Transcriptional Regulation of Autophagy Genes via Stage-Specific Activation of CEBPB and PPARG during Adipogenesis: A Systematic Study Using Public Gene Expression and Transcription Factor Binding Datasets
Abstract
1. Introduction
2. Results
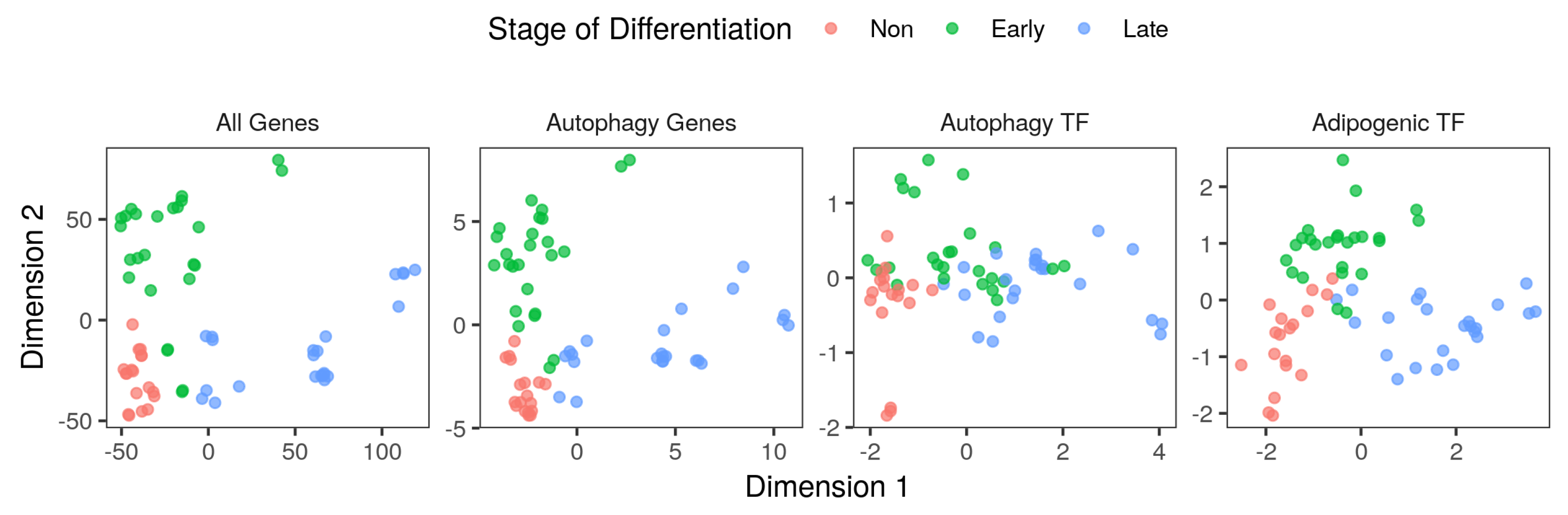
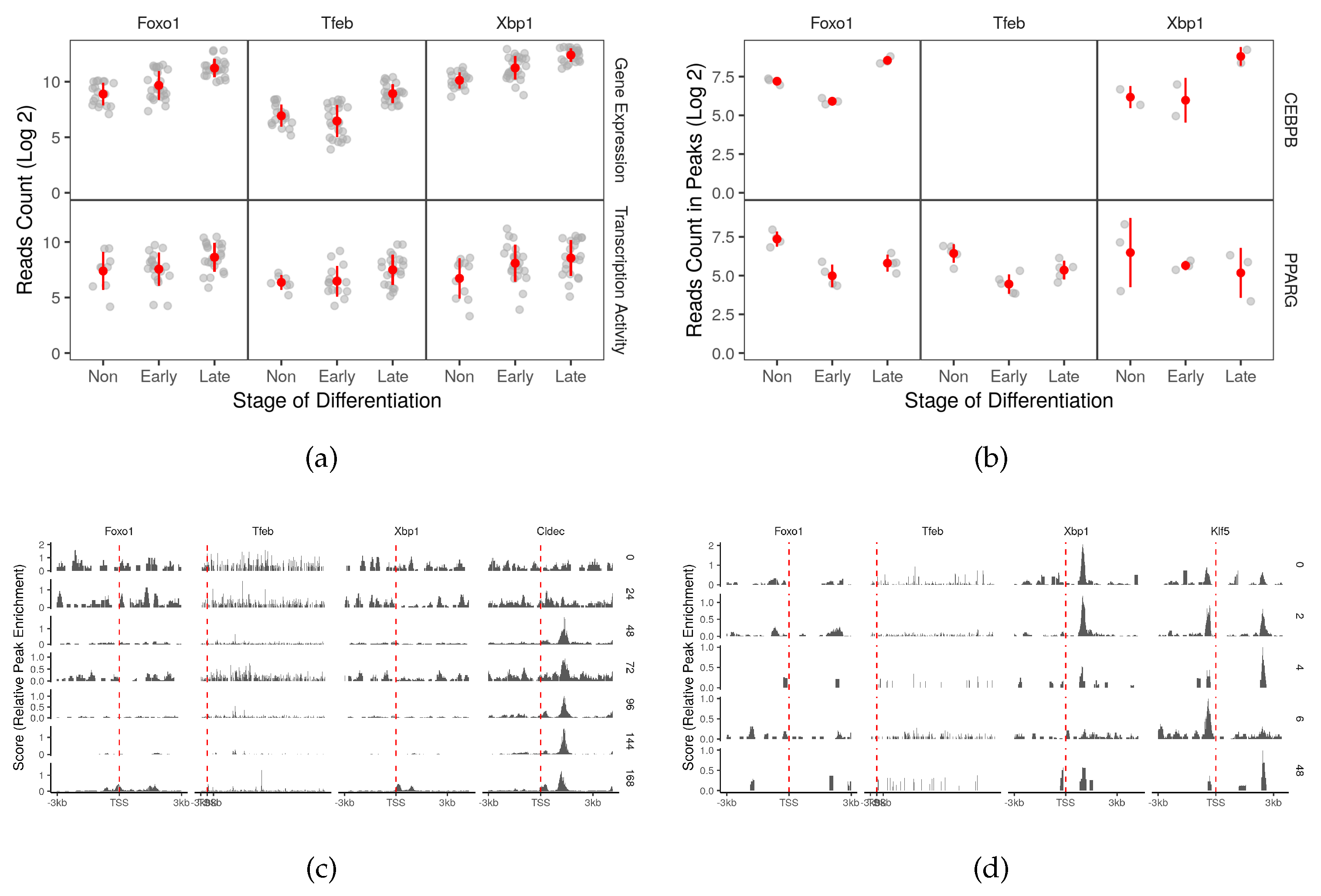
2.1. Autophagy Genes are Regulated as Part of the Transcriptional Program of the Adipocyte Differentiation
2.2. Adipogenic Transcription Regulators Correlate with the Expression of Autophagy Genes
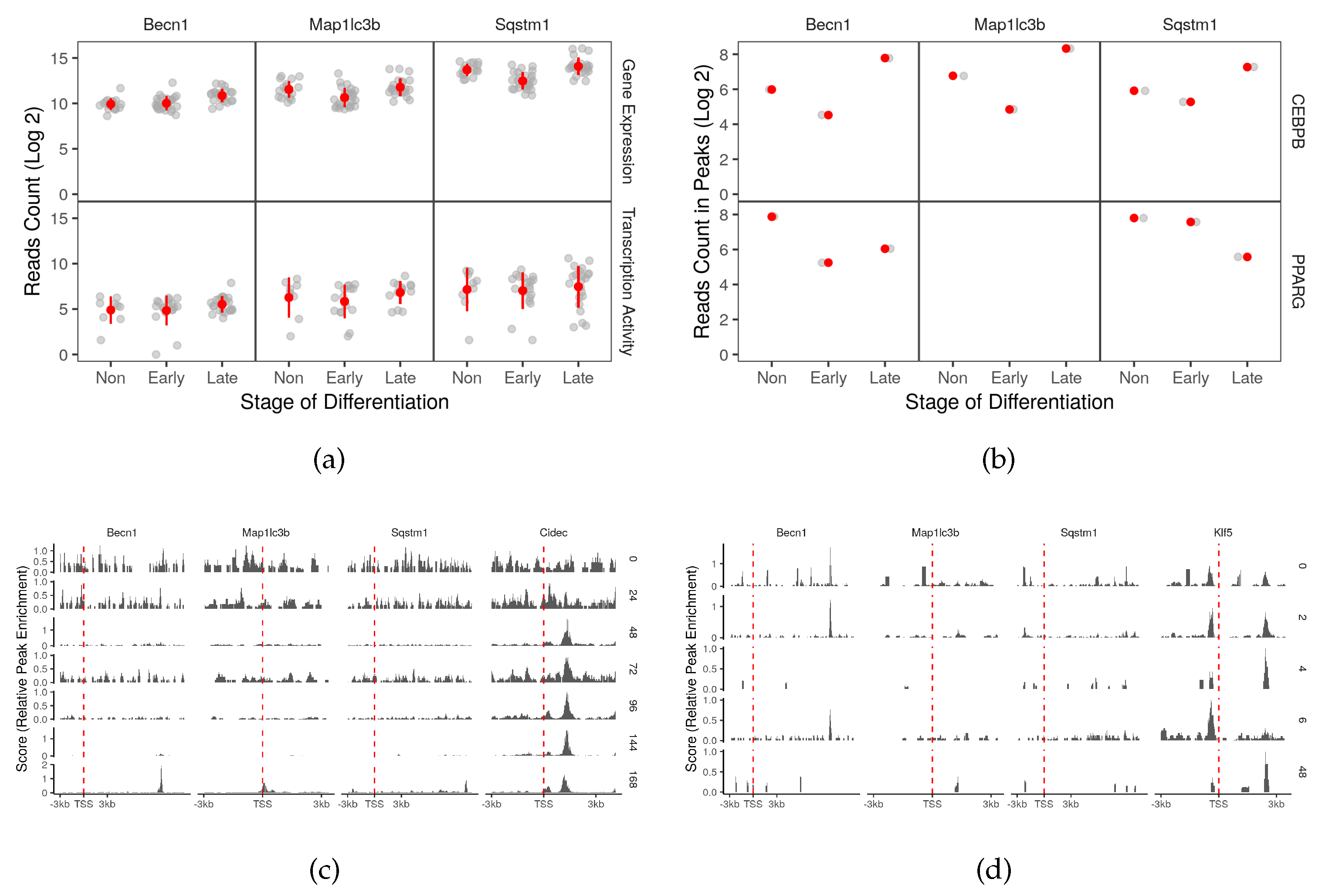
2.3. Master Adipocyte Regulators CEBPB and PPARG Directly Target Key Autophagy Genes
2.4. CEBPB and PPARG Target Autophagy Transcription Factors Genes
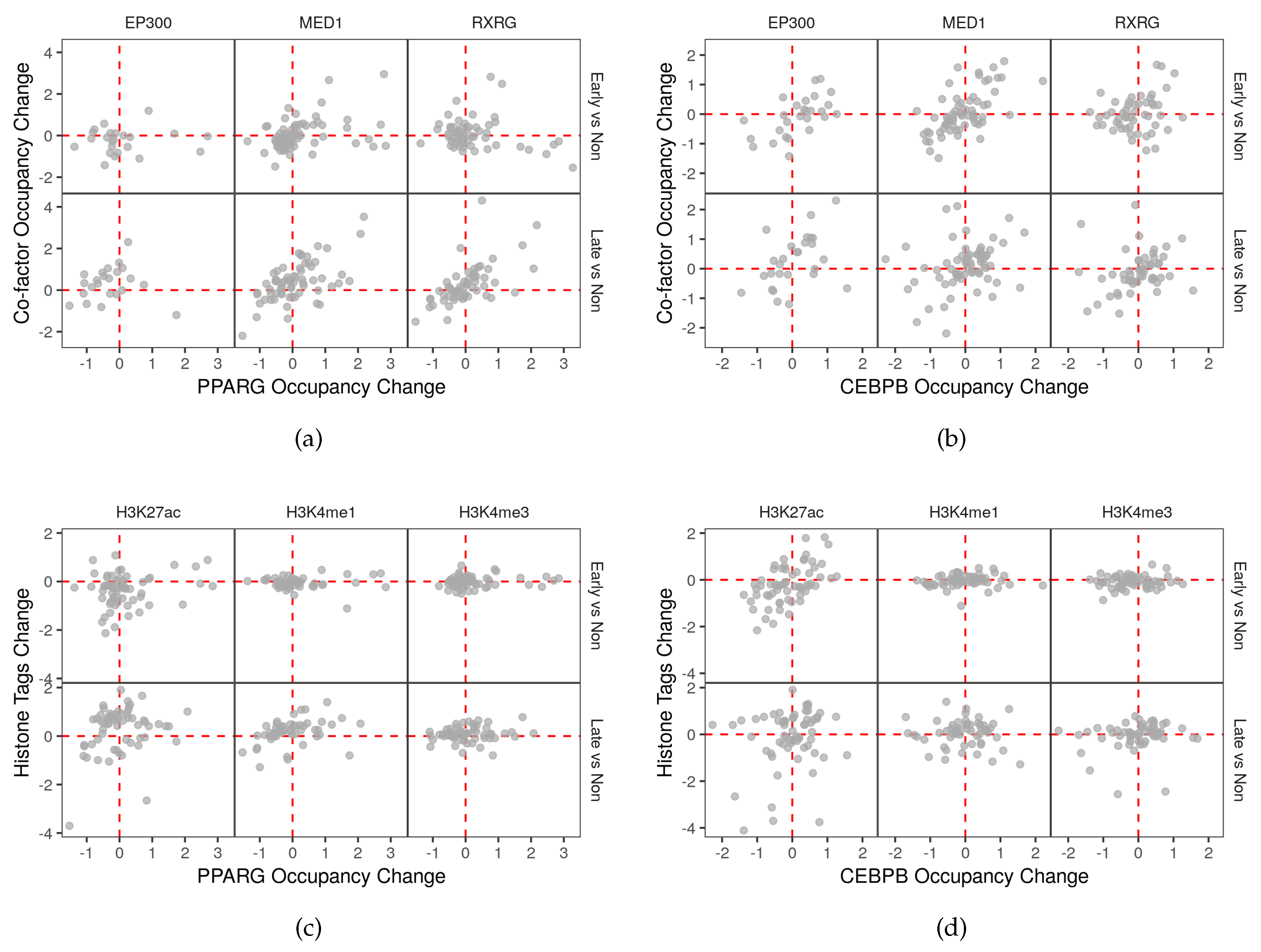
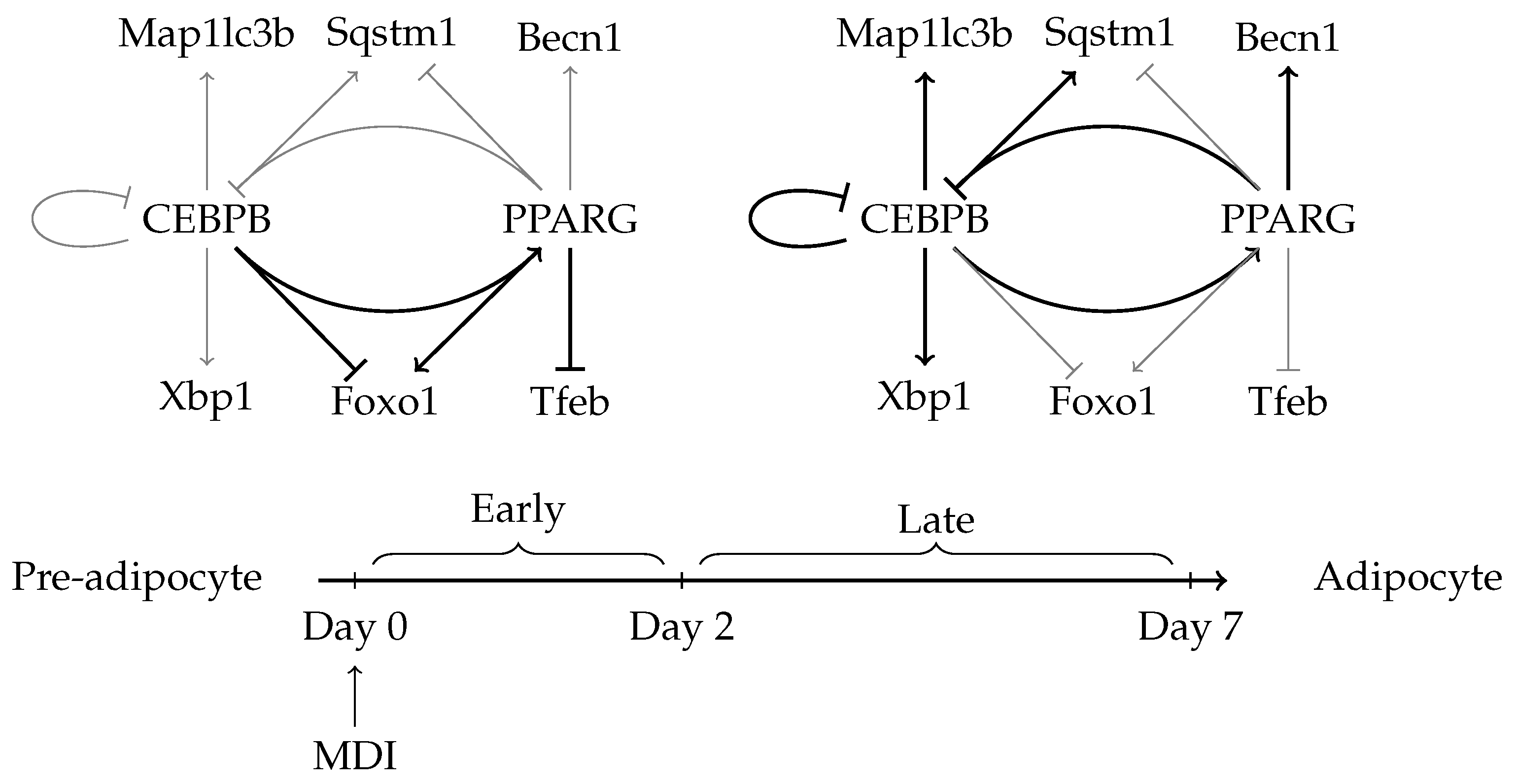
2.5. Auto-Regulation and Transcriptional Feedback Loops May Affect the Regulation of Autophagy
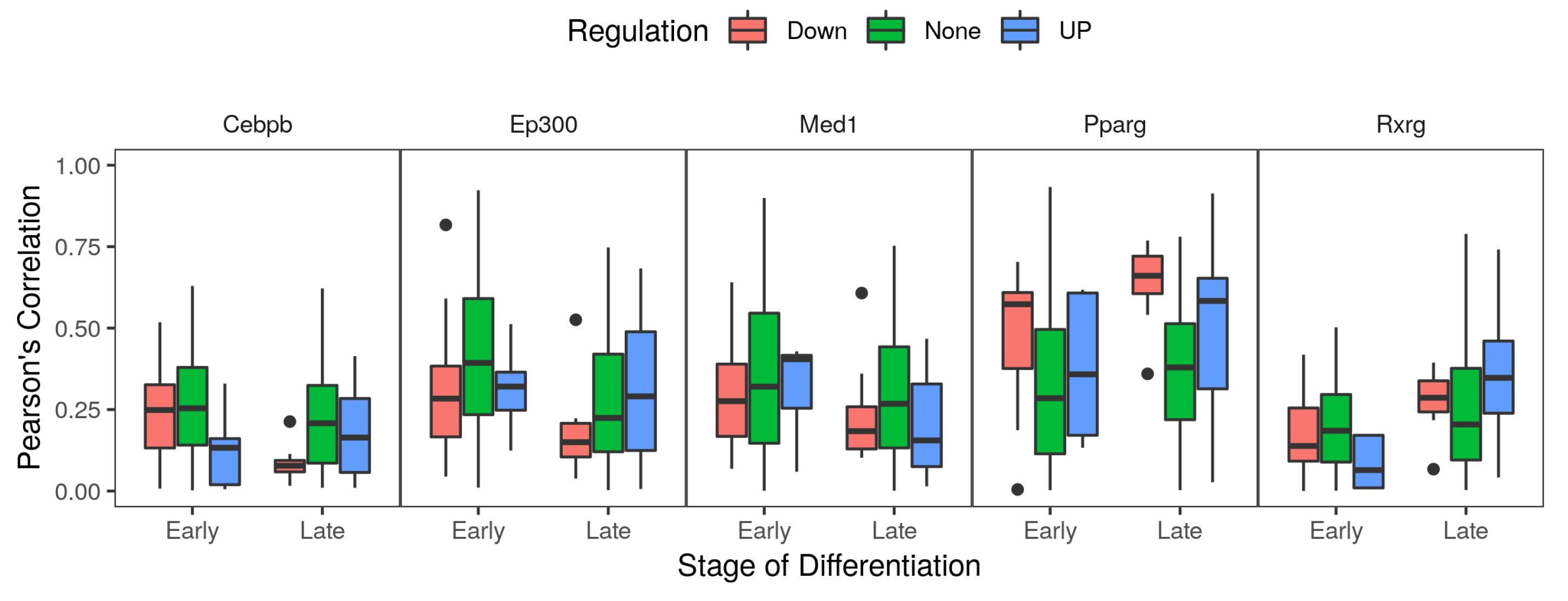
2.6. Co-Factors and Histone Modifications Modulate the Functions of Adipogenic Transcription Factors
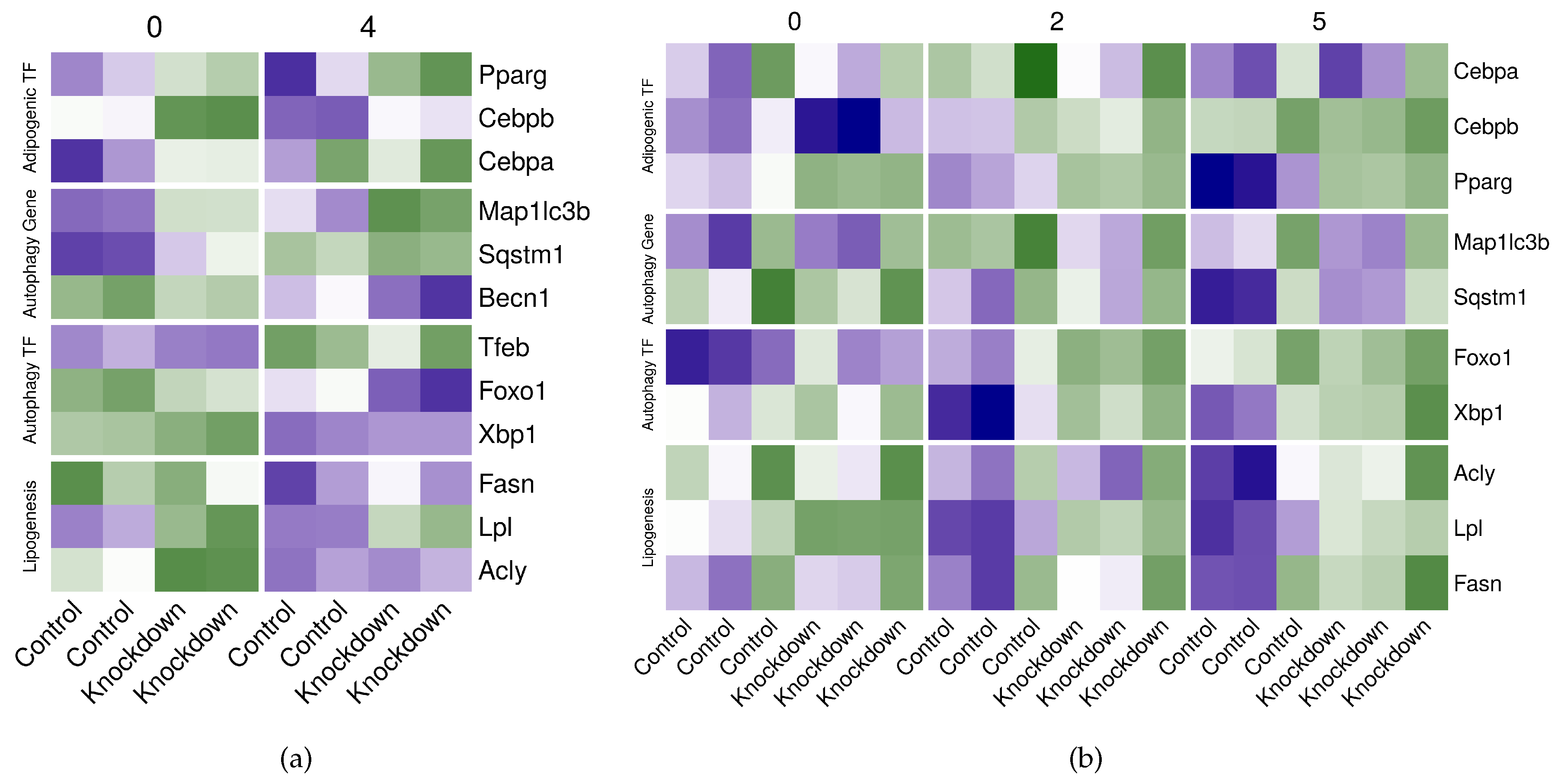
2.7. Perturbing the Adipogenic Factors in Differentiating Adipocytes Disturbs the Expression of Autophagy Genes
3. Discussion
4. Materials and Methods
4.1. Pre-Adipocyte 3T3-L1 Differentiation Protocol
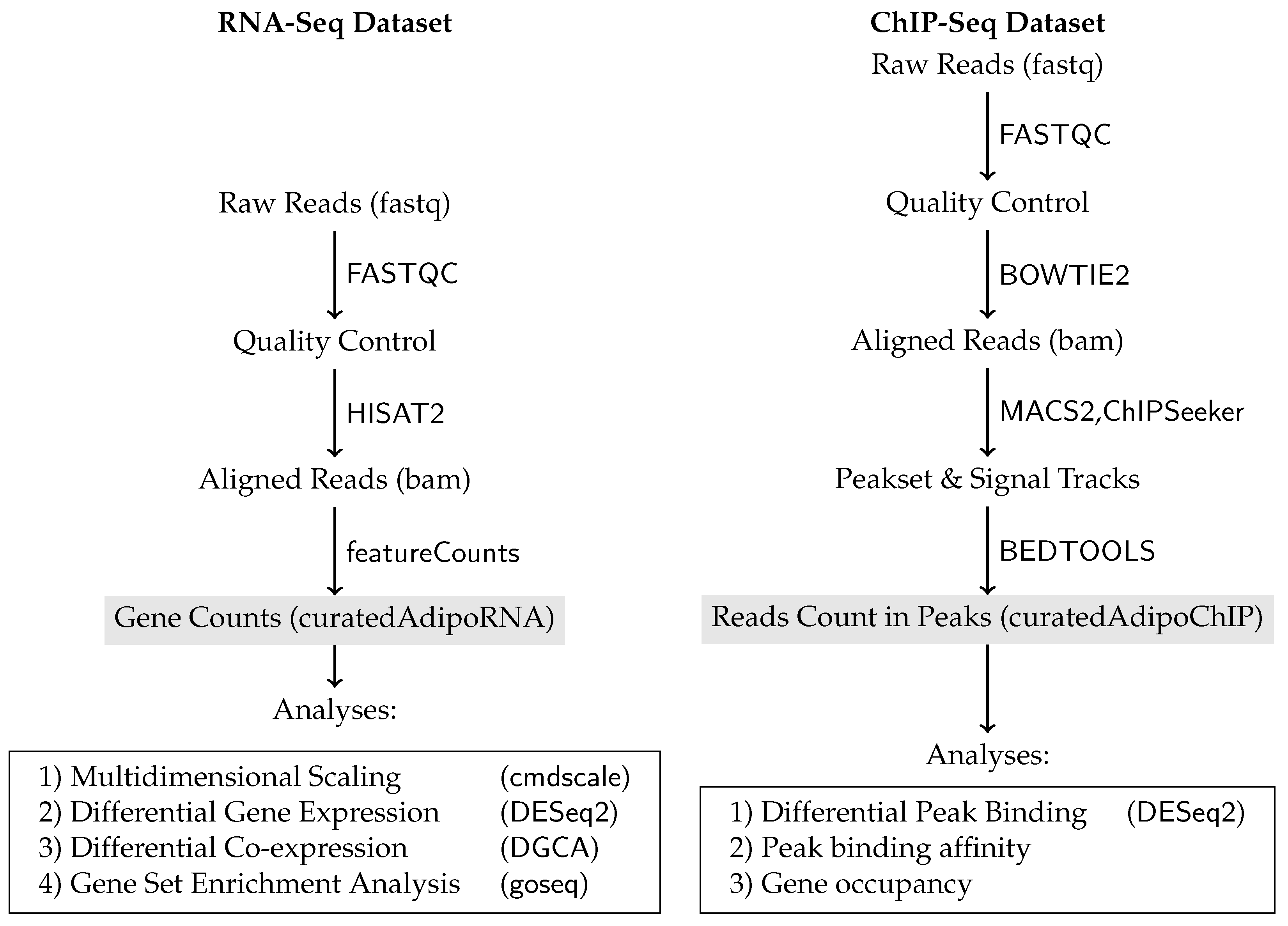
4.2. Data Collection, Pre-Processing, and Processing
