The Role of Adenine Nucleotide Translocase in the Assembly of Respiratory Supercomplexes in Cardiac Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. siRNA Transfection
2.4. Mitochondrial Oxygen Consumption Rate and ATP Production
2.5. Isolation of Mitochondria
2.6. Enzymatic Activity of ETC Complexes in Cultured Cells
2.7. Total and Mitochondrial ROS Production in Cultured Cells
2.8. Mitochondrial Membrane Potential and Total ATP
2.9. SDS-PAGE and Western Blotting
2.10. Co-Immunoprecipitation
2.11. Analysis of RCS by Blue Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
2.12. Statistical Analysis
3. Results
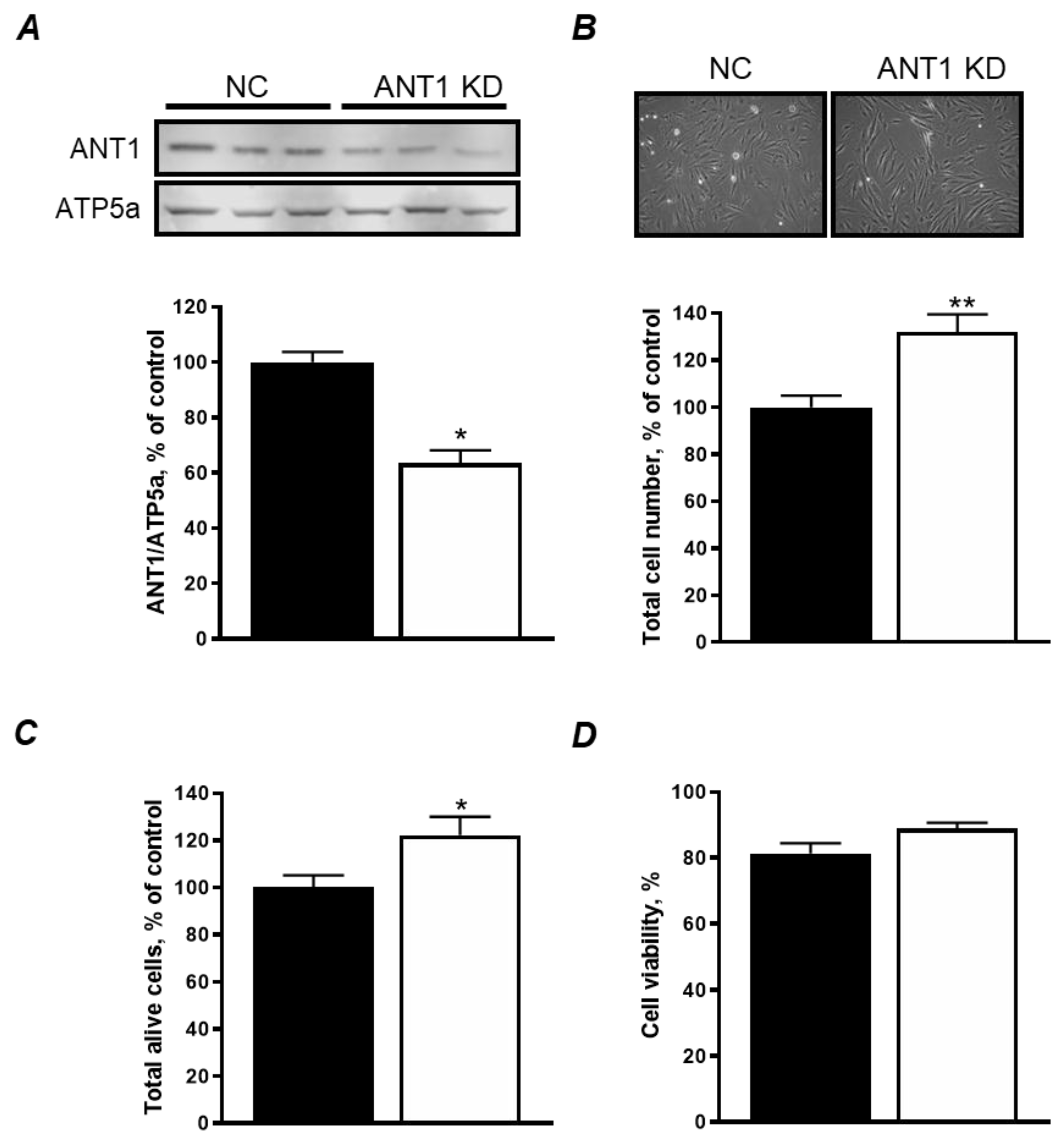
3.1. ANT1 KD Increases Cellular Proliferation Without Affecting Cell Viability
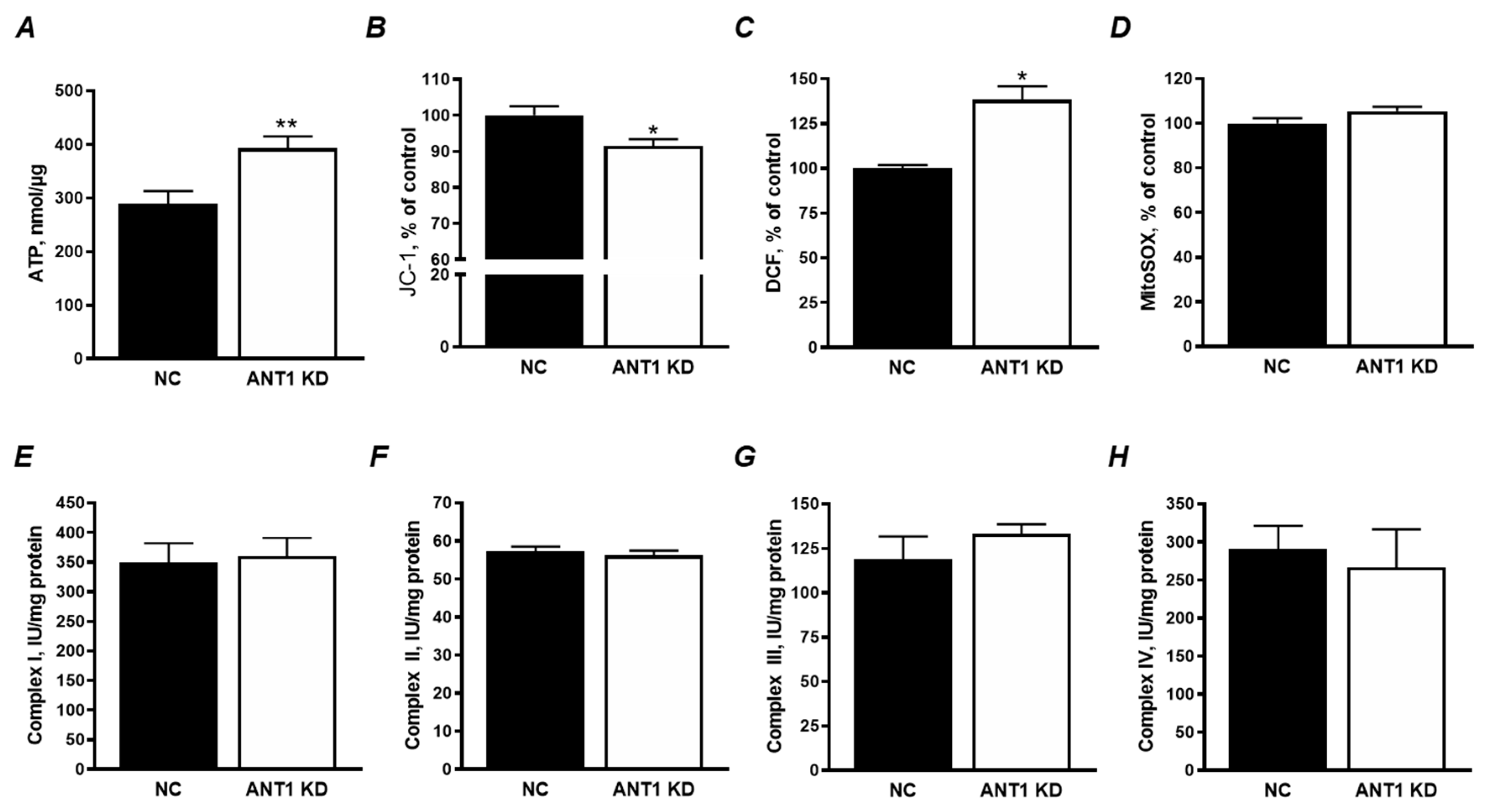
3.2. ANT1 KD Increased Total ATP and ROS Levels with no Effect on the ETC Activity and mtROS Production
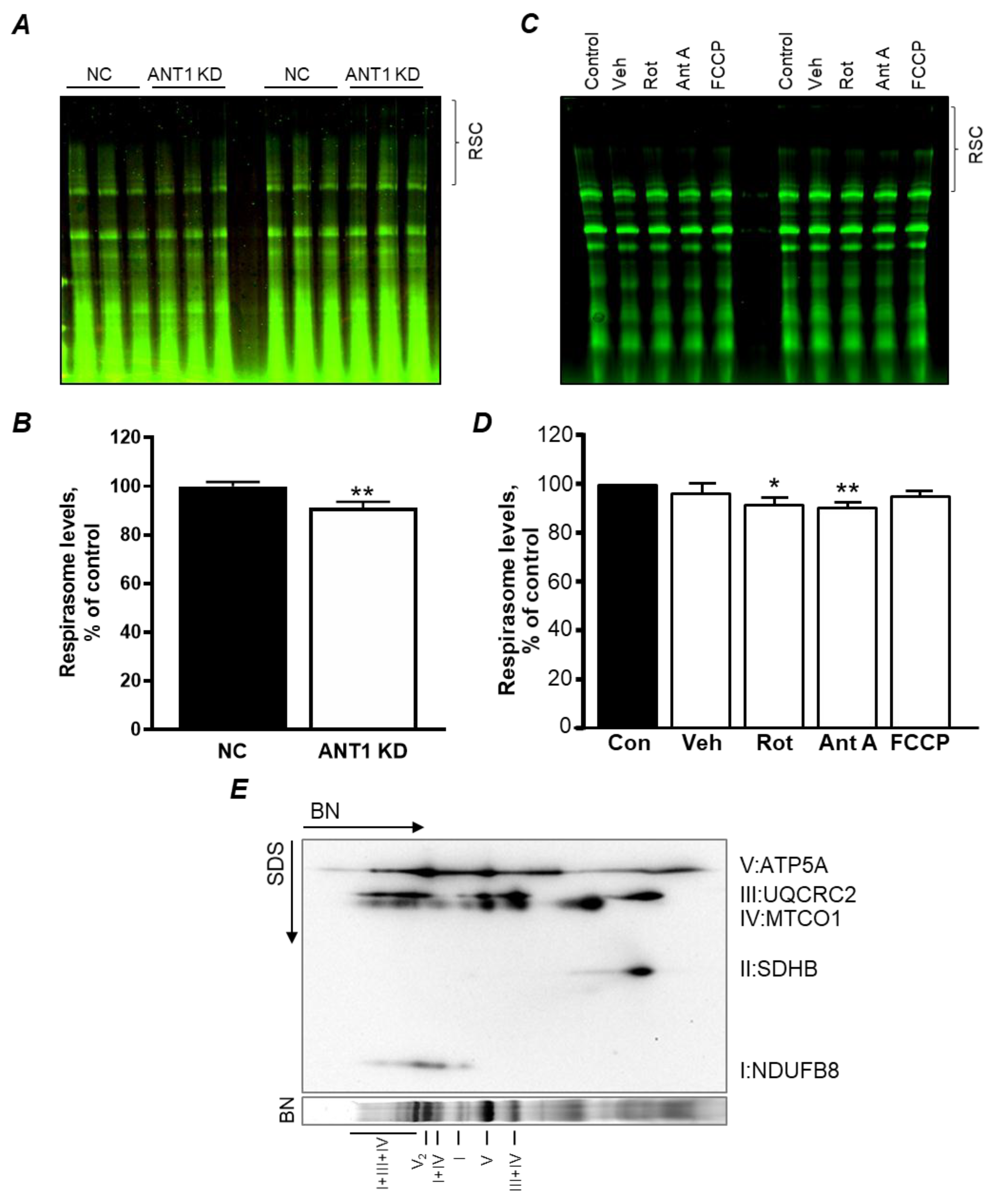
3.3. Mitochondrial Oxygen Consumption Rate and OXPHOS is not Affected by ANT1 Downregulation
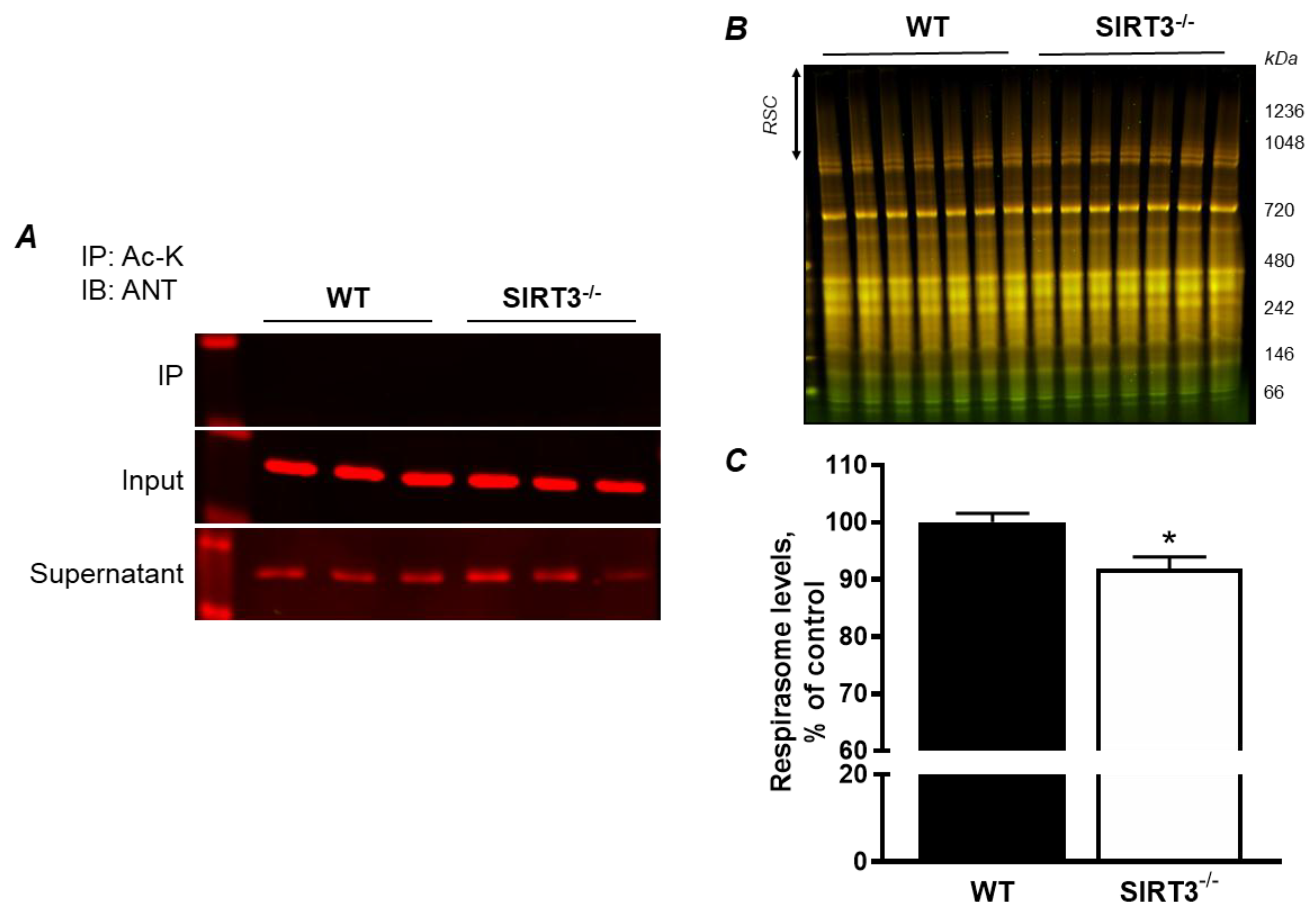
3.4. ANT1 KD in H9c2 Cells Induce RCS Dissociation: the Role of Acetylation
4. Discussion
5. Limitations of the Study
Author Contributions
Funding
Conflicts of Interest
References
- Stepien, G.; Torroni, A.; Chung, A.B.; Hodge, J.A.; Wallace, D.C. Differential expression of adenine nucleotide translocator isoforms in mammalian tissues and during muscle cell differentiation. J. Biol. Chem. 1992, 267, 14592–14597. [Google Scholar] [PubMed]
