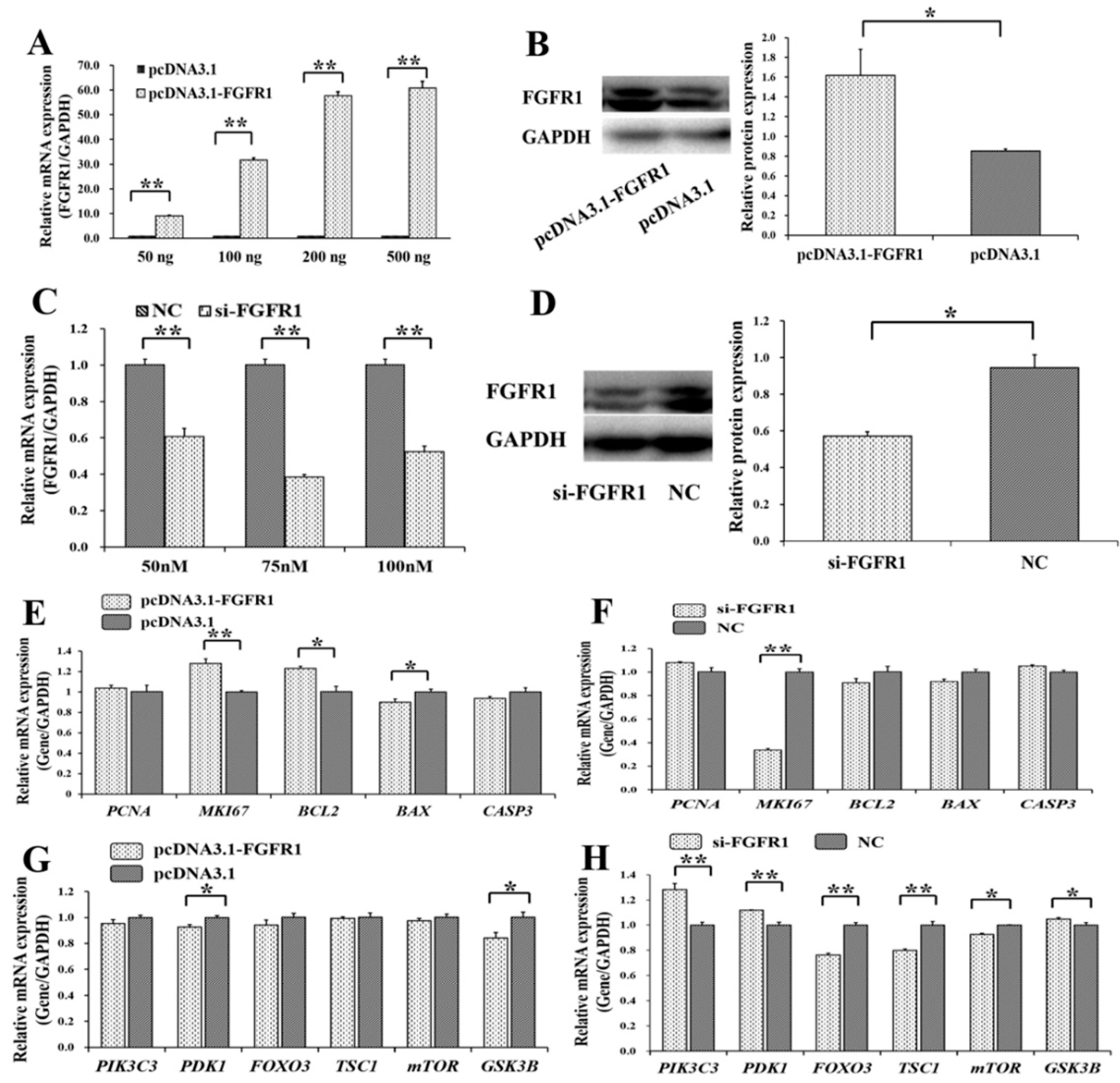
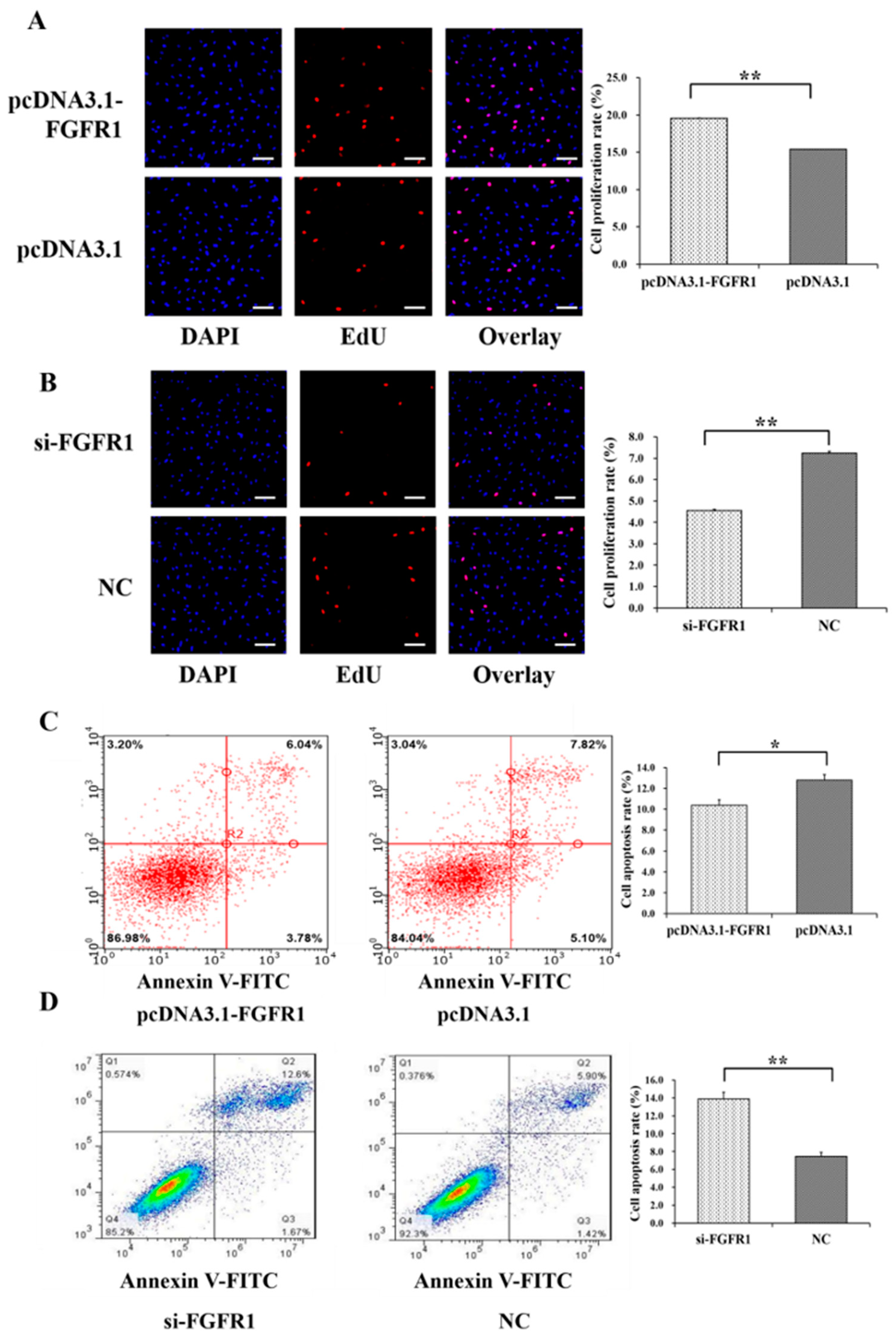
3.4. Pro-Proliferation and Anti-Apoptotic Effects of FGFR1 on GCs
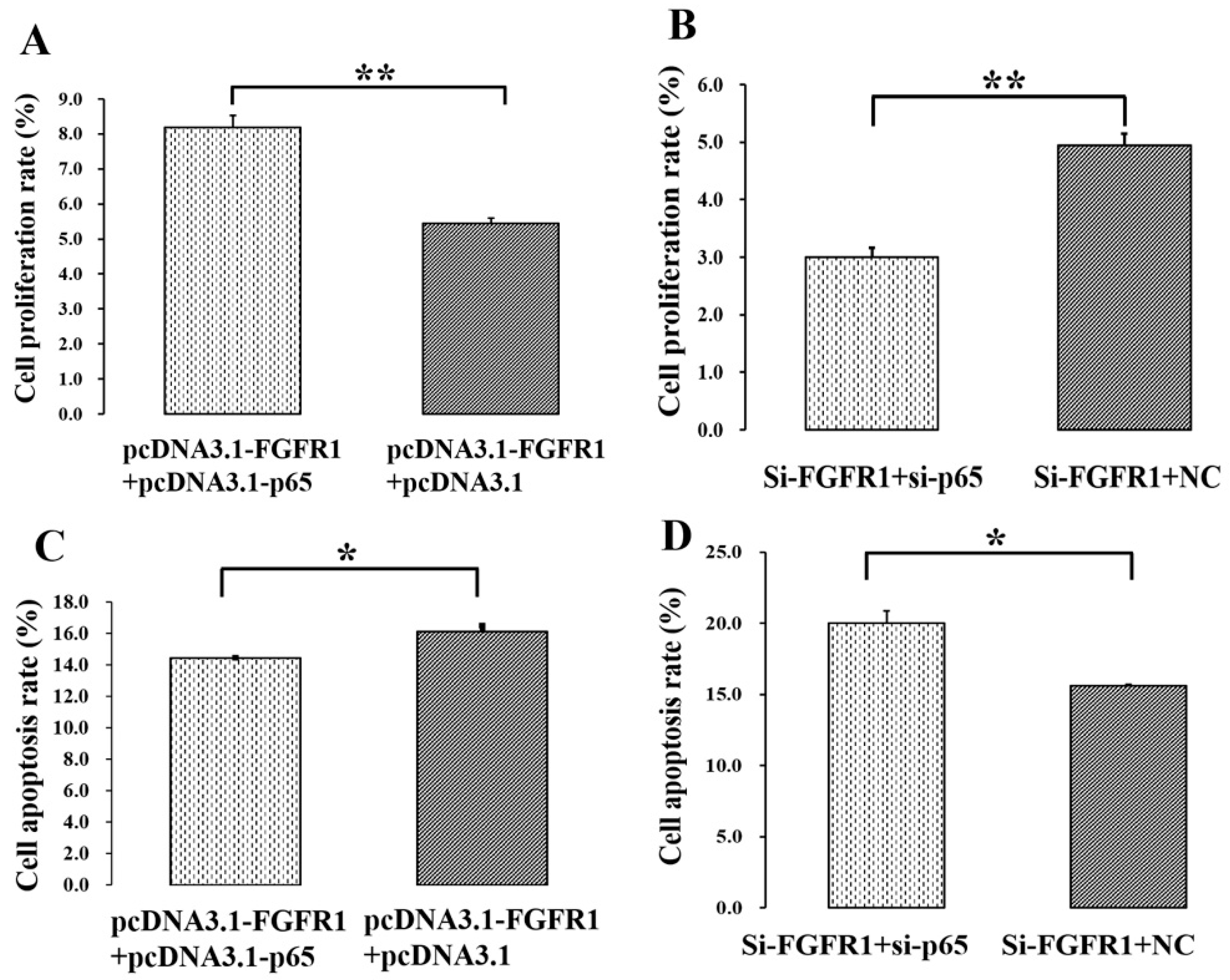
3.5. p65 Enhances the Pro-Proliferation and Anti-Apoptotic Effects of FGFR1 in GCs
4. Discussion
Author Contributions
Funding
Data Availability Statements
Conflicts of Interest
References
- Hunter, M.G. Oocyte maturation and ovum quality in pigs. Rev. Reprod. 2000, 5, 122–130. [Google Scholar] [CrossRef] [PubMed]
- McGee, E.A.; Hsueh, A.J. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Soede, N.M.; Langendijk, P.; Kemp, B. Reproductive cycles in pigs. Anim. Reprod. Sci. 2011, 124, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef]
- Manabe, N.; Goto, Y.; Matsuda-Minehata, F.; Inoue, N.; Maeda, A.; Sakamaki, K.; Miyano, T. Regulation mechanism of selective atresia in porcine follicles: Regulation of granulosa cell apoptosis during atresia. J. Reprod. Dev. 2004, 50, 493–514. [Google Scholar] [CrossRef]
- Shao, Y.Y.; Chang, Z.P.; Cheng, Y.; Wang, X.C.; Zhang, J.P.; Feng, X.J.; Guo, Y.T.; Liu, J.J.; Hou, R.G. Shaoyao-Gancao Decoction alleviated hyperandrogenism in a letrozole-induced rat model of polycystic ovary syndrome by inhibition of NF-kappaB activation. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Owens, L.A.; Kristensen, S.G.; Lerner, A.; Christopoulos, G.; Lavery, S.; Hanyaloglu, A.C.; Hardy, K.; Yding Andersen, C.; Franks, S. Gene expression in granulosa cells from small antral follicles from women with or without polycystic ovaries. J. Clin. Endocrinol. Metab. 2019. [Google Scholar] [CrossRef]
- Pfieffer, M.L. Polycystic ovary syndrome: An update. Nursing 2019, 49, 34–40. [Google Scholar] [CrossRef]
- Mishra, S.R.; Thakur, N.; Somal, A.; Parmar, M.S.; Reshma, R.; Rajesh, G.; Yadav, V.P.; Bharti, M.K.; Bharati, J.; Paul, A.; et al. Expression and localization of fibroblast growth factor (FGF) family in buffalo ovarian follicle during different stages of development and modulatory role of FGF2 on steroidogenesis and survival of cultured buffalo granulosa cells. Res. Vet. Sci. 2016, 108, 98–111. [Google Scholar] [CrossRef]
