Comprehensive Multi-Omics Analysis Reveals Aberrant Metabolism of Epstein–Barr-Virus-Associated Gastric Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Consideration
2.2. Chemicals and Reagents
2.3. Sample Collection
2.4. Sample Preparation for Untargeted Lipidomics
2.5. Sample Preparation for Pseudotargeted and Untargeted Metabolomics
2.6. Lipid Analysis by UPLC-QToF MS
2.7. Metabolite Analysis by HPLC-QqQ MS
2.8. Metabolite Analysis by GC-MS
2.9. Data Exploration and Visualization
2.10. Survival Analysis
2.11. Pathway Enrichment Analysis
2.12. Statistical Analysis
2.13. Data Availability
2.14. Quantitative Reverse Transcription–Polymerase Chain Reaction (RT-PCR)
3. Results
3.1. EBVaGC Model Establishment and Validation
3.2. Metabolic Associated Genes Are Downregulated in EBVaGC
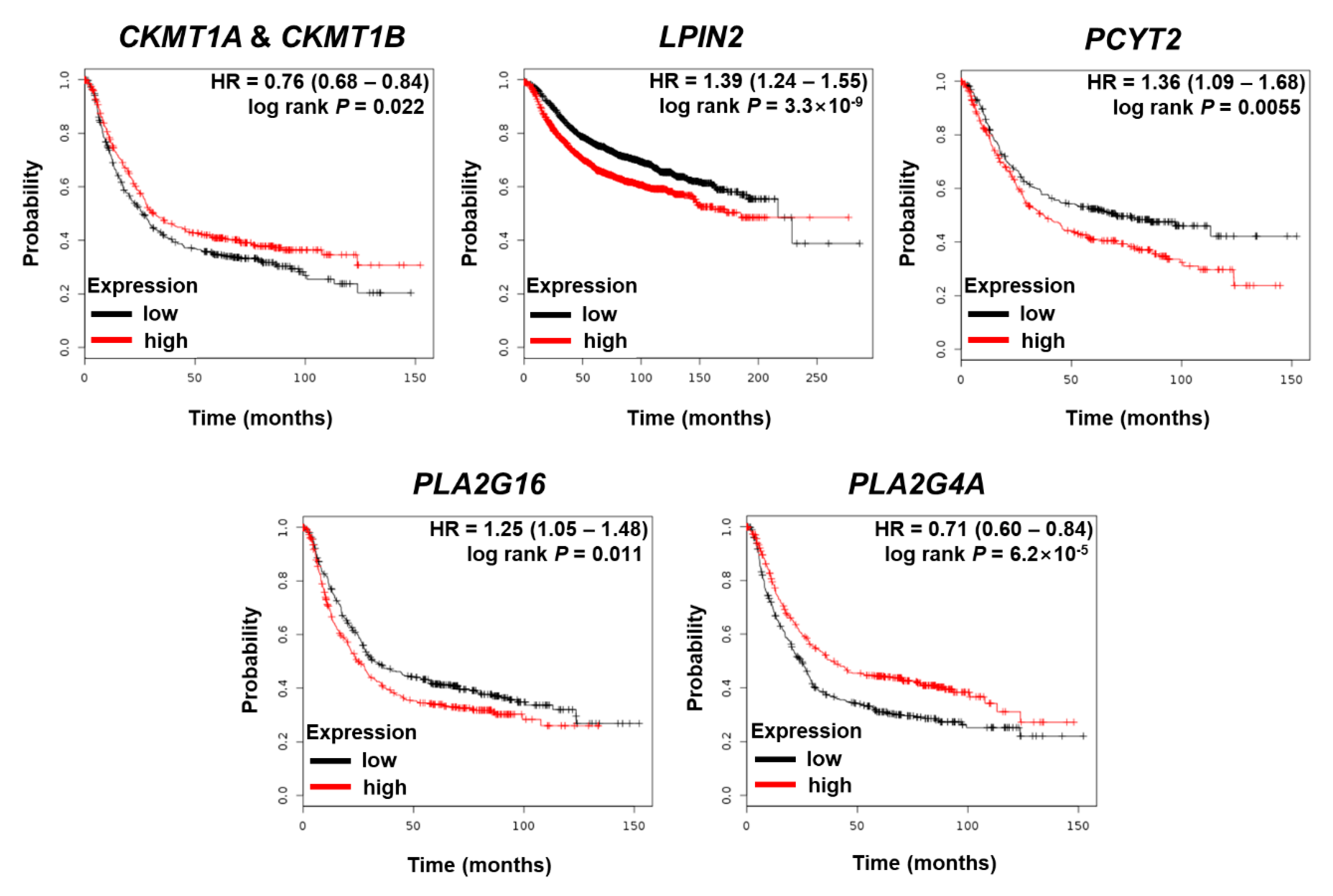
3.3. The Effects of Dysregulated Metabolic Regulator Coding Genes on the Survival of GC Patients
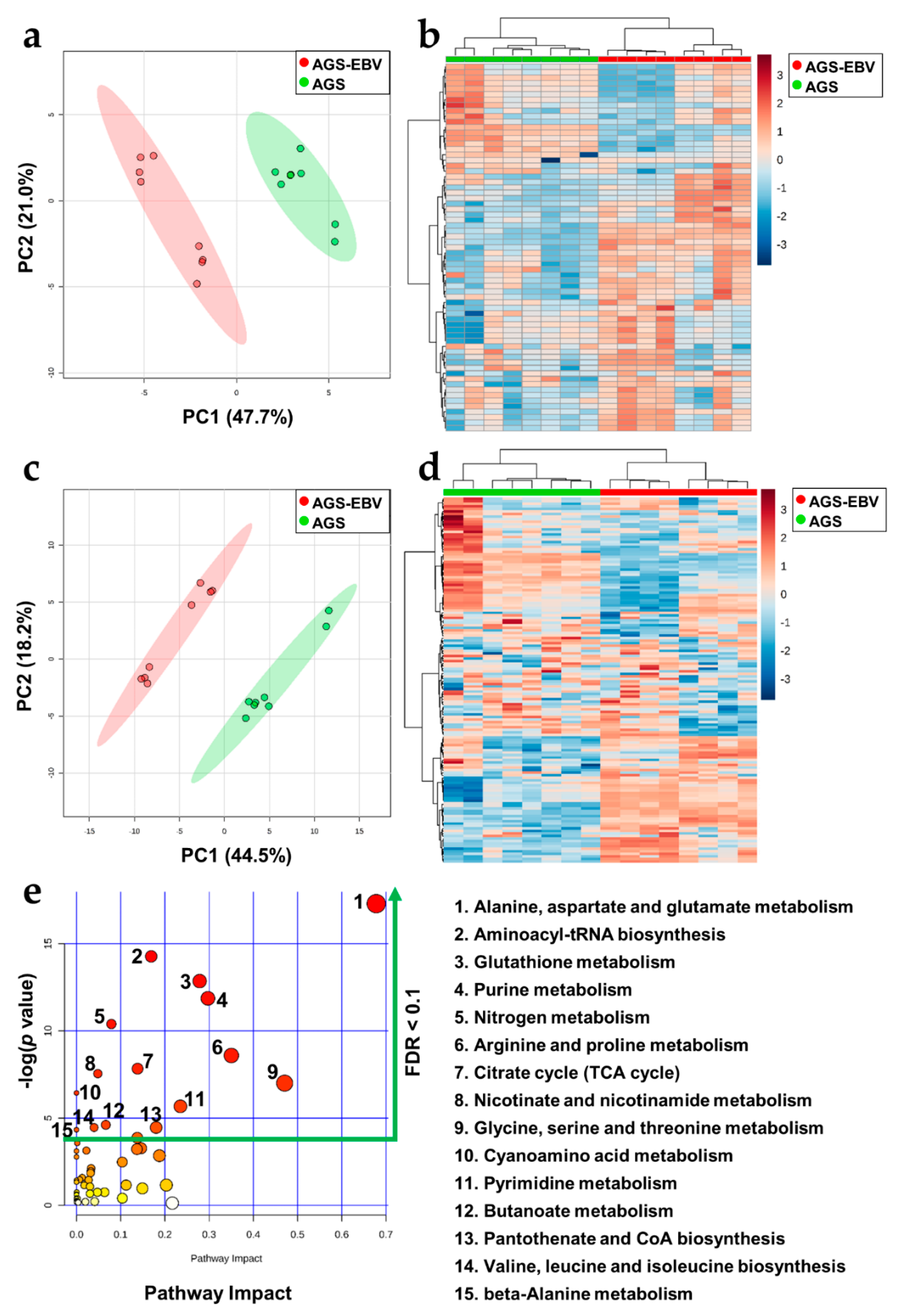
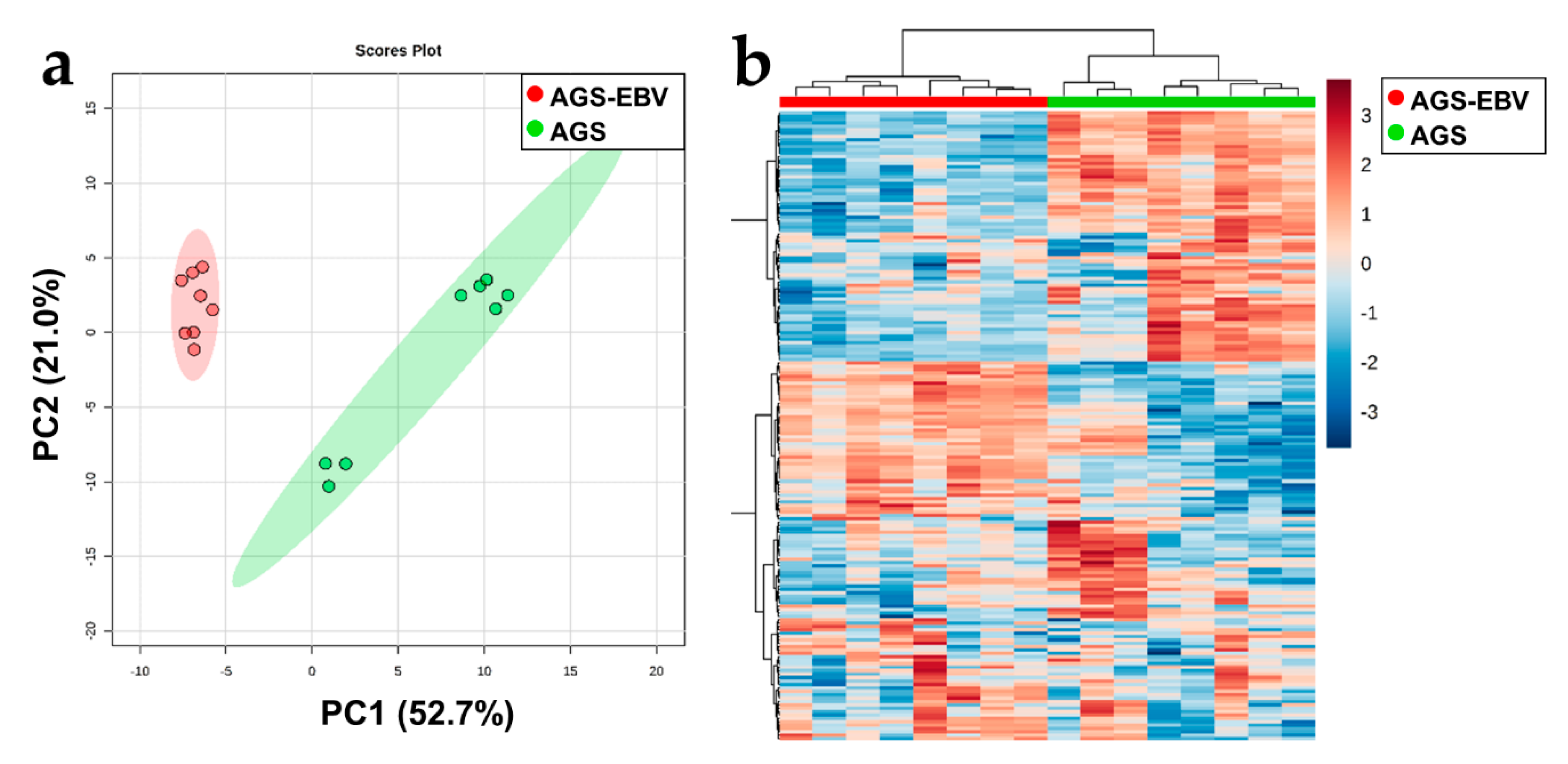
3.4. Significant Metabolic Alterations in EBVaGC
3.5. Significant Lipid Metabolism of EBVaGC
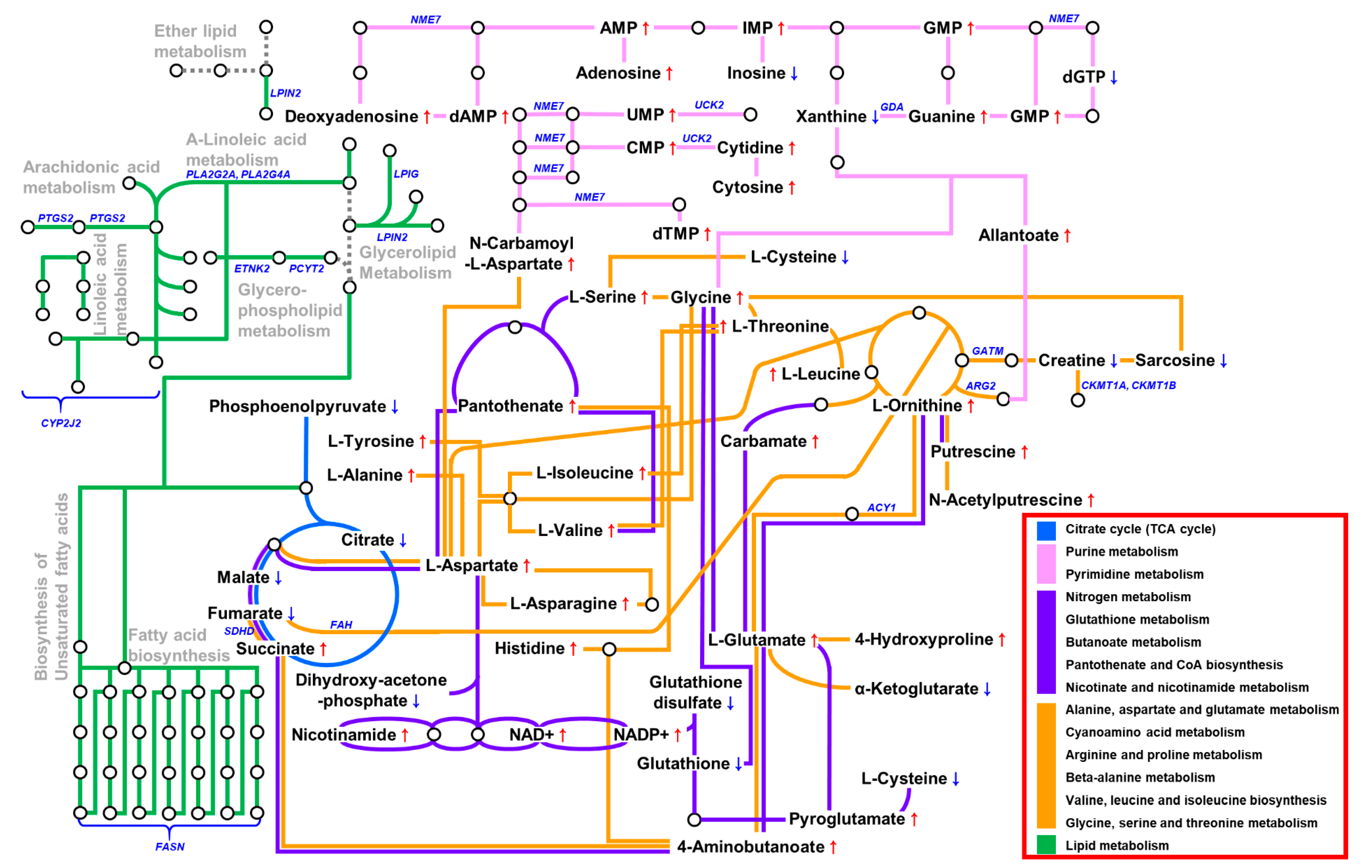
3.6. The Metabolic Landscape of EBVaGC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Karimi, P.; Islami, F.; Anandasabapathy, S.; Freedman, N.D.; Kamangar, F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Prev. Biomark. 2014, 23, 700–713. [Google Scholar] [CrossRef]
- Crew, K.D.; Neugut, A.I. Epidemiology of gastric cancer. World J. Gastroenterol. 2006, 12, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Boysen, T.; Mohammadi, M.; Melbye, M.; Hamilton-Dutoit, S.; Vainer, B.; Hansen, A.V.; Wohlfahrt, J.; Friborg, J. EBV-associated gastric carcinoma in high- and low-incidence areas for nasopharyngeal carcinoma. Br. J. Cancer 2009, 101, 530–533. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Naseem, M.; Barzi, A.; Brezden-Masley, C.; Puccini, A.; Berger, M.D.; Tokunaga, R.; Battaglin, F.; Soni, S.; McSkane, M.; Zhang, W.; et al. Outlooks on Epstein-Barr virus associated gastric cancer. Cancer Treat. Rev. 2018, 66, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.H. EBV and human cancer. Exp. Mol. Med. 2015, 47, 130. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.; Iizasa, H.; Yoshiyama, H.; Shimokuri, K.; Kobayashi, Y.; Sasaki, S.; Nakamura, M.; Yanai, H.; Sakai, K.; Suehiro, Y.; et al. Clinical Importance of Epstein-Barr Virus-Associated Gastric Cancer. Cancers 2018, 10, 167. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.M.; Kim, E.H. Epstein-Barr Virus and Gastric Cancer Risk: A Meta-analysis With Meta-regression of Case-control Studies. J. Prev. Med. Public Health 2016, 49, 97–107. [Google Scholar] [CrossRef]
- Camargo, M.C.; Kim, W.H.; Chiaravalli, A.M.; Kim, K.M.; Corvalan, A.H.; Matsuo, K.; Yu, J.; Sung, J.J.; Herrera-Goepfert, R.; Meneses-Gonzalez, F.; et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: An international pooled analysis. Gut 2014, 63, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Kohlruss, M.; Grosser, B.; Krenauer, M.; Slotta-Huspenina, J.; Jesinghaus, M.; Blank, S.; Novotny, A.; Reiche, M.; Schmidt, T.; Ismani, L.; et al. Prognostic implication of molecular subtypes and response to neoadjuvant chemotherapy in 760 gastric carcinomas: Role of Epstein-Barr virus infection and high- and low-microsatellite instability. J. Pathol. Clin. Res. 2019. [Google Scholar] [CrossRef]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.A.; Lee, J.; Sano, T.; Janjigian, Y.Y.; Fan, D.; Song, S. Gastric adenocarcinoma. Nat. Rev. Dis. Prim. 2017, 3, 17036. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Kang, Y.P.; Yoon, J.H.; Long, N.P.; Koo, G.B.; Noh, H.J.; Oh, S.J.; Lee, S.B.; Kim, H.M.; Hong, J.Y.; Lee, W.J.; et al. Spheroid-Induced Epithelial-Mesenchymal Transition Provokes Global Alterations of Breast Cancer Lipidome: A Multi-Layered Omics Analysis. Front. Oncol. 2019, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- Long, N.P.; Jung, K.H.; Anh, N.H.; Yan, H.H.; Nghi, T.D.; Park, S.; Yoon, S.J.; Min, J.E.; Kim, H.M.; Lim, J.H.; et al. An Integrative Data Mining and Omics-Based Translational Model for the Identification and Validation of Oncogenic Biomarkers of Pancreatic Cancer. Cancers 2019, 11, 155. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Serra, O.; Galan, M.; Ginesta, M.M.; Calvo, M.; Sala, N.; Salazar, R. Comparison and applicability of molecular classifications for gastric cancer. Cancer Treat. Rev. 2019, 77, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Antonowicz, S.; Kumar, S.; Wiggins, T.; Markar, S.R.; Hanna, G.B. Diagnostic Metabolomic Blood Tests for Endoluminal Gastrointestinal Cancer—A Systematic Review and Assessment of Quality. Cancer Epidemiol. Prev. Biomark. 2016, 25, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Abbassi-Ghadi, N.; Kumar, S.; Huang, J.; Goldin, R.; Takats, Z.; Hanna, G.B. Metabolomic profiling of oesophago-gastric cancer: A systematic review. Eur. J. Cancer 2013, 49, 3625–3637. [Google Scholar] [CrossRef]
- Chan, A.W.; Gill, R.S.; Schiller, D.; Sawyer, M.B. Potential role of metabolomics in diagnosis and surveillance of gastric cancer. World J. Gastroenterol. 2014, 20, 12874–12882. [Google Scholar] [CrossRef]
- Sohn, B.H.; Hwang, J.E.; Jang, H.J.; Lee, H.S.; Oh, S.C.; Shim, J.J.; Lee, K.W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.N.; Chae, H.S.; Oh, S.T.; Kang, J.H.; Park, C.H.; Park, W.S.; Takada, K.; Lee, J.M.; Lee, W.K.; Lee, S.K. Expression of viral microRNAs in Epstein-Barr virus-associated gastric carcinoma. J. Virol. 