Serine–Arginine Protein Kinase SRPK2 Modulates the Assembly of the Active Zone Scaffolding Protein CAST1/ERC2
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Plasmid Construction
2.3. Cell Culture and Transfection
2.4. Rat Hippocampal Neuron Culture
2.5. Immunofluorescence Imaging and Quantification of Fluorescence Intensity
2.6. Co-Immunoprecipitation
2.7. Subcellular Fractionation
2.8. Immunoblotting
2.9. Imaging Synaptic Proteins
2.10. Statistical Analysis
2.11. Ethical Approval
3. Results
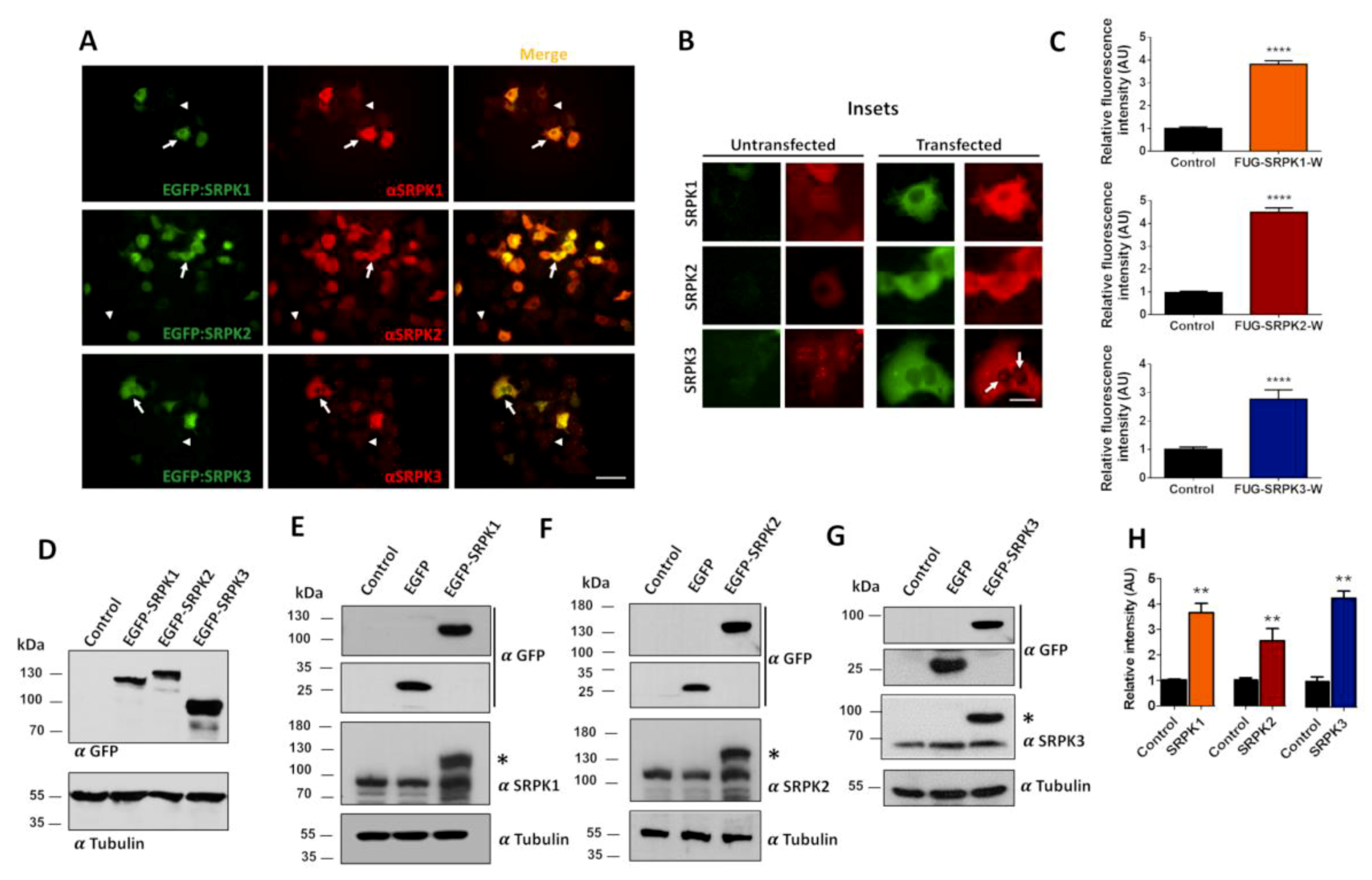
3.1. Establishing a Model to Study SRPKs in Heterologous Cells
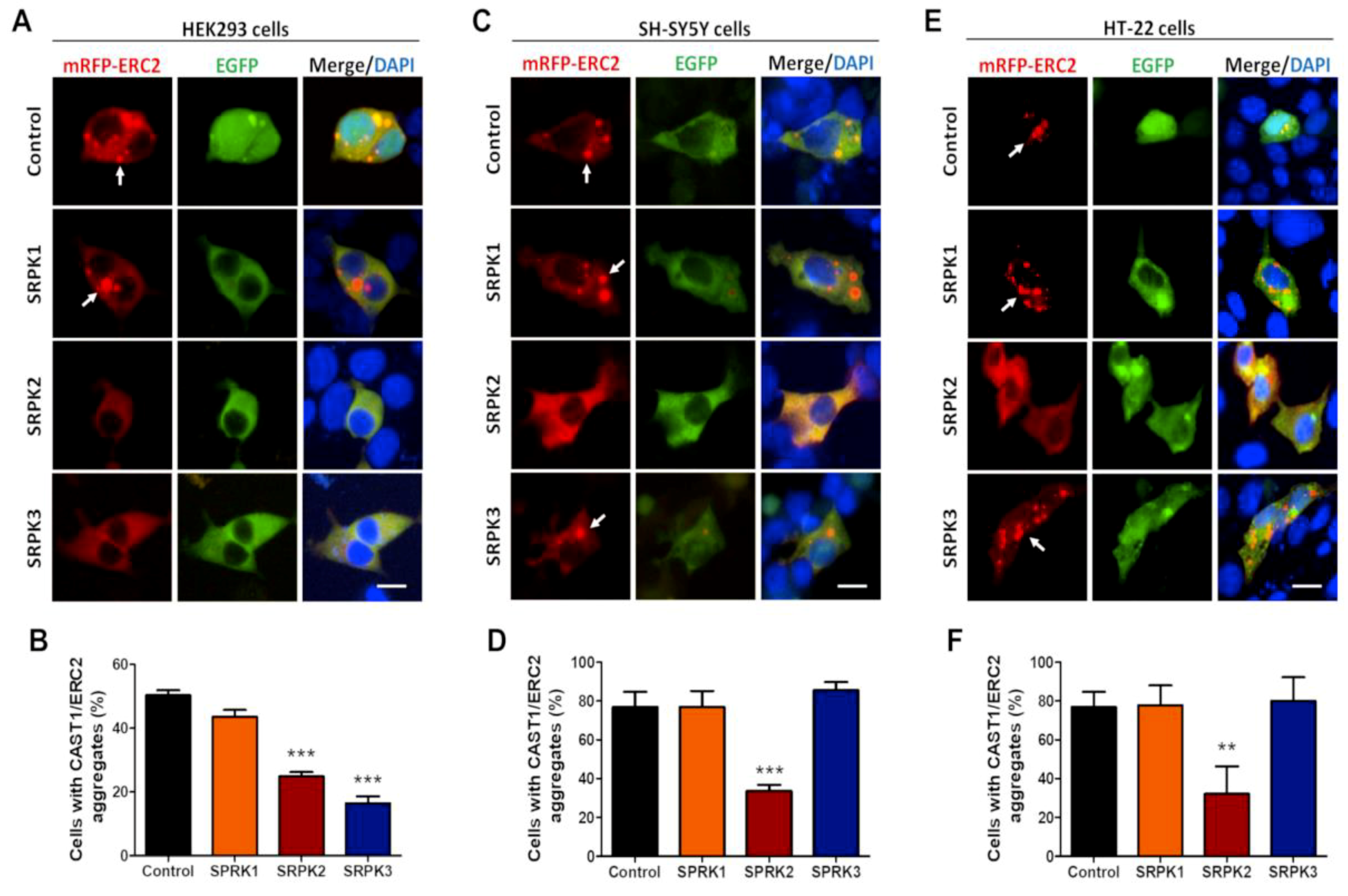
3.2. Effect of SRPKs on CAST1/ERC2 Aggregation in Heterologous Cells
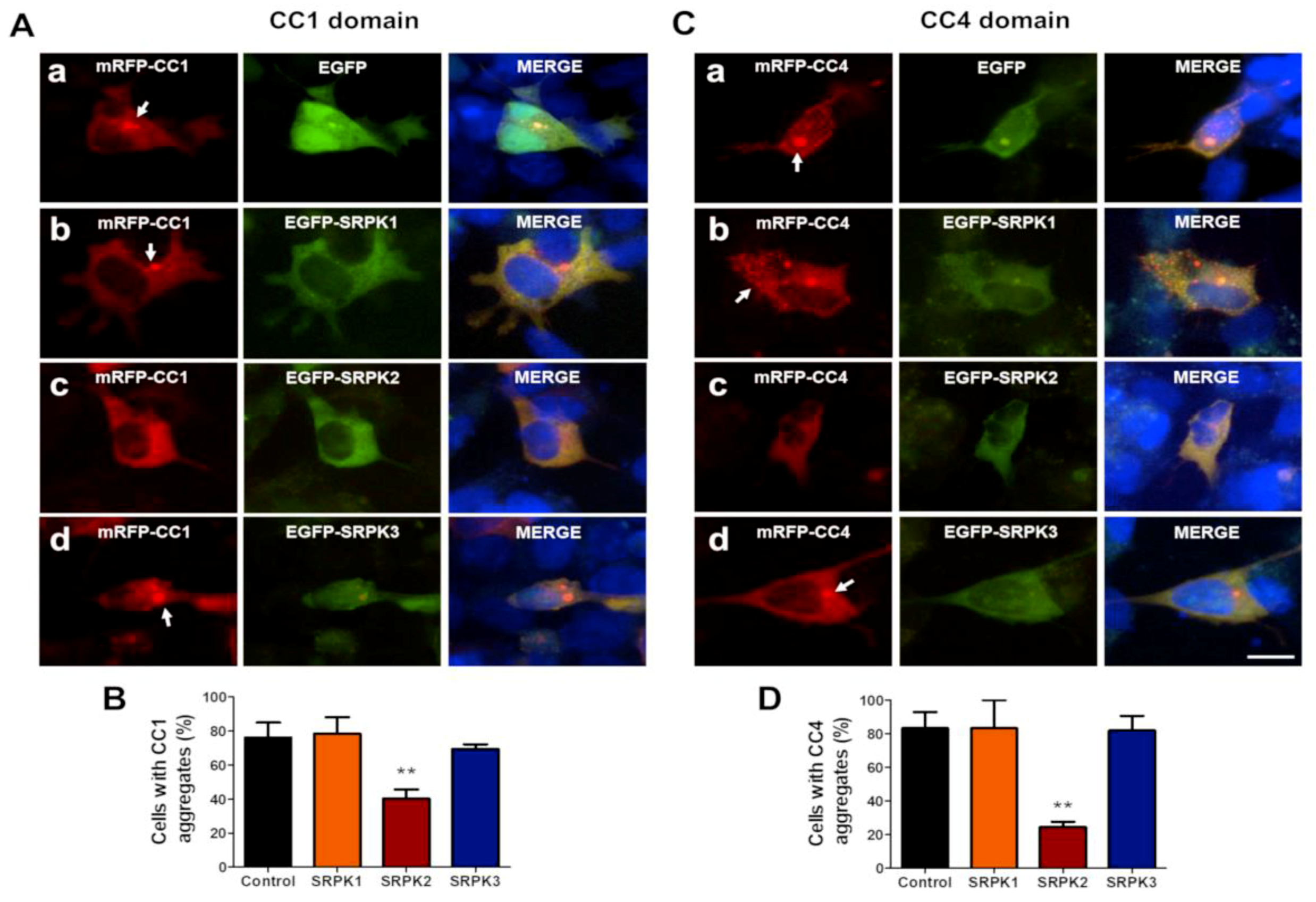
3.3. SRPKs Affect Self-Aggregation of Coiled-Coil Domains 1 and 4 of CAST1/ERC2
3.4. Exploring Whether SRPK2 Forms a Complex with CAST1/ERC2
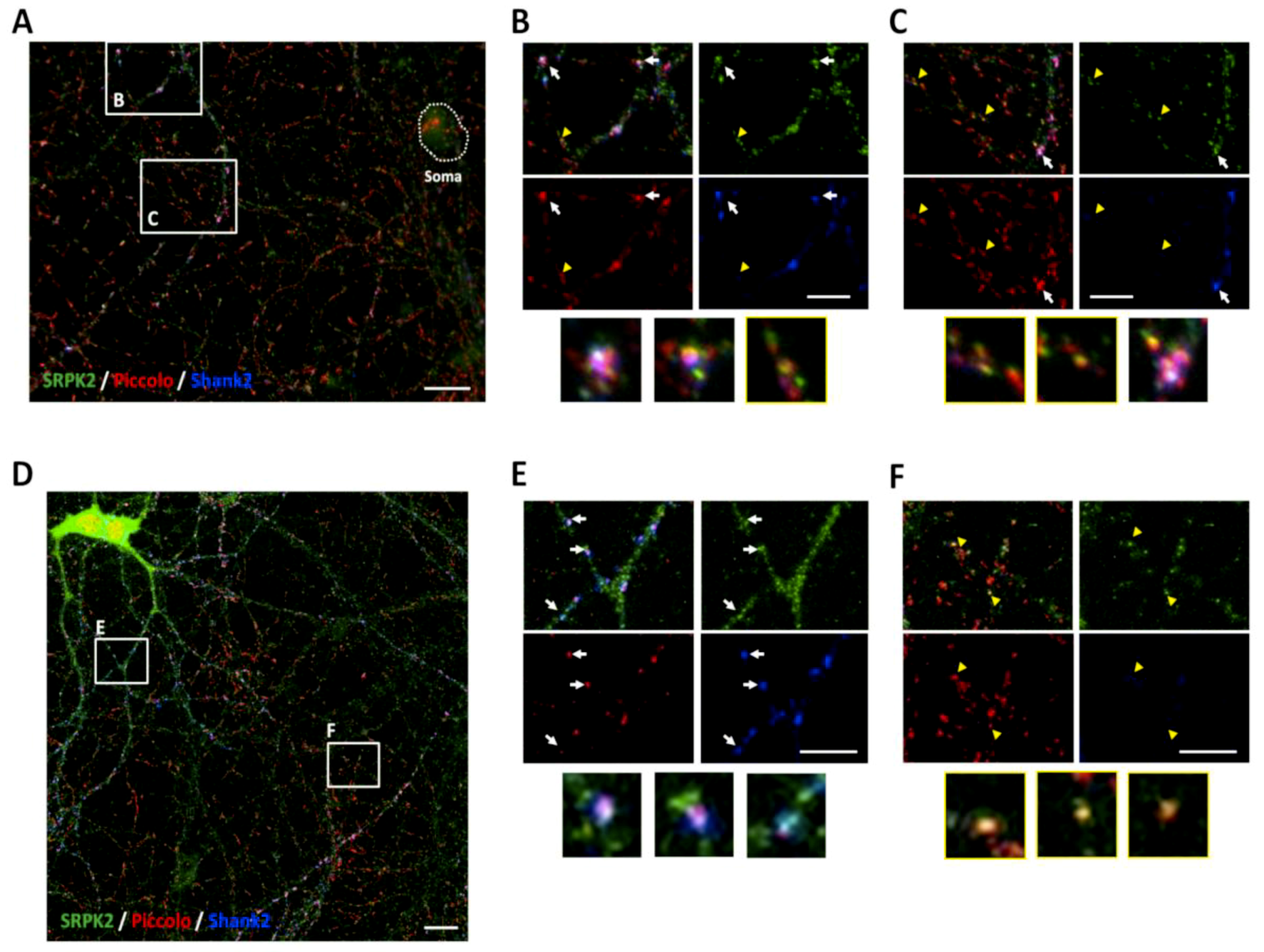
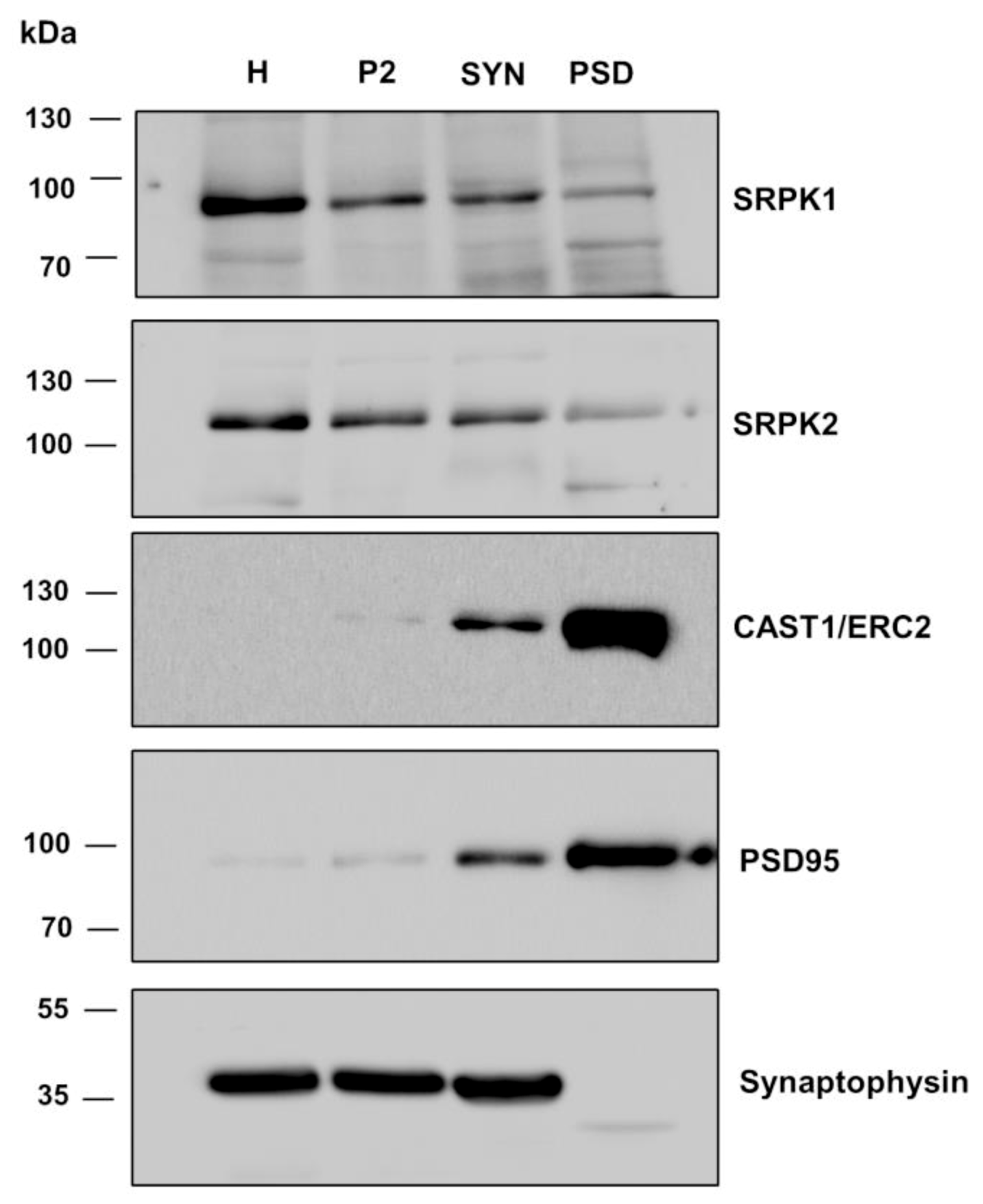
3.5. SRPK2 Is Localized in Synapses of Hippocampal Neurons in Culture
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Walsh, C.T.; Gatto, G.J.; Garneau-Tsodikova, S.; Garneau-Tsodikova, S.; Garneau-Tsodikova, S. Protein Posttranslational Modifications: The Chemistry of Proteome Diversifications. Angew. Chem. Int. Ed. 2005, 44, 7342–7372. [Google Scholar] [CrossRef] [PubMed]
- Garner, C.C.; Zhai, R.G.; Gundelfinger, E.D.; Ziv, N.E. Molecular mechanisms of CNS synaptogenesis. Trends Neurosci. 2002, 25, 243–250. [Google Scholar] [CrossRef]