- Berisha, B.; Schams, D.; Kosmann, M.; Amselgruber, W.; Einspanier, R. Expression and localisation of vascular endothelial growth factor and basic fibroblast growth factor during the final growth of bovine ovarian follicles. J. Endocrinol. 2000, 167, 371–382. [Google Scholar] [CrossRef]
- Lin, X.; Ma, Y.; Qian, T.; Yao, J.; Mi, Y.; Zhang, C. Basic fibroblast growth factor promotes prehierarchical follicle growth and yolk deposition in the chicken. Theriogenology 2019, 139, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhang, G.; Lin, X.; Zhao, D.; Zhang, C.; Mi, Y. Reciprocal stimulating effects of bFGF and FSH on chicken primordial follicle activation through AKT and ERK pathway. Theriogenology 2019, 132, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu, A.K. Update on the Genetics of Idiopathic Hypogonadotropic Hypogonadism. J. Clin. Res. Pediatr. Endocrinol. 2017, 9, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.R. The Genetic Basis of Delayed Puberty. Front. Endocrinol. 2019, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Berisha, B.; Sinowatz, F.; Schams, D. Expression and localization of fibroblast growth factor (FGF) family members during the final growth of bovine ovarian follicles. Mol. Reprod. Dev. 2004, 67, 162–171. [Google Scholar] [CrossRef]
- Lin, J.; Jia, Y.; Zeng, W.; Mi, Y.; Zhang, C. Basic FGF promotes proliferation of ovarian granulosa cells in the laying chickens via FGFR1 and PKC pathway. Reprod. Domest. Anim. 2012, 47, 135–142. [Google Scholar] [CrossRef]
- Pavlova, S.; Klucska, K.; Vasicek, D.; Kotwica, J.; Sirotkin, A.V. Transcription factor NF-kappaB (p50/p50, p65/p65) controls porcine ovarian cells functions. Anim. Reprod. Sci. 2011, 128, 73–84. [Google Scholar] [CrossRef]