2007, 81, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, J.; Kiss, C.; Imai, S.; Takada, K.; Okita, K.; Klein, G.; Szekely, L. Upregulation of the truncated basic hair keratin 1(hHb1-ΔN) in carcinoma cells by Epstein-Barr virus (EBV). Int. J. Cancer 2003, 107, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.T.; Seo, J.S.; Moon, U.Y.; Kang, K.H.; Shin, D.J.; Yoon, S.K.; Kim, W.H.; Park, J.G.; Lee, S.K. A naturally derived gastric cancer cell line shows latency I Epstein-Barr virus infection closely resembling EBV-associated gastric cancer. Virology 2004, 320, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Yi, T.; Ahn, S.H.; Lim, D.K.; Kim, S.N.; Lee, H.J.; Cho, Y.K.; Lim, J.Y.; Sung, J.H.; Yun, J.H.; et al. Comparative study on metabolite level in tissue-specific human mesenchymal stem cells by an ultra-performance liquid chromatography quadrupole time of flight mass spectrometry. Anal. Chim. Acta 2018, 1024, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Smilowitz, J.T.; Fiehn, O. Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal. Chem. 2017, 89, 12360–12368. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.J.; Long, N.P.; Jung, K.H.; Kim, H.M.; Hong, Y.J.; Fang, Z.; Kim, S.J.; Kim, T.J.; Anh, N.H.; Hong, S.S.; et al. Systemic and Local Metabolic Alterations in Sleep-Deprivation-Induced Stress: A Multiplatform Mass-Spectrometry-Based Lipidomics and Metabolomics Approach. J. Proteome Res. 2019. [Google Scholar] [CrossRef]
- Yuan, M.; Breitkopf, S.B.; Yang, X.; Asara, J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012, 7, 872–881. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, 486–494. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Szasz, A.M.; Lanczky, A.; Nagy, A.; Forster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabo, A.; Gyorffy, B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, M.R.; Jeucken, A.; Wassenaar, T.A.; van de Lest, C.H.A.; Brouwers, J.F.; Helms, J.B. LION/web: A web-based ontology enrichment tool for lipidomic data analysis. GigaScience 2019, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Jeong, I.; Kang, S.K.; Kwon, W.S.; Kim, H.J.; Kim, K.H.; Kim, H.M.; Lee, A.; Lee, S.K.; Bogenrieder, T.; Chung, H.C.; et al. Regulation of proliferation and invasion by the IGF signalling pathway in Epstein-Barr virus-positive gastric cancer. J. Cell. Mol. Med. 2018, 22, 5899–5908. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Park, C.; Kim, H.J.; Park, J.; Hwang, J.; Kim, J.I.; Choi, M.G.; Kim, S.; Kim, K.M.; Kang, M.S. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology 2015, 148, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Mun, D.G.; Bhin, J.; Kim, S.; Kim, H.; Jung, J.H.; Jung, Y.; Jang, Y.E.; Park, J.M.; Kim, H.; Jung, Y.; et al. Proteogenomic Characterization of Human Early-Onset Gastric Cancer. Cancer Cell 2019, 35, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Patel, K.; Singhi, A.D.; Ren, B.; Zhu, B.; Shaikh, F.; Sun, W. Programmed Death-Ligand 1 Expression is Common in Gastric Cancer Associated with Epstein-Barr Virus or Microsatellite Instability. Am. J. Surg. Pathol. 2016, 40, 1496–1506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Vucetic, M.; Cormerais, Y.; Parks, S.K.; Pouyssegur, J. The Central Role of Amino Acids in Cancer Redox Homeostasis: Vulnerability Points of the Cancer Redox Code. Front. Oncol. 2017, 7, 319. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Choi, B.H.; Coloff, J.L. The Diverse Functions of Non-Essential Amino Acids in Cancer. Cancers 2019, 11, 675. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Z.; Wang, J.; Chen, C.; Tang, X.; Zhu, J.; Liu, J. Metabolic reprogramming results in abnormal glycolysis in gastric cancer: A review. OncoTargets Ther. 2019, 12, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Zhou, L. Gastric cancer: Metabolic and metabolomics perspectives Review. Int. J. Oncol. 2017, 51, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Takehara, H.; Yoshida, K.; Nishi, M.; Miyake, H.; Kita, Y.; Komi, N. Increased aspartate and glutamate levels in both gastric and colon cancer tissues. Tokushima J. Exp. Med. 1993, 40, 19–25. [Google Scholar] [PubMed]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Kumar, R.; Sheets, E.D.; Benkovic, S.J. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 2008, 320, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Maehara, Y.; Kusumoto, T.; Sakaguchi, Y.; Kusumoto, H.; Kido, Y.; Anai, H.; Sugimachi, K. Pyrimidine nucleotide synthesis is more extensive in poorly differentiated than in well-differentiated human gastric carcinoma. Cancer 1989, 63, 96–101. [Google Scholar] [CrossRef]
- Linder, N.; Haglund, C.; Lundin, M.; Nordling, S.; Ristimaki, A.; Kokkola, A.; Mrena, J.; Wiksten, J.P.; Lundin, J. Decreased xanthine oxidoreductase is a predictor of poor prognosis in early-stage gastric cancer. J. Clin. Pathol. 2006, 59, 965–971. [Google Scholar] [CrossRef]
- Long, J.; Zhang, C.J.; Zhu, N.; Du, K.; Yin, Y.F.; Tan, X.; Liao, D.F.; Qin, L. Lipid metabolism and carcinogenesis, cancer development. Am. J. Cancer Res. 2018, 8, 778–791. [Google Scholar]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, 189. [Google Scholar] [CrossRef]
- Wassenaar, T.A.; Ingolfsson, H.I.; Bockmann, R.A.; Tieleman, D.P.; Marrink, S.J. Computational Lipidomics with insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 2015, 11, 2144–2155. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D. Structural and thermodynamic determinants of chain-melting transition temperatures for phospholipid and glycolipids membranes. Biochim. Biophys. Acta 2010, 1798, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Dev, J.; Mezhyrova, J.; Pan, L.; Piai, A.; Chou, J.J. The Unusual Transmembrane Partition of the Hexameric Channel of the Hepatitis C Virus. Structure 2018, 26, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Menendez, J.A.; Lupu, R. Fatty acid synthase regulates estrogen receptor-alpha signaling in breast cancer cells. Oncogenesis 2017, 6, 299. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Chen, L.; Zhou, M.; Zhang, J.; Sun, L.; Huang, N.; Bin, J.; Liao, Y.; Liao, W. MACC1 decreases the chemosensitivity of gastric cancer cells to oxaliplatin by regulating FASN expression. Oncol. Rep. 2017, 37, 2583–2592. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dai, J.; Shen, J.; Pan, W.; Shen, S.; Das, U.N. Effects of polyunsaturated fatty acids on the growth of gastric cancer cells in vitro. Lipids Health Dis. 2013, 12, 71. [Google Scholar] [CrossRef] [PubMed]

| LION ID | Pathways | Annotated | p-Value | FDR* | |
|---|---|---|---|---|---|
| 1 | LION:0080973 | Below average bilayer thickness | 22 | 3.50 × 10−10 | 1.67 × 10−8 |
| 2 | LION:0001741 | Below average transition temperature | 24 | 6.30 × 10−10 | 1.67 × 10−8 |
| 3 | LION:0080982 | Above average lateral diffusion | 21 | 1.60 × 10−9 | 2.83 × 10−8 |
| 4 | LION:0080968 | Very low bilayer thickness | 15 | 1.80 × 10−6 | 1.91 × 10−6 |
| 5 | LION:0080980 | Very high lateral diffusion | 15 | 5.10 × 10−6 | 1.91 × 10−6 |
| 6 | LION:0001735 | Very low transition temperature | 12 | 3.50 × 10−10 | 4.51 × 10−5 |
| 7 | LION:0000030 | Diacylglycerophosphocholines | 20 | 2.00 × 10−4 | 1.51 × 10−3 |
| 8 | LION:0080979 | High lateral diffusion | 8 | 6.90 × 10−4 | 4.12 × 10−3 |