- Graham, B.H.; Waymire, K.G.; Cottrell, B.; Trounce, I.A.; MacGregor, G.R.; Wallace, D.C. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 1997, 16, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Echaniz-Laguna, A.; Chassagne, M.; Ceresuela, J.; Rouvet, I.; Padet, S.; Acquaviva, C.; Nataf, S.; Vinzio, S.; Bozon, D.; Mousson de Camaret, B. Complete loss of expression of the ANT1 gene causing cardiomyopathy and myopathy. J. Med. Genet. 2012, 49, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Narula, N.; Zaragoza, M.V.; Sengupta, P.P.; Li, P.; Haider, N.; Verjans, J.; Waymire, K.; Vannan, M.; Wallace, D.C. Adenine nucleotide translocase 1 deficiency results in dilated cardiomyopathy with defects in myocardial mechanics, histopathological alterations, and activation of apoptosis. JACC Cardiovasc. Imaging 2011, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Alberio, S.; Pisano, I.; Lodi, T.; Meznaric-Petrusa, M.; Zidar, J.; Santoro, A.; Scarcia, P.; Fontanesi, F.; Lamantea, E.; et al. Complete loss-of-function of the heart/muscle-specific adenine nucleotide translocator is associated with mitochondrial myopathy and cardiomyopathy. Hum. Mol. Genet. 2005, 14, 3079–3088. [Google Scholar] [CrossRef] [PubMed]
- Esposito, L.A.; Melov, S.; Panov, A.; Cottrell, B.A.; Wallace, D.C. Mitochondrial disease in mouse results in increased oxidative stress. Proc. Natl. Acad. Sci. USA 1999, 96, 4820–4825. [Google Scholar] [CrossRef] [PubMed]
- Klumpe, I.; Savvatis, K.; Westermann, D.; Tschope, C.; Rauch, U.; Landmesser, U.; Schultheiss, H.P.; Dorner, A. Transgenic overexpression of adenine nucleotide translocase 1 protects ischemic hearts against oxidative stress. J. Mol. Med. (Berl) 2016, 94, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.W.; Acoba, M.G.; Selvaraju, K.; Huang, T.C.; Nirujogi, R.S.; Sathe, G.; Pandey, A.; Claypool, S.M. Human adenine nucleotide translocases physically and functionally interact with respirasomes. Mol. Biol Cell 2017, 28, 1489–1506. [Google Scholar] [CrossRef] [PubMed]
- Claypool, S.M.; Oktay, Y.; Boontheung, P.; Loo, J.A.; Koehler, C.M. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 2008, 182, 937–950. [Google Scholar] [CrossRef]
- Schagger, H.; Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000, 19, 1777–1783. [Google Scholar] [CrossRef]
- Lobo-Jarne, T.; Ugalde, C. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin. Cell Dev. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Javadov, S. Elucidating the contribution of ETC complexes I and II to the respirasome formation in cardiac mitochondria. Sci. Rep. 2018, 8, 17732. [Google Scholar] [CrossRef] [PubMed]
- Cimen, H.; Han, M.J.; Yang, Y.; Tong, Q.; Koc, H.; Koc, E.C. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry 2010, 49, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.T.; Lee, H.C.; Liao, C.C.; Wei, Y.H. Regulation of mitochondrial F(o)F(1)ATPase activity by Sirt3-catalyzed deacetylation and its deficiency in human cells harboring 4977bp deletion of mitochondrial DNA. Biochim. Biophys. Acta 2013, 1832, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.S.; Barreto-Torres, G.; Kuznetsov, A.V.; Khuchua, Z.; Javadov, S. Crosstalk between AMPK activation and angiotensin II-induced hypertrophy in cardiomyocytes: The role of mitochondria. J. Cell Mol. Med. 2014, 18, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Lewis, T.S.; Powers, C.; Khuchua, Z.; Baines, C.P.; Wipf, P.; Javadov, S. Elucidating Mitochondrial Electron Transport Chain Supercomplexes in the Heart During Ischemia-Reperfusion. Antioxid. Redox. Signal. 