- Baron, M.K.; Boeckers, T.M.; Vaida, B.; Faham, S.; Gingery, M.; Sawaya, M.R.; Salyer, D.; Gundelfinger, E.D.; Bowie, J.U. An Architectural Framework That May Lie at the Core of the Postsynaptic Density. Science 2006, 311, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Gundelfinger, E.D.; Boeckers, T.M.; Baron, M.K.; Bowie, J.U. A role for zinc in postsynaptic density asSAMbly and plasticity? Trends Biochem. Sci. 2006, 31, 366–373. [Google Scholar] [CrossRef]
- Arons, M.H.; Lee, K.; Thynne, C.J.; Kim, S.A.; Schob, C.; Kindler, S.; Montgomery, J.M.; Garner, C.C. Shank3 Is Part of a Zinc-Sensitive Signaling System That Regulates Excitatory Synaptic Strength. J. Neurosci. 2016, 36, 9124–9134. [Google Scholar] [CrossRef]
- Fritschy, J.-M.; Harvey, R.J.; Schwarz, G. Gephyrin: Where do we stand, where do we go? Trends Neurosci. 2008, 31, 257–264. [Google Scholar] [CrossRef]
- Calamai, M.; Specht, C.G.; Heller, J.; Alcor, D.; Machado, P.; Vannier, C.; Triller, A. Gephyrin Oligomerization Controls GlyR Mobility and Synaptic Clustering. J. Neurosci. 2009, 29, 7639–7648. [Google Scholar] [CrossRef]
- Gundelfinger, E.D.; Reissner, C.; Garner, C.C. Role of Bassoon and Piccolo in Assembly and Molecular Organization of the Active Zone. Front. Synaptic Neurosci. 2016, 7, 923. [Google Scholar] [CrossRef]
- Torres, V.I.; Inestrosa, N.C. Vertebrate Presynaptic Active Zone Assembly: A Role Accomplished by Diverse Molecular and Cellular Mechanisms. Mol. Neurobiol. 2018, 55, 4513–4528. [Google Scholar] [CrossRef]
- Wang, X.; Hu, B.; Zieba, A.; Neumann, N.G.; Kasper-Sonnenberg, M.; Honsbein, A.; Hultqvist, G.; Conze, T.; Witt, W.; Limbach, C.; et al. A Protein Interaction Node at the Neurotransmitter Release Site: Domains of Aczonin/Piccolo, Bassoon, CAST, and Rim Converge on the N-Terminal Domain of Munc13-1. J. Neurosci. 2009, 29, 12584–12596. [Google Scholar] [CrossRef]