4.3. Gene Expression Analysis
4.4. Gene Set Enrichment Analysis
4.5. Gene Co-Expression Analysis
4.6. Peak Binding Analysis
4.7. Occupancy and Affinity Analyses
4.8. Data Management, Transformation and Visualization
4.9. Source Code and Reproducibility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FDR | False Discovery Rate |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| MDI | 1-Methyl-3-isobutylxanthine, Dexamethasone and Insulin |
| MDS | Multidimensional Scaling |
| PCC | Pearson’s Correlation Coefficient |
| SRA | Sequence Read Archive |
| TSS | Transcription Start Site |
| UTR | Un-translated Region |
References
- Dong, H.; Czaja, M.J. Regulation of lipid droplets by autophagy. Trends Endocrinol. Metab. TEM 2011, 22, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Xiang, Y.; Wang, Y.; Baikati, K.; Cuervo, A.M.; Luu, Y.K.; Tang, Y.; Pessin, J.E.; Schwartz, G.J.; Czaja, M.J. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Investig. 2009, 119, 3329–3339. [Google Scholar] [CrossRef] [PubMed]
- Green, H.; Kehinde, O. An established preadipose cell line and its differentiation in culture II. Factors affecting the adipose conversion. Cell 1975, 5, 19–27. [Google Scholar] [CrossRef]
- Guo, L.; Huang, J.X.; Liu, Y.Y.; Li, X.; Zhou, S.R.; Qian, S.W.; Liu, Y.Y.; Zhu, H.; Huang, H.Y.; Dang, Y.J.; et al. Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Mol. Cell. Biol. 2013, 33, 3180–3190. [Google Scholar] [CrossRef]