2017, 27, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Parodi-Rullan, R.M.; Chapa-Dubocq, X.R.; Javadov, S. Acetylation of Mitochondrial Proteins in the Heart: The Role of SIRT3. Front. Physiol. 2018, 9, 1094. [Google Scholar] [CrossRef]
- Salvatori, I.; Valle, C.; Ferri, A.; Carri, M.T. SIRT3 and mitochondrial metabolism in neurodegenerative diseases. Neurochem. Int. 2017, 109, 184–192. [Google Scholar] [CrossRef]
- McDonnell, E.; Peterson, B.S.; Bomze, H.M.; Hirschey, M.D. SIRT3 regulates progression and development of diseases of aging. Trends Endocrinol. Metab. 2015, 26, 486–492. [Google Scholar] [CrossRef]
- Parodi-Rullan, R.M.; Chapa-Dubocq, X.; Rullan, P.J.; Jang, S.; Javadov, S. High Sensitivity of SIRT3 Deficient Hearts to Ischemia-Reperfusion Is Associated with Mitochondrial Abnormalities. Front. Pharmacol. 2017, 8, 275. [Google Scholar] [CrossRef]
- Ogunbona, O.B.; Baile, M.G.; Claypool, S.M. Cardiomyopathy-associated mutation in the ADP/ATP carrier reveals translation-dependent regulation of cytochrome c oxidase activity. Mol. Biol. Cell 2018, 29, 1449–1464. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, N.R.; Gupta, M.; Kim, G.; Rajamohan, S.B.; Isbatan, A.; Gupta, M.P. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Investig. 2009, 119, 2758–2771. [Google Scholar] [CrossRef] [PubMed]