- Wagh, D.A.; Rasse, T.M.; Asan, E.; Hofbauer, A.; Schwenkert, I.; Dürrbeck, H.; Buchner, S.; Dabauvalle, M.-C.; Schmidt, M.; Qin, G.; et al. Bruchpilot, a Protein with Homology to ELKS/CAST, Is Required for Structural Integrity and Function of Synaptic Active Zones in Drosophila. Neuron 2006, 49, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Kittel, R.J.; Wichmann, C.; Rasse, T.M.; Fouquet, W.; Schmidt, M.; Schmid, A.; Wagh, D.A.; Pawlu, C.; Kellner, R.R.; Willig, K.I.; et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 2006, 312, 1051–1054. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.L.; Fetter, R.D.; Davis, G.W. Negative regulation of active zone assembly by a newly identified SR protein kinase. PLoS Boil. 2009, 7, e1000193. [Google Scholar] [CrossRef]
- Nieratschker, V.; Schubert, A.; Jauch, M.; Bock, N.; Bucher, D.; Dippacher, S.; Krohne, G.; Asan, E.; Buchner, S.; Buchner, E. Bruchpilot in Ribbon-Like Axonal Agglomerates, Behavioral Defects, and Early Death in SRPK79D Kinase Mutants of Drosophila. PLoS Genet. 2009, 5, e1000700. [Google Scholar] [CrossRef] [PubMed]
- Beck, K.; Brodsky, B. Supercoiled Protein Motifs: The Collagen Triple-Helix and the α-Helical Coiled Coil. J. Struct. Boil. 1998, 122, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.A.; Squire, J.M. Fibrous Proteins: New Structural and Functional Aspects Revealed. Membrane Proteins 2005, 70, 1–10. [Google Scholar] [CrossRef]
- Truebestein, L.; Leonard, T.A. Coiled-coils: The long and short of it. BioEssays 2016, 38, 903–916. [Google Scholar] [CrossRef]
- Ohtsuka, T.; Takao-Rikitsu, E.; Inoue, E.; Inoue, M.; Takeuchi, M.; Matsubara, K.; Deguchi-Tawarada, M.; Satoh, K.; Morimoto, K.; Nakanishi, H.; et al. Cast: A novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13-1. J. Cell Biol. 2002, 158, 577–590. [Google Scholar] [CrossRef]