| 9 | LION:0001736 | Low transition temperature | 12 | 7.00 × 10−4 | 4.12 × 10−3 |
| 10 | LION:0012010 | Membrane component | 63 | 9.70 × 10−4 | 5.14 × 10−3 |
| 11 | LION:0080969 | Low bilayer thickness | 7 | 2.58 × 10−3 | 5.14 × 10−3 |
| 12 | LION:0000095 | Headgroup with positive charge/zwitter ion | 61 | 3.94 × 10−3 | 1.74 × 10−2 |
| 13 | LION:0000084 | Ceramide phosphocholines (sphingomyelins) | 8 | 0.012 | 4.86 × 10−2 |
| 14 | LION:0000038 | Diacylglycerophosphoethanolamines | 9 | 0.015 | 5.69 × 10−2 |
| 15 | LION:0000465 | Neutral intrinsic curvature | 33 | 0.022 | 7.93 × 10−2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, S.J.; Kim, J.Y.; Long, N.P.; Min, J.E.; Kim, H.M.; Yoon, J.H.; Anh, N.H.; Park, M.C.; Kwon, S.W.; Lee, S.K. Comprehensive Multi-Omics Analysis Reveals Aberrant Metabolism of Epstein–Barr-Virus-Associated Gastric Carcinoma. Cells 2019, 8, 1220. https://doi.org/10.3390/cells8101220
Yoon SJ, Kim JY, Long NP, Min JE, Kim HM, Yoon JH, Anh NH, Park MC, Kwon SW, Lee SK. Comprehensive Multi-Omics Analysis Reveals Aberrant Metabolism of Epstein–Barr-Virus-Associated Gastric Carcinoma. Cells. 2019; 8(10):1220. https://doi.org/10.3390/cells8101220
Chicago/Turabian StyleYoon, Sang Jun, Jun Yeob Kim, Nguyen Phuoc Long, Jung Eun Min, Hyung Min Kim, Jae Hee Yoon, Nguyen Hoang Anh, Myung Chan Park, Sung Won Kwon, and Suk Kyeong Lee. 2019. "Comprehensive Multi-Omics Analysis Reveals Aberrant Metabolism of Epstein–Barr-Virus-Associated Gastric Carcinoma" Cells 8, no. 10: 1220. https://doi.org/10.3390/cells8101220
APA StyleYoon, S. J., Kim, J. Y., Long, N. P., Min, J. E., Kim, H. M., Yoon, J. H., Anh, N. H., Park, M. C., Kwon, S. W., & Lee, S. K. (2019). Comprehensive Multi-Omics Analysis Reveals Aberrant Metabolism of Epstein–Barr-Virus-Associated Gastric Carcinoma. Cells, 8(10), 1220. https://doi.org/10.3390/cells8101220