- Pillai, V.B.; Samant, S.; Sundaresan, N.R.; Raghuraman, H.; Kim, G.; Bonner, M.Y.; Arbiser, J.L.; Walker, D.I.; Jones, D.P.; Gius, D.; et al. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat. Commun. 2015, 6, 6656. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, J.; Li, N.; Wang, S.; Liu, H.; Li, J.; Zhang, Y.; Bu, P. Mouse SIRT3 attenuates hypertrophy-related lipid accumulation in the heart through the deacetylation of LCAD. PLoS ONE 2015, 10, e0118909. [Google Scholar] [CrossRef] [PubMed]
- Porter, G.A.; Urciuoli, W.R.; Brookes, P.S.; Nadtochiy, S.M. SIRT3 deficiency exacerbates ischemia-reperfusion injury: Implication for aged hearts. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H1602–H1609. [Google Scholar] [CrossRef] [PubMed]
- Koentges, C.; Pfeil, K.; Schnick, T.; Wiese, S.; Dahlbock, R.; Cimolai, M.C.; Meyer-Steenbuck, M.; Cenkerova, K.; Hoffmann, M.M.; Jaeger, C.; et al. SIRT3 deficiency impairs mitochondrial and contractile function in the heart. Basic Res. Cardiol. 2015, 110, 36. [Google Scholar] [CrossRef] [PubMed]
- Parodi-Rullan, R.; Barreto-Torres, G.; Ruiz, L.; Casasnovas, J.; Javadov, S. Direct renin inhibition exerts an anti-hypertrophic effect associated with improved mitochondrial function in post-infarction heart failure in diabetic rats. Cell Physiol. Biochem. 2012, 29, 841–850. [Google Scholar] [CrossRef]
- Horton, J.L.; Martin, O.J.; Lai, L.; Riley, N.M.; Richards, A.L.; Vega, R.B.; Leone, T.C.; Pagliarini, D.J.; Muoio, D.M.; Bedi, K.C., Jr.; et al. Mitochondrial protein hyperacetylation in the failing heart. JCI Insight 2016, 2. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Javadov, S.; Sickinger, S.; Frotschnig, S.; Grimm, M. H9c2 and HL-1 cells demonstrate distinct features of energy metabolism, mitochondrial function and sensitivity to hypoxia-reoxygenation. Biochim. Biophys. Acta 2015, 1853, 276–284. [Google Scholar] [CrossRef]