- Takao-Rikitsu, E.; Mochida, S.; Inoue, E.; Deguchi-Tawarada, M.; Inoue, M.; Ohtsuka, T.; Takai, Y. Physical and functional interaction of the active zone proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J. Cell Boil. 2004, 164, 301–311. [Google Scholar] [CrossRef]
- Gui, J.F.; Tronchere, H.; Chandler, S.D.; Fu, X.D. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. 1994, 91, 10824–10828. [Google Scholar] [CrossRef]
- Mytilinaios, D.; Tsamis, K.; Nikolakaki, E.; Giannakouros, T. Distribution of SRPK1 in human brain. J. Chem. Neuroanat. 2012, 43, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Kuroyanagi, N.; Onogi, H.; Wakabayashi, T.; Hagiwara, M. Novel SR-Protein-Specific Kinase, SRPK2, Disassembles Nuclear Speckles. Biochem. Biophys. Res. Commun. 1998, 242, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Lin, W.; Dyck, J.A.; Yeakley, J.M.; Songyang, Z.; Cantley, L.C.; Fu, X.-D. SRPK2: A Differentially Expressed SR Protein-specific Kinase Involved in Mediating the Interaction and Localization of Pre-mRNA Splicing Factors in Mammalian Cells. J. Cell Boil. 1998, 140, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, O.; Arnold, M.; Nakagawa, M.; Hamada, H.; Shelton, J.M.; Kusano, H.; Harris, T.M.; Childs, G.; Campbell, K.P.; Richardson, J.A.; et al. Centronuclear myopathy in mice lacking a novel muscle-specific protein kinase transcriptionally regulated by MEF2. Genome Res. 2005, 19, 2066–2077. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yu, W.; Xiong, Y.; Xie, H.; Ren, Z.; Xu, D.; Lei, M.; Zuo, B.; Feng, X. Molecular characterization and expression patterns of serine/arginine-rich specific kinase 3 (SPRK3) in porcine skeletal muscle. Mol. Biol. Rep. 2011, 38, 2903–2909. [Google Scholar] [CrossRef]