- Xu, J.J.; Wang, G.; Luo, X.; Wang, L.J.; Bao, Y.; Yang, X. Role of nuclear factor-kappaB pathway in the transition of mouse secondary follicles to antral follicles. J. Cell Physiol. 2019, 234, 22565–22580. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Y.; Xu, S.; Qi, G.; Wu, X. Bioinformatics identification of microRNAs involved in polycystic ovary syndrome based on microarray data. Mol. Med. Rep. 2019, 20, 281–291. [Google Scholar] [CrossRef]
- Peluso, J.J.; Pru, C.A.; Liu, X.; Kelp, N.C.; Pru, J.K. Progesterone receptor membrane component 1 and 2 regulate granulosa cell mitosis and survival through a NFKappaB-dependent mechanismdagger. Biol. Reprod. 2019, 100, 1571–1580. [Google Scholar] [CrossRef]
- Gao, H.; Lin, L.; Haq, I.U.; Zeng, S.M. Inhibition of NF-kappaB promotes autophagy via JNK signaling pathway in porcine granulosa cells. Biochem. Biophys. Res. Commun. 2016, 473, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhou, X.; He, Y.; Zhong, Y.; Zhang, A.; Zhang, Z.; Zhang, H.; Li, J. C/EBPbeta Promotes STAT3 Expression and Affects Cell Apoptosis and Proliferation in Porcine Ovarian Granulosa Cells. Genes (Basel) 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Deng, X.; Zhou, X.; Zhang, A.; Xing, Y.; Zhang, Z.; Zhang, H.; Li, J. MiR-126-3p promotes the cell proliferation and inhibits the cell apoptosis by targeting TSC1 in the porcine granulosa cells. In Vitro Cell Dev. Biol. Anim. 2018, 54, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Men, Y.; Fan, Y.H.; Shen, Y.Y.; Lu, L.G.; Kallen, A.N. The Steroidogenic Acute Regulatory Protein (StAR) Is Regulated by the H19/let-7 Axis. Endocrinology 2017, 158, 402–409. [Google Scholar] [CrossRef]
- Wang, S.; Yang, S.; Lai, Z.; Ding, T.; Shen, W.; Shi, L.; Jiang, J.; Ma, L.; Tian, Y.; Du, X.; et al. Effects of culture and transplantation on follicle activation and early follicular growth in neonatal mouse ovaries. Cell Tissue Res. 2013, 354, 609–621. [Google Scholar] [CrossRef]