- Liu, J.; Yao, Q.; Xiao, L.; Ma, W.; Li, F.; Lai, B.; Wang, N. PPAR γ induces NEDD 4 gene expression to promote autophagy and insulin action. FEBS J. 2019. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, J.; Liao, W.; Liu, X.; Zhang, H.; Wang, S.; Wang, D.; Feng, J.; Yu, L.; Zhu, W.G. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 2010, 12, 665–675. [Google Scholar] [CrossRef]
- Nakae, J.; Kitamura, T.; Kitamura, Y.; Biggs, W.H.; Arden, K.C.; Accili, D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 2003, 4, 119–129. [Google Scholar] [CrossRef]
- Armoni, M.; Harel, C.; Karni, S.; Chen, H.; Bar-Yoseph, F.; Ver, M.R.; Quon, M.J.; Karnieli, E. FOXO1 Represses Peroxisome Proliferator-activated Receptor-γ1 and -γ2 Gene Promoters in Primary Adipocytes. J. Biol. Chem. 2006, 281, 19881–19891. [Google Scholar] [CrossRef]
- Ahmed, M.; Quoc Nguyen, H.; Seok Hwang, J.; Zada, S.; Huyen Lai, T.; Soo Kang, S.; Ryong Kim, D. Systematic characterization of autophagy-related genes during the adipocyte differentiation using public-access data. Oncotarget 2018, 9. [Google Scholar] [CrossRef]
- Baerga, R.; Zhang, Y.; Chen, P.H.; Goldman, S.; Jin, S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 2009, 5, 1118–1130. [Google Scholar] [CrossRef]