- Bullock, N.; Oltean, S. The many faces of SRPK1. J. Pathol. 2017, 241, 437–440. [Google Scholar] [CrossRef]
- Li, Z.; Xu, Y.; Lin, Y. Transcriptome analyses reveal genes of alternative splicing associated with muscle development in chickens. Gene 2018, 676, 146–155. [Google Scholar] [CrossRef]
- Jang, S.-W.; Liu, X.; Fu, H.; Rees, H.; Yepes, M.; Levey, A.; Ye, K. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 Mediates Cell Cycle and Cell Death in Neurons*. J. Boil. Chem. 2009, 284, 24512–24525. [Google Scholar] [CrossRef]
- Hong, Y.; Chan, C.B.; Kwon, I.-S.; Li, X.; Song, M.; Lee, H.-P.; Liu, X.; Sompol, P.; Jin, P.; Lee, H.-G.; et al. SRPK2 phosphorylates tau and mediates the cognitive defects in Alzheimer’s disease. J. Neurosci. 2012, 32, 17262–17272. [Google Scholar] [CrossRef]
- Chan, C.B.; Ye, K. Serine-arginine protein kinases: New players in neurodegenerative diseases? Rev. Neurosci. 2013, 24, 401–413. [Google Scholar] [CrossRef]
- Wang, Z.-H.; Liu, P.; Liu, X.; Manfredsson, F.P.; Sandoval, I.M.; Yu, S.P.; Wang, J.-Z.; Ye, K. Delta-Secretase Phosphorylation by SRPK2 Enhances Its Enzymatic Activity, Provoking Pathogenesis in Alzheimer’s Disease. Mol. Cell 2017, 67, 812–825.e5. [Google Scholar] [CrossRef] [PubMed]
- Leal-Ortiz, S.; Waites, C.L.; Terry-Lorenzo, R.; Zamorano, P.; Gundelfinger, E.D.; Garner, C.C. Piccolo modulation of Synapsin1a dynamics regulates synaptic vesicle exocytosis. J. Cell Boil. 2008, 181, 831–846. [Google Scholar] [CrossRef] [PubMed]
- Lira, M.; Arancibia, D.; Orrego, P.R.; Montenegro-Venegas, C.; Cruz, Y.; García, J.; Leal-Ortiz, S.; Godoy, J.A.; Gundelfinger, E.D.; Inestrosa, N.C. The Exocyst Component Exo70 Modulates Dendrite Arbor Formation, Synapse Density, and Spine Maturation in Primary Hippocampal Neurons. Mol. Neurobiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kaech, S.; Banker, G. Culturing hippocampal neurons. Nat. Protoc. 2006, 1, 2406–2415. [Google Scholar] [CrossRef] [PubMed]