- Asselin, E.; Xiao, C.W.; Wang, Y.F.; Tsang, B.K. Mammalian follicular development and atresia: Role of apoptosis. Biol. Signals Recept. 2000, 9, 87–95. [Google Scholar] [CrossRef]
- Phoophitphong, D.; Srisuwatanasagul, S.; Koonjaenak, S.; Tummaruk, P. Apoptotic Cell Localization in Preantral and Antral Follicles in Relation to Non-cyclic and Cyclic Gilts. Reprod. Domest. Anim. 2016, 51, 400–406. [Google Scholar] [CrossRef]
- Jiang, J.Y.; Cheung, C.K.; Wang, Y.; Tsang, B.K. Regulation of cell death and cell survival gene expression during ovarian follicular development and atresia. Front. Biosci. 2003, 8, d222–d237. [Google Scholar] [CrossRef]
- Wang, Y.; Chan, S.; Tsang, B.K. Involvement of inhibitory nuclear factor-kappaB (NFkappaB)-independent NFkappaB activation in the gonadotropic regulation of X-linked inhibitor of apoptosis expression during ovarian follicular development in vitro. Endocrinology 2002, 143, 2732–2740. [Google Scholar] [CrossRef][Green Version]
- Vasudevan, K.M.; Gurumurthy, S.; Rangnekar, V.M. Suppression of PTEN expression by NF-kappa B prevents apoptosis. Mol. Cell Biol. 2004, 24, 1007–1021. [Google Scholar] [CrossRef]
- Simon, P.S.; Sharman, S.K.; Lu, C.; Yang, D.; Paschall, A.V.; Tulachan, S.S.; Liu, K. The NF-kappaB p65 and p50 homodimer cooperate with IRF8 to activate iNOS transcription. BMC Cancer 2015, 15, 770. [Google Scholar] [CrossRef] [PubMed]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: From the known and the unknown. J. Cell Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Uri-Belapolsky, S.; Miller, I.; Shaish, A.; Levi, M.; Harats, D.; Ninio-Many, L.; Kamari, Y.; Shalgi, R. Interleukin 1-alpha deficiency increases the expression of Follicle-stimulating hormone receptors in granulosa cells. Mol. Reprod. Dev. 2017, 84, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Pelusi, C.; Ikeda, Y.; Zubair, M.; Parker, K.L. Impaired follicle development and infertility in female mice lacking steroidogenic factor 1 in ovarian granulosa cells. Biol. Reprod. 