The Role of Stiffness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration
Abstract
1. Introduction
2. The Mechanisms of Stiffness Sensing
3. Mechanosensitive Signaling Pathways
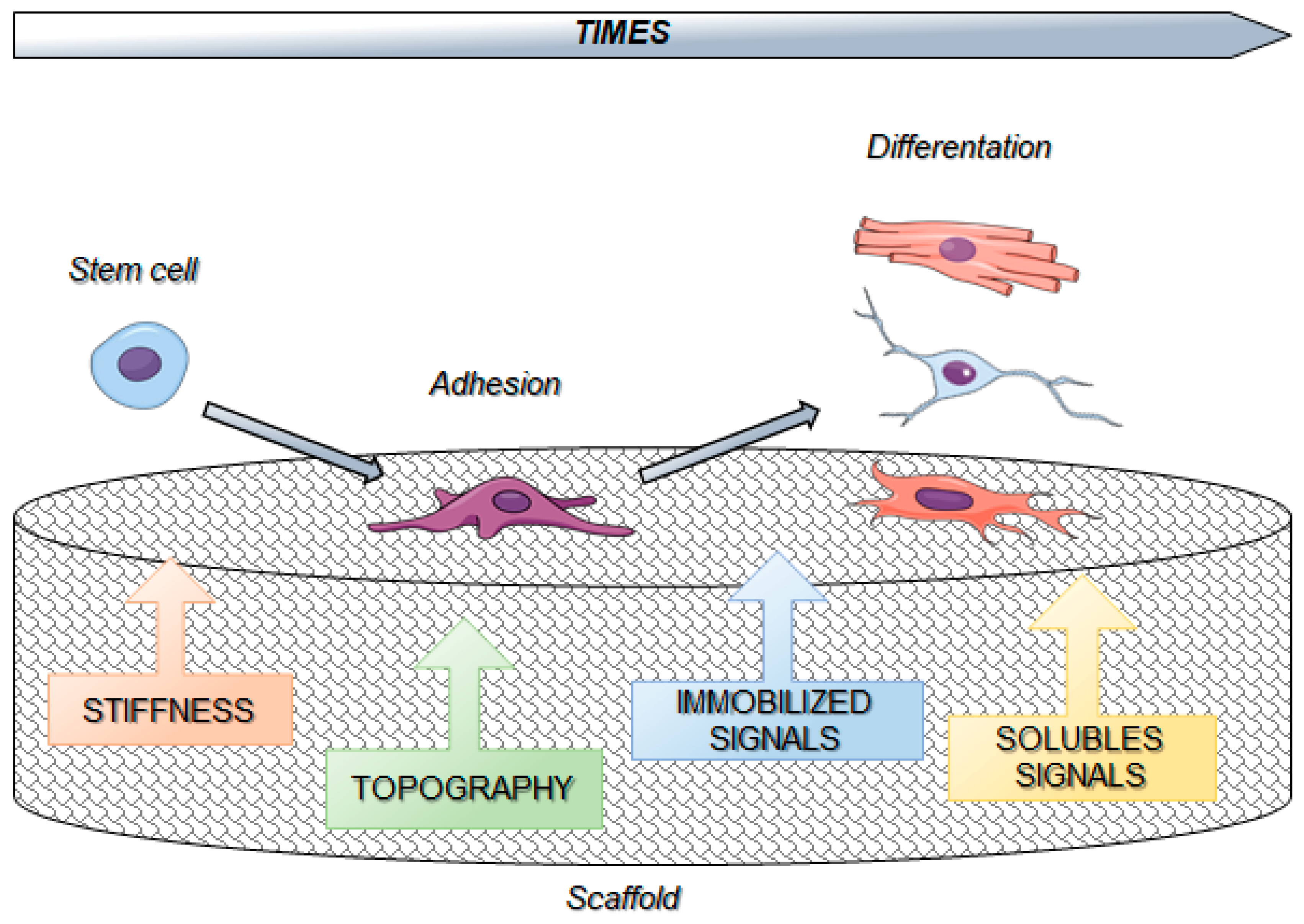
4. Biomaterials
5. Mechanical Properties of Biomaterials
5.1. Viscoelastic Properties
5.2. Effects of Stiffness
5.3. 2D and 3D Hydrogels
5.4. 3D Fibrillar Matrices
5.5. Effects of Topography
6. Biomaterials and Cell Reprogramming
7. Biomaterials and Stem Cells Interactions
8. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Isomursu, A.; Lerche, M.; Taskinen, M.E.; Ivaska, J.; Peuhu, E. Integrin signaling and mechanotransduction in regulation of somatic stem cells. Exp. Cell Res. 2019, 378, 217–225. [Google Scholar] [CrossRef]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.-J. The Role of the Extracellular Matrix in Cancer Stemness. Front. Cell Dev. Boil. 2019, 7, 86. [Google Scholar] [CrossRef]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef]
- Liu, W.F.; Chen, C.S. Engineering biomaterials to control cell function. Mater. Today 2005, 8, 28–35. [Google Scholar] [CrossRef]
- Handorf, A.M.; Zhou, Y.; A Halanski, M.; Li, W.-J. Tissue Stiffness Dictates Development, Homeostasis, and Disease Progression. Organogenesis 2015, 11, 1–15. [Google Scholar] [CrossRef]
- Barnes, J.M.; Przybyla, L.; Weaver, V.M. Tissue mechanics regulate brain development, homeostasis and disease. J. Cell Sci. 2017, 130, 71–82. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.D.P.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.-W.; Tewari, M.; et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef]
- Macri-Pellizzeri, L.; De-Juan-Pardo, E.M.; Prosper, F.; Pelacho, B. Role of substrate biomechanics in controlling (stem) cell fate: Implications in regenerative medicine. J. Tissue Eng. Regener. Med. 2018, 12, 1012–1019. [Google Scholar] [CrossRef]
- Lo, C.-M.; Wang, H.-B.; Dembo, M.; Wang, Y.-L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000, 79, 144–152. [Google Scholar] [CrossRef]
- DuChez, B.J.; Doyle, A.D.; Dimitriadis, E.K.; Yamada, K.M. Durotaxis by Human Cancer Cells. Biophys. J. 2019, 116, 670–683. [Google Scholar] [CrossRef]
- Thompson, A.J.; Pillai, E.K.; Dimov, I.B.; Foster, S.K.; E Holt, C.; Franze, K. Rapid changes in tissue mechanics regulate cell behaviour in the developing embryonic brain. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Wang, H.-B.; Dembo, M.; Wang, Y.-L. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Physiol. 2000, 279, 279. [Google Scholar] [CrossRef]
- Ghassemi, S.; Meacci, G.; Liu, S.; Gondarenko, A.A.; Mathur, A.; Roca-Cusachs, P.; Sheetz, M.P.; Hone, J. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl. Acad. Sci. USA 2012, 109, 5328–5333. [Google Scholar] [CrossRef]