- Smalla, K.-H.; Klemmer, P.; Wyneken, U. Isolation of the Postsynaptic Density: A Specialization of the Subsynaptic Cytoskeleton; Humana Press: Totowa, NJ, USA, 2013; pp. 265–280. [Google Scholar]
- Ko, J.; Na, M.; Kim, S.; Lee, J.-R.; Kim, E. Interaction of the ERC Family of RIM-binding Proteins with the Liprin-α Family of Multidomain Proteins. J. Boil. Chem. 2003, 278, 42377–42385. [Google Scholar] [CrossRef]
- Nakata, T.; Kitamura, Y.; Shimizu, K.; Tanaka, S.; Fujimori, M.; Yokoyama, S.; Ito, K.; Emi, M. Fusion of a novel gene, ELKS, toRET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosom. Cancer 1999, 25, 97–103. [Google Scholar] [CrossRef]
- Yokota, T.; Nakata, T.; Minami, S.; Inazawa, J.; Emi, M. Genomic organization and chromosomal mapping of ELKS, a gene rearranged in a papillary thyroid carcinoma. J. Hum. Genet. 2000, 45, 6–11. [Google Scholar] [CrossRef]
- Deguchi-Tawarada, M.; Inoue, E.; Takao-Rikitsu, E.; Inoue, M.; Ohtsuka, T.; Takai, Y.; Deguchi-Tawarada, M.; Takao-Rikitsu, E. CAST2: Identification and characterization of a protein structurally related to the presynaptic cytomatrix protein CAST. Genes Cells 2004, 9, 15–23. [Google Scholar] [CrossRef]
- Ghosh, S.; Kundu, A.; Chattopadhyay, K. Small Molecules Attenuate the Interplay between Conformational Fluctuations, Early Oligomerization and Amyloidosis of Alpha Synuclein. Sci. Rep. 2018, 8, 5481. [Google Scholar] [CrossRef]
- Dieck, S.; Sanmartí-Vila, L.; Langnaese, K.; Richter, K.; Kindler, S.; Soyke, A.; Wex, H.; Smalla, K.H.; Kämpf, U.; Fränzer, J.T. Gundelfinger, Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J. Cell Biol. 1998, 142, 499–509. [Google Scholar] [CrossRef]