- Zhang, Y.; Goldman, S.; Baerga, R.; Zhao, Y.; Komatsu, M.; Jin, S. Adipose-specific deletion of autophagy- related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 19860–19865. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Durán, A.; Selloum, M.; Champy, M.F.; Diez-Guerra, F.J.; Flores, J.M.; Serrano, M.; Auwerx, J.; Diaz-Meco, M.T.; Moscat, J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006, 3, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Pfluger, P.T.; Kim, J.Y.; Nogueiras, R.; Duran, A.; Pagès, G.; Pouysségur, J.; Tschöp, M.H.; Diaz-Meco, M.T.; Moscat, J. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010, 11, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Panda, S.; Lin, J.D. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011, 30, 4642–4651. [Google Scholar] [CrossRef]
- Khan, R.; Raza, S.H.A.; Junjvlieke, Z.; Wang, X.; Garcia, M.; Elnour, I.E.; Wang, H.; Linsen, Z. Function and Transcriptional Regulation of Bovine TORC2 Gene in Adipocytes: Roles of C/EBP, XBP1, INSM1 and ZNF263. Int. J. Mol. Sci. 2019, 20, 4338. [Google Scholar] [CrossRef] [PubMed]
- Vlahakis, A.; Powers, T. A role for TOR complex 2 signaling in promoting autophagy. Autophagy 2014. [Google Scholar] [CrossRef]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.S.U.; Erdin, S.S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef]