2008, 79, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Sudo, H.; Minami, A. Regulation of apoptosis in nucleus pulposus cells by optimized exogenous Bcl-2 overexpression. J. Orthop. Res. 2010, 28, 1608–1613. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, G.; Yuan, Z.; Dong, Y.; Wang, J.; Lu, W. Effect of kisspeptin on the proliferation and apoptosis of bovine granulosa cells. Anim. Reprod. Sci. 2017, 185, 1–7. [Google Scholar] [CrossRef]
- Bildik, G.; Akin, N.; Seyhan, A.; Esmaeilian, Y.; Yakin, K.; Keles, I.; Balaban, B.; Ata, B.; Urman, B.; Oktem, O. Luteal granulosa cells from natural cycles are more capable of maintaining their viability, steroidogenic activity and LH receptor expression than those of stimulated IVF cycles. Hum. Reprod. 2019, 34, 345–355. [Google Scholar] [CrossRef]
- Chen, Z.; Lei, L.; Wen, D.; Yang, L. Melatonin attenuates palmitic acid-induced mouse granulosa cells apoptosis via endoplasmic reticulum stress. J. Ovarian Res. 2019, 12, 43. [Google Scholar] [CrossRef]
- Lei, L.; Han, F.; Cui, Q.; Liao, W.; Liu, H.; Guan, G.; Yang, L. IRS2 depletion inhibits cell proliferation and decreases hormone secretion in mouse granulosa cells. J. Reprod. Dev. 2018, 64, 409–416. [Google Scholar] [CrossRef]
- Zhu, W.; Yang, M.; Shang, J.; Xu, Y.; Wang, Y.; Tao, Q.; Zhang, L.; Ding, Y.; Chen, Y.; Zhao, D.; et al. MiR-222 inhibits apoptosis in porcine follicular granulosa cells by targeting the THBS1 gene. Anim. Sci. J. 2019, 90, 719–727. [Google Scholar] [CrossRef]
- Lobach, V.N.; Casalechi, M.; Dela Cruz, C.; Pereira, M.T.; Del Puerto, H.L.; Reis, F.M. Caspase-3 gene expression in human luteinized granulosa cells is inversely correlated with the number of oocytes retrieved after controlled ovarian stimulation. Hum. Fertil. 2019, 22, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Makker, A.; Goel, M.M.; Mahdi, A.A. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: An update. J. Mol. Endocrinol. 2014, 53, R103–R118. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Nagaraju, G.; Liu, Z.; Liu, K. Functional roles of the phosphatidylinositol 3-kinases (PI3Ks) signaling in the mammalian ovary. Mol. Cell Endocrinol. 2012, 356, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Furlong, H.C.; Stampfli, M.R.; Gannon, A.M.; Foster, W.G. Cigarette smoke exposure triggers the autophagic cascade via activation of the AMPK pathway in mice. Biol. Reprod. 2015, 93, 93. [Google Scholar] [CrossRef]
- Xin, X.; Li, Z.; Zhong, Y.; Li, Q.; Wang, J.; Zhang, H.; Yuan, X.; Li, J.; Zhang, Z. KISS1 Suppresses Apoptosis and Stimulates the Synthesis of E2 in Porcine Ovarian Granulosa Cells. Anim. (Basel) 2019, 9. [Google Scholar] [CrossRef]
- Liu, Z.; Castrillon, D.H.; Zhou, W.; Richards, J.S. FOXO1/3 depletion in granulosa cells alters follicle growth, death and regulation of pituitary FSH. Mol. Endocrinol. 2013, 27, 238–252. [Google Scholar] [CrossRef]
- Yamamoto, H.; Yamashita, Y.; Saito, N.; Hayashi, A.; Hayashi, M.; Terai, Y.; Ohmichi, M. Lower FOXO3 mRNA expression in granulosa cells is involved in unexplained infertility. J. Obs. Gynaecol. Res. 2017, 43, 1021–1028. [Google Scholar] [CrossRef]
- Huang, L.; Wang, Z.B.; Jiang, Z.Z.; Hu, M.W.; Lin, F.; Zhang, Q.H.; Luo, Y.B.; Hou, Y.; Zhao, Y.; Fan, H.Y.; et al. Specific disruption of Tsc1 in ovarian granulosa cells promotes ovulation and causes progressive accumulation of corpora lutea. PLoS ONE 2013, 8, e54052. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, T.; Guo, Y.; Sun, T.; Li, H.; Zhang, X.; Yin, H.; Cao, G.; Yin, Y.; Wang, H.; et al. Oocyte stage-specific effects of MTOR determine granulosa cell fate and oocyte quality in mice. Proc. Natl. Acad. Sci. USA 2018, 115, E5326–E5333. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, Y.H.; Kim, H.M.; Park, H.O.; Ha, N.C.; Kim, T.H.; Park, M.; Lee, K.; Bae, J. FOXL2 posttranslational modifications mediated by GSK3 beta determine the growth of granulosa cell tumours. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