- Maas, C.; Torres, V.I.; Altrock, W.D.; Leal-Ortiz, S.; Wagh, D.; Terry-Lorenzo, R.T.; Fejtova, A.; Gundelfinger, E.D.; Ziv, N.E.; Garner, C.C. Formation of Golgi-derived active zone precursor vesicles. J. Neurosci. 2012, 32, 11095–11108. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.M.; Kang, Y. Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 2007, 17, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Lucido, A.L.; Sanchez, F.S.; Thostrup, P.; Kwiatkowski, A.V.; Leal-Ortiz, S.; Gopalakrishnan, G.; Liazoghli, D.; Belkaid, W.; Lennox, R.B.; Grütter, P.; et al. Rapid assembly of functional presynaptic boutons triggered by adhesive contacts. J. Neurosci. 2009, 29, 12449–12466. [Google Scholar] [CrossRef] [PubMed]
- Dresbach, T.; Torres, V.; Wittenmayer, N.; Altrock, W.D.; Zamorano, P.; Zuschratter, W.; Nawrotzki, R.; Ziv, N.E.; Garner, C.C.; Gundelfinger, E.D. Assembly of Active Zone Precursor Vesicles: Obligatory trafficking of presynaptic cytomatrix proteins bassoon and piccolo via a trans-golgi compartment. J. Biol. Chem. 2005, 281, 6038–6047. [Google Scholar] [CrossRef] [PubMed]
- Mochida, S.; Hida, Y.; Tanifuji, S.; Hagiwara, A.; Hamada, S.; Abe, M.; Ma, H.; Yasumura, M.; Kitajima, I.; Sakimura, K.; et al. SAD-B Phosphorylation of CAST Controls Active Zone Vesicle Recycling for Synaptic Depression. Cell Rep. 2016, 16, 2901–2913. [Google Scholar] [CrossRef]
- Driller, J.H.; Lützkendorf, J.; Depner, H.; Siebert, M.; Kuropka, B.; Weise, C.; Piao, C.; Petzoldt, A.G.; Lehmann, M.; Stelzl, U.; et al. Phosphorylation of the Bruchpilot N-terminus in Drosophila unlocks axonal transport of active zone building blocks. J. Cell Sci. 2019, 132, jcs225151. [Google Scholar] [CrossRef]