- Borutaite, V.; Mildaziene, V.; Katiliute, Z.; Kholodenko, B.; Toleikis, A. The function of ATP/ADP translocator in the regulation of mitochondrial respiration during development of heart ischemic injury. Biochim. Biophys. Acta 1993, 1142, 175–180. [Google Scholar] [CrossRef]
- Murdock, D.G.; Boone, B.E.; Esposito, L.A.; Wallace, D.C. Up-regulation of nuclear and mitochondrial genes in the skeletal muscle of mice lacking the heart/muscle isoform of the adenine nucleotide translocator. J. Biol. Chem. 1999, 274, 14429–14433. [Google Scholar] [CrossRef] [PubMed]
- Chevrollier, A.; Loiseau, D.; Gautier, F.; Malthiery, Y.; Stepien, G. ANT2 expression under hypoxic conditions produces opposite cell-cycle behavior in 143B and HepG2 cancer cells. Mol. Carcinog. 2005, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.N.; DeHart, D.N.; Patnaik, J.; Klatt, S.C.; Gooz, M.B.; Lemasters, J.J. ATP/ADP turnover and import of glycolytic ATP into mitochondria in cancer cells is independent of the adenine nucleotide translocator. J. Biol. Chem. 2017, 292, 16969. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid. Med. Cell Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Blaza, J.N.; Larsson, N.G.; Hirst, J. The Enigma of the Respiratory Chain Supercomplex. Cell Metab. 2017, 25, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Rosca, M.G.; Vazquez, E.J.; Kerner, J.; Parland, W.; Chandler, M.P.; Stanley, W.; Sabbah, H.N.; Hoppel, C.L. Cardiac mitochondria in heart failure: Decrease in respirasomes and oxidative phosphorylation. Cardiovasc. Res. 2008, 80, 30–39. [Google Scholar] [CrossRef]