- Fiorentino, L.; Cavalera, M.; Menini, S.; Marchetti, V.; Mavilio, M.; Fabrizi, M.; Conserva, F.; Casagrande, V.; Menghini, R.; Pontrelli, P.; et al. Loss of TIMP3 underlies diabetic nephropathy via FoxO1/STAT1 interplay. EMBO Mol. Med. 2013, 5, 441–455. [Google Scholar] [CrossRef]
- Margariti, A.; Li, H.; Chen, T.; Martin, D.; Vizcay-Barrena, G.; Alam, S.; Karamariti, E.; Xiao, Q.; Zampetaki, A.; Zhang, Z.; et al. XBP1 mRNA splicing triggers an autophagic response in endothelial cells through BECLIN-1 transcriptional activation. J. Biol. Chem. 2013, 288, 859–872. [Google Scholar] [CrossRef]
- Wu, Z.; Bucher, N.L.; Farmer, S.R. Induction of peroxisome proliferator-activated receptor gamma during the conversion of 3T3 fibroblasts into adipocytes is mediated by {C}/{EBPbeta}, {C}/{EBPdelta}, and glucocorticoids. Mol. Cell. Biol. 1996, 16, 4128–4136. [Google Scholar] [CrossRef]
- Wu, Z.; Rosen, E.D.; Brun, R.; Hauser, S.; Adelmant, G.; Troy, A.E.; McKeon, C.; Darlington, G.J.; Spiegelman, B.M. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell 1999, 3, 151–158. [Google Scholar] [CrossRef]
- Tanaka, T.; Yoshida, N.; Kishimoto, T.; Akira, S. Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 1997, 16, 7432–7443. [Google Scholar] [CrossRef] [PubMed]