- Caiazzo, M.; Okawa, Y.; Ranga, A.; Piersigilli, A.; Tabata, Y.; Lutolf, M.P. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater. 2016, 15, 344–352. [Google Scholar] [CrossRef]
- Iskratsch, T.; Wolfenson, H.; Sheetz, M.P. Appreciating force and shape — the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Boil. 2014, 15, 825–833. [Google Scholar] [CrossRef]
- Bershadsky, A.D.; Balaban, N.Q.; Geiger, B. Adhesion-Dependent Cell Mechanosensitivity. Annu. Rev. Cell Dev. Boil. 2003, 19, 677–695. [Google Scholar] [CrossRef]
- Chen, C.S.; Tan, J.; Tien, J. Mechanotransduction at Cell-Matrix and Cell-Cell Contacts. Annu. Rev. Biomed. Eng. 2004, 6, 275–302. [Google Scholar] [CrossRef]
- Geiger, B.; Bershadsky, A.; Pankov, R.; Yamada, K.M. Transmembrane crosstalk between the extracellular matrix--cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001, 2, 793–805. [Google Scholar] [CrossRef]
- Sastry, S.K.; Burridge, K. Focal Adhesions: A Nexus for Intracellular Signaling and Cytoskeletal Dynamics. Exp. Cell Res. 2000, 261, 25–36. [Google Scholar] [CrossRef]
- Bangasser, B.L.; Shamsan, G.A.; Chan, C.E.; Opoku, K.N.; Tüzel, E.; Schlichtmann, B.W.; Kasim, J.A.; Fuller, B.J.; McCullough, B.R.; Rosenfeld, S.S.; et al. Shifting the optimal stiffness for cell migration. Nat. Commun. 2017, 8, 15313. [Google Scholar] [CrossRef]
- Chan, C.E.; Odde, D.J. Traction Dynamics of Filopodia on Compliant Substrates. Science. 2008, 322, 1687–1691. [Google Scholar] [CrossRef]
- Elosegui-Artola, A.; Oria, R.; Chen, Y.; Kosmalska, A.; Pérez-González, C.; Castro, N.; Zhu, C.; Trepat, X.; Roca-Cusachs, P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nature 2016, 18, 540–548. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Geiger, B. The switchable integrin adhesome. J. Cell Sci. 2010, 123, 1385–1388. [Google Scholar] [CrossRef]
- Atherton, P.; Stutchbury, B.; Jethwa, D.; Ballestrem, C. Mechanosensitive components of integrin adhesions: Role of vinculin. Exp. Cell Res. 2016, 343, 21–27. [Google Scholar] [CrossRef]
- Gattazzo, F.; Urciuolo, A.; Bonaldo, P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta (BBA) Gen. Subj. 2014, 1840, 2506–2519. [Google Scholar] [CrossRef]
- Speight, P.; Kofler, M.; Szaszi, K.; Kapus, A. Context-dependent switch in chemo/mechanotransduction via multilevel crosstalk among cytoskeleton-regulated MRTF and TAZ and TGFβ-regulated Smad3. Nat. Commun. 2016, 7, 11642. [Google Scholar] [CrossRef]
- Low, B.C.; Pan, C.Q.; Shivashankar, G.; Bershadsky, A.; Sudol, M.; Sheetz, M. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014, 588, 2663–2670. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Alan, J.K.; A Lundquist, E. Mutationally activated Rho GTPases in cancer. Small GTPases 2013, 4, 159–163. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.-Y.; Yu, J.; Guan, K.-L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef]