| Name | Sequence | Product (bp) | Accession Number |
|---|---|---|---|
| CDS-FGFR1 | F: CGGGATCCGCTCACCAACGAGGATCAA R: GCTCTAGAGTCACAGCTAAGGATGGAGT | 2597 | XM_005671767.3 |
| CDS-P65 | F: GGGGTACCATGGACGACCTCTTCCCCCT R: GCTCTAGATTAGGAGCTGATCTGACTCA | 1662 | NM_001114281.1 |
| ChIP-FGFR1 | F: GACTCAGTTTAGCGCATTGC R: GAGAAAAGTCCTCAGGCTCC | 92 | XM_005671767.3 |
| ChIP-GAPDH | F: GATGTCCTGAGCCCCTACAG R: GGTAGGTGATGGGGACTGAG | 102 | NM_001206359.1 |
| qRT-PCR-FGFR1 | F: GGCTACAAGGTCCGTTATG R: CAATCTTACTCCCATTCACC | 289 | XM_005671767.3 |
| qRT-PCR-P65 | F: CATGCGCTTCCGCTACAAG R: GGTCCCGCTTCTTTACACAC | 284 | NM_001114281.1 |
| qRT-PCR-GAPDH | F: TCGGAGTGAACGGATTTG R: TCACCCCATTTGATGTTGG | 250 | NM_001206359.1 |
| qRT-PCR-PCNA | F: ATGCAGACACCTTGGCACTA R: TTTCCGAGTTCTCCACTTGC | 247 | NM_001291925.1 |
| qRT-PCR-CASP3 | F: AGGAGCAGTTTTATTTGCGTGC R: AGTCCCACTGTCCGTCTCAA | 204 | NM_214131.1 |
| qRT-PCR-GSK3B | F: TAGTCGAGCCAAACAGACGC R: ATTGGGTTCTCCTCGGACCA | 202 | NM_001128443.1 |
| qRT-PCR-BAX | F: AGCGCATTGGAGATGAACTG R: AAGTAGAAAAGCGCGACCAC | 157 | XM_003127290.5 |
| qRT-PCR-BCL2 | F: TTGCCGAGATGTCCAGCCAG R: TCAGTCATCCACAGGGCGAT | 202 | XM_021099593.1 |
| qRT-PCR-PIK3C3 | F: AACGGGCTTTGAGATAGTGAA R: AAGTTGCTTGGTTGGTGGATA | 184 | NM_001012956.2 |
| qRT-PCR-PDK1 | F: ACATGTACTCCACTGCACCC R: AGGCGTGATATGGGCAATCC | 91 | NM_001159608.1 |
| qRT-PCR-FOXO3 | F: ACAAACGGCTCACTCTGTCCCA R: GAACTGTTGCTGTCGCCCTTATC | 85 | NM_001135959.1 |
| qRT-PCR-TSC1 | F: AGCCGTCGTGGAAGCATAAG R: GTGCAGGTGGACGAGATAGA | 245 | XM_021070814.1 |
| qRT-PCR-mTOR | F: CCGTCATCCCCTTATCCACC R: GGTCGTAGTCTGGAGCCATC | 153 | XM_003127584.6 |
| qRT-PCR-MKI67 | F: ACAACAGGAGGAGGAAGTGCT R: TTGACCTAGACGCGGGGAT | 300 | XM_021073740.1 |
| Name | Sequences | Product (bp) | Accession Number |