- Zheng, Y.; Fu, X.-D.; Ou, J.-H.J. Suppression of hepatitis B virus replication by SRPK1 and SRPK2 via a pathway independent of the phosphorylation of the viral core protein. Virology 2005, 342, 150–158. [Google Scholar] [CrossRef][Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arancibia, D.; Lira, M.; Cruz, Y.; Barrera, D.P.; Montenegro-Venegas, C.; Godoy, J.A.; Garner, C.C.; Inestrosa, N.C.; Gundelfinger, E.D.; Zamorano, P.; et al. Serine–Arginine Protein Kinase SRPK2 Modulates the Assembly of the Active Zone Scaffolding Protein CAST1/ERC2. Cells 2019, 8, 1333. https://doi.org/10.3390/cells8111333
Arancibia D, Lira M, Cruz Y, Barrera DP, Montenegro-Venegas C, Godoy JA, Garner CC, Inestrosa NC, Gundelfinger ED, Zamorano P, et al. Serine–Arginine Protein Kinase SRPK2 Modulates the Assembly of the Active Zone Scaffolding Protein CAST1/ERC2. Cells. 2019; 8(11):1333. https://doi.org/10.3390/cells8111333
Chicago/Turabian StyleArancibia, Duxan, Matias Lira, Yocelin Cruz, Daniela P. Barrera, Carolina Montenegro-Venegas, Juan A. Godoy, Craig C. Garner, Nibaldo C. Inestrosa, Eckart D. Gundelfinger, Pedro Zamorano, and et al. 2019. "Serine–Arginine Protein Kinase SRPK2 Modulates the Assembly of the Active Zone Scaffolding Protein CAST1/ERC2" Cells 8, no. 11: 1333. https://doi.org/10.3390/cells8111333
APA StyleArancibia, D., Lira, M., Cruz, Y., Barrera, D. P., Montenegro-Venegas, C., Godoy, J. A., Garner, C. C., Inestrosa, N. C., Gundelfinger, E. D., Zamorano, P., & Torres, V. I. (2019). Serine–Arginine Protein Kinase SRPK2 Modulates the Assembly of the Active Zone Scaffolding Protein CAST1/ERC2. Cells, 8(11), 1333. https://doi.org/10.3390/cells8111333