- Jang, S.; Javadov, S. Association between ROS production, swelling and the respirasome integrity in cardiac mitochondria. Arch. Biochem. Biophys. 2017, 630, 1–8. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
M. Parodi-Rullán, R.; Chapa-Dubocq, X.; Guzmán-Hernández, R.; Jang, S.; A. Torres-Ramos, C.; Ayala-Peña, S.; Javadov, S. The Role of Adenine Nucleotide Translocase in the Assembly of Respiratory Supercomplexes in Cardiac Cells. Cells 2019, 8, 1247. https://doi.org/10.3390/cells8101247
M. Parodi-Rullán R, Chapa-Dubocq X, Guzmán-Hernández R, Jang S, A. Torres-Ramos C, Ayala-Peña S, Javadov S. The Role of Adenine Nucleotide Translocase in the Assembly of Respiratory Supercomplexes in Cardiac Cells. Cells. 2019; 8(10):1247. https://doi.org/10.3390/cells8101247
Chicago/Turabian StyleM. Parodi-Rullán, Rebecca, Xavier Chapa-Dubocq, Roberto Guzmán-Hernández, Sehwan Jang, Carlos A. Torres-Ramos, Sylvette Ayala-Peña, and Sabzali Javadov. 2019. "The Role of Adenine Nucleotide Translocase in the Assembly of Respiratory Supercomplexes in Cardiac Cells" Cells 8, no. 10: 1247. https://doi.org/10.3390/cells8101247
APA StyleM. Parodi-Rullán, R., Chapa-Dubocq, X., Guzmán-Hernández, R., Jang, S., A. Torres-Ramos, C., Ayala-Peña, S., & Javadov, S. (2019). The Role of Adenine Nucleotide Translocase in the Assembly of Respiratory Supercomplexes in Cardiac Cells. Cells, 8(10), 1247. https://doi.org/10.3390/cells8101247