- Haakonsson, A.K.; Stahl Madsen, M.; Nielsen, R.; Sandelin, A.; Mandrup, S. Acute Genome-Wide Effects of Rosiglitazone on PPARγ Transcriptional Networks in Adipocytes. Mol. Endocrinol. 2013, 27, 1536–1549. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Rada-Iglesias, A. Is H3K4me1 at enhancers correlative or causative? Nat. Genet. 2018, 50, 4–5. [Google Scholar] [CrossRef]
- Sarusi Portuguez, A.; Schwartz, M.; Siersbaek, R.; Nielsen, R.; Sung, M.H.; Mandrup, S.; Kaplan, T.; Hakim, O. Hierarchical role for transcription factors and chromatin structure in genome organization along adipogenesis. FEBS J. 2017, 284, 3230–3244. [Google Scholar] [CrossRef]
- Glenn, K.C.; Shieh, J.J.; Laird, D.M. Characterization of 3T3-L1 storage lipid metabolism: Effect of somatotropin and insulin on specific pathways. Endocrinology 1992. [Google Scholar] [CrossRef]
- Sarjeant, K.; Stephens, J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008417. [Google Scholar] [CrossRef]
- Thomson, M.J.; Williams, M.G.; Frost, S.C. Development of insulin resistance in 3T3-L1 adipocytes. J. Biol. Chem. 1997. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIP seeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Java Package. 2018. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 4 October 2019).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Cox, T.F.; Cox, M. Multidimensional Scaling, 2nd ed.; Chapman and Hall: New York, NY, USA, 2000; p. 328. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Eds.; Springer: New York, NY, USA, 2005; Chapter 23; pp. 397–420. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Carlson, M. org.Mm.eg.db: Genome Wide Annotation for Mouse. R Package Version 3.8.2. 2019. Available online: https://doi.org/doi:10.18129/B9.bioc.org.Mm.eg.db (accessed on 4 October 2019).
- Team, B.C.; Maintainer, B.P. TxDb.Mmusculus.UCSC.mm10.knownGene: Annotation Package for TxDb Object(s). R Package Version 3.4.7. 2019. Available online: https://doi.org/doi:10.18129/B9.bioc.TxDb.Mmusculus.UCSC.mm10.knownGene (accessed on 4 October 2019).
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- McKenzie, A.T.; Katsyv, I.; Song, W.M.; Wang, M.; Zhang, B. DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Syst. Biol. 2016, 10, 1–25. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Obenchain, V.; Hester, J.; Pagès, H. SummarizedExperiment: SummarizedExperiment Container. R Package Version 1.14.1. 2019. Available online: https://doi.org/doi:10.18129/B9.bioc.SummarizedExperiment (accessed on 4 October 2019).
- Sean, D.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. tidyverse: Easily Install and Load the “Tidyverse”. R Package Version 1.2.1. 2017. Available online: https://cran.r-project.org/package=tidyverse (accessed on 4 October 2019).