|---|---|---|---|
| p1 (−2044/+401) | F: GGGGTACCAAATTAGGGGACAAGGTTATCT | 2445 | XM_005671767.3 |
| R: GAAGATCTGAGTTGGCGGAAAAGTTGGG | |||
| p2 (−1652/+401) | F: GGGGTACCATAGCCTGATTCCTCAAGTCTG | 2053 | XM_005671767.3 |
| ditto | |||
| p3 (−1069/+401) | F: GGGGTACCGTTGCGCTGCCTGTGGTGTA | 1470 | XM_005671767.3 |
| ditto | |||
| p4 (−815/+401) | F: GGGGTACCACTTCAGGGCTACAGCGTCT | 1216 | XM_005671767.3 |
| ditto | |||
| p5 (−700/+401) | F: GGGGTACCAGCCAGAACGCAGGAAAGGA | 1101 | XM_005671767.3 |
| ditto | |||
| p6 (−377/+401) | F: GGGGTACCGACTCAGTTTAGCGCATTGC | 778 | XM_005671767.3 |
| ditto | |||
| p7 (−84/+401) | F: GGGGTACCGCTTCGGCTCCATTGTTCC | 485 | XM_005671767.3 |
| ditto | |||
| P6 deletion | F: GGCGACCTCG****CCGCGGGCGCGCGCTGCATC | 748 | XM_005671767.3 |
| R: GAAGATCTGAGTTGGCGGAAAAGTTGGG | |||
| P6 deletion | F: GGGGTACCGACTCAGTTTAGCGCATTGC | 50 | |
| R: GATGCAGCGCGCGCCCGCGG****CGAGGTCGCC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, X.; Li, Z.; Kong, Y.; Zhong, Y.; He, Y.; Zhang, A.; Zhou, X.; Jiang, Y.; Zhang, Z.; Zhang, H.; et al. P65 Targets FGFR1 to Regulate the Survival of Ovarian Granulosa Cells. Cells 2019, 8, 1334. https://doi.org/10.3390/cells8111334
Yuan X, Li Z, Kong Y, Zhong Y, He Y, Zhang A, Zhou X, Jiang Y, Zhang Z, Zhang H, et al. P65 Targets FGFR1 to Regulate the Survival of Ovarian Granulosa Cells. Cells. 2019; 8(11):1334. https://doi.org/10.3390/cells8111334
Chicago/Turabian StyleYuan, Xiaolong, Zhonghui Li, Yaru Kong, Yuyi Zhong, Yingting He, Ailing Zhang, Xiaofeng Zhou, Yao Jiang, Zhe Zhang, Hao Zhang, and et al. 2019. "P65 Targets FGFR1 to Regulate the Survival of Ovarian Granulosa Cells" Cells 8, no. 11: 1334. https://doi.org/10.3390/cells8111334
APA StyleYuan, X., Li, Z., Kong, Y., Zhong, Y., He, Y., Zhang, A., Zhou, X., Jiang, Y., Zhang, Z., Zhang, H., & Li, J. (2019). P65 Targets FGFR1 to Regulate the Survival of Ovarian Granulosa Cells. Cells, 8(11), 1334. https://doi.org/10.3390/cells8111334