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Lawrence, M.; Gentleman, R.; Carey, V. rtracklayer: An R package for interfacing with genome browsers. Bioinformatics 2009, 25, 1841–1842. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2; Springer: New York, NY, USA, 2011. [Google Scholar] [CrossRef]
- Schloerke, B.; Crowley, J.; Cook, D.; Briatte, F.; Marbach, M.; Thoen, E.; Elberg, A.; Larmarange, J. GGally: Extension to “ggplot2”. R Package Version 1.4.0. 2018. Available online: https://cran.r-project.org/package=GGally (accessed on 4 October 2019).
- Dahl, D.B.; Scott, D.; Roosen, C.; Magnusson, A.; Swinton, J. xtable: Export Tables to LaTeX or HTML. R Package Version 1.8-4. 2018. Available online: https://cran.r-project.org/package=xtable (accessed on 4 October 2019).
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M.; Ishaque, N. EnrichedHeatmap: An R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genom. 2018, 19, 234. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]


| Category | Gene | Early vs. Non | Late vs. Non | Late vs. Early | |||
|---|---|---|---|---|---|---|---|
| FC | SE | FC | SE | FC | SE | ||
| Adipogenic TF | Cebpb | 1.5 | 0.19 | −1.3 | 0.18 | ||
| Med1 | 0.3 | 0.1 | 0.28 | 0.1 | |||
| Pparg | 1.55 | 0.25 | 2.76 | 0.25 | 1.21 | 0.23 | |
| Rxrg | 7.89 | 0.72 | 6.89 | 0.63 | |||
| Autophagy TF | Foxo1 | 1.1 | 0.22 | 2.03 | 0.22 | 0.92 | 0.2 |
| Tfeb | 1.66 | 0.26 | 1.65 | 0.24 | |||
| Trp53 | −0.52 | 0.18 | −1.25 | 0.19 | −0.73 | 0.17 | |
| Xbp1 | 1.44 | 0.17 | 1.87 | 0.17 | 0.44 | 0.16 | |
| Zkscan3 | 0.59 | 0.12 | 0.41 | 0.11 | |||
| Autophagy Gene | Atg4b | 0.38 | 0.07 | −0.46 | 0.06 | ||
| Becn1 | 0.31 | 0.08 | 0.44 | 0.09 | 0.13 | 0.08 | |
| Map1lc3a | −0.5 | 0.14 | 0.62 | 0.13 | |||
| Map1lc3b | −0.72 | 0.17 | −0.27 | 0.17 | 0.45 | 0.15 | |
| Sqstm1 | −1.02 | 0.15 | 0.91 | 0.14 | |||
| Ulk1 | 0.74 | 0.15 | 0.55 | 0.14 | |||
| Category | Gene | Cebpb KD vs. Control | Pparg KD vs. Control | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 4 h | 0 d | 2 d | 5 d | |||||||
| FC | SE | FC | SE | FC | SE | FC | SE | FC | SE | ||
| Adipogenic TF | Cebpb | −2.61 | 0.17 | −1.94 | 0.17 | −0.28 | 0.11 | ||||
| Pparg | −0.65 | 0.17 | −1.19 | 0.27 | −2.27 | 0.18 | −1.92 | 0.19 | −2.53 | 0.1 | |
| Cebpa | 0.29 | 0.11 | |||||||||
| Autophagy Gene | Map1lc3b | −0.54 | 0.12 | −0.67 | 0.19 | 0.93 | 0.3 | ||||
| Sqstm1 | −0.58 | 0.11 | |||||||||
| Autophagy TF | Foxo1 | 0.41 | 0.19 | 0.84 | 0.2 | −0.81 | 0.26 | ||||
| Xbp1 | −0.3 | 0.15 | −0.88 | 0.09 | −0.62 | 0.08 | |||||
| Lipogenesis | Acly | −0.75 | 0.11 | −0.74 | 0.14 | ||||||
| Lpl | −2.54 | 0.37 | −2.25 | 0.19 | −2.18 | 0.21 | −1.72 | 0.08 | −1.36 | 0.1 | |
| Fasn | −0.91 | 0.31 | |||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, M.; Lai, T.H.; Hwang, J.S.; Zada, S.; Pham, T.M.; Kim, D.R. Transcriptional Regulation of Autophagy Genes via Stage-Specific Activation of CEBPB and PPARG during Adipogenesis: A Systematic Study Using Public Gene Expression and Transcription Factor Binding Datasets. Cells 2019, 8, 1321. https://doi.org/10.3390/cells8111321
Ahmed M, Lai TH, Hwang JS, Zada S, Pham TM, Kim DR. Transcriptional Regulation of Autophagy Genes via Stage-Specific Activation of CEBPB and PPARG during Adipogenesis: A Systematic Study Using Public Gene Expression and Transcription Factor Binding Datasets. Cells. 2019; 8(11):1321. https://doi.org/10.3390/cells8111321
Chicago/Turabian StyleAhmed, Mahmoud, Trang Huyen Lai, Jin Seok Hwang, Sahib Zada, Trang Minh Pham, and Deok Ryong Kim. 2019. "Transcriptional Regulation of Autophagy Genes via Stage-Specific Activation of CEBPB and PPARG during Adipogenesis: A Systematic Study Using Public Gene Expression and Transcription Factor Binding Datasets" Cells 8, no. 11: 1321. https://doi.org/10.3390/cells8111321
APA StyleAhmed, M., Lai, T. H., Hwang, J. S., Zada, S., Pham, T. M., & Kim, D. R. (2019). Transcriptional Regulation of Autophagy Genes via Stage-Specific Activation of CEBPB and PPARG during Adipogenesis: A Systematic Study Using Public Gene Expression and Transcription Factor Binding Datasets. Cells, 8(11), 1321. https://doi.org/10.3390/cells8111321