- Mana-Capelli, S.; Paramasivam, M.; Dutta, S.; Mccollum, D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol. Biol. Cell 2014, 25, 1676–1685. [Google Scholar] [CrossRef]
- Ohgushi, M.; Minaguchi, M.; Sasai, Y. Rho-Signaling-Directed YAP/TAZ Activity Underlies the Long-Term Survival and Expansion of Human Embryonic Stem Cells. Cell Stem Cell 2015, 17, 448–461. [Google Scholar] [CrossRef]
- Feng, X.; Degese, M.S.; Iglesias-Bartolome, R.; Vaqué, J.P.; Molinolo, A.A.; Rodrigues, M.; Zaidi, M.R.; Ksander, B.R.; Merlino, G.; Sodhi, A.; et al. Hippo-Independent Activation of YAP by the GNAQ Uveal Melanoma Oncogene through a Trio-regulated Rho GTPase Signaling Circuitry. Cancer Cell 2014, 25, 831–845. [Google Scholar] [CrossRef]
- Rammensee, S.; Kang, M.S.; Georgiou, K.; Kumar, S.; Schaffer, D.V. Dynamics of Mechanosensitive Neural Stem Cell Differentiation: Dynamics of Mechanosensitive NSC Differentiation. Stem Cells 2017, 35, 497–506. [Google Scholar] [CrossRef]
- Oliver-De La Cruz, J.; Nardone, G.; Vrbsky, J.; Pompeiano, A.; Perestrelo, A.R.; Capradossi, F.; Melajová, K.; Filipensky, P.; Forte, G. Substrate mechanics controls adipogenesis through YAP phosphorylation by dictating cell spreading. Biomaterials 2019, 205, 64–80. [Google Scholar] [CrossRef]
- Vartiainen, M.K.; Guettler, S.; Larijani, B.; Treisman, R. Nuclear Actin Regulates Dynamic Subcellular Localization and Activity of the SRF Cofactor MAL. Science 2007, 316, 1749–1752. [Google Scholar] [CrossRef]
- Esnault, C.; Stewart, A.; Gualdrini, F.; East, P.; Horswell, S.; Matthews, N.; Treisman, R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genome Res. 2014, 28, 943–958. [Google Scholar] [CrossRef]
- Miano, J.M.; Long, X.; Fujiwara, K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Physiol. 2007, 292, C70–C81. [Google Scholar] [CrossRef]
- Bialik, J.F.; Ding, M.; Speight, P.; Dan, Q.; Miranda, M.Z.; Di Ciano-Oliveira, C.; Kofler, M.M.; Rotstein, O.D.; Pedersen, S.F.; Szászi, K.; et al. Profibrotic epithelial phenotype: a central role for MRTF and TAZ. Sci. Rep. 2019, 9, 4323. [Google Scholar] [CrossRef]
- Fan, L.; Sebe, A.; Peterfi, Z.; Masszi, A.; Thirone, A.C.; Rotstein, O.D.; Nakano, H.; McCulloch, C.A.; Szászi, K.; Mucsi, I.; et al. Cell Contact–dependent Regulation of Epithelial–Myofibroblast Transition via the Rho-Rho Kinase-Phospho-Myosin Pathway. Mol. Boil. Cell 2007, 18, 1083–1097. [Google Scholar] [CrossRef]
- Masszi, A.; Speight, P.; Charbonney, E.; Lodyga, M.; Nakano, H.; Szászi, K.; Kapus, A. Fate-determining mechanisms in epithelial–myofibroblast transition: major inhibitory role for Smad3. J. Exp. Med. 2010, 207, i5. [Google Scholar] [CrossRef]
- Rozycki, M.; Bialik, J.F.; Speight, P.; Dan, Q.; Knudsen, T.E.T.; Szeto, S.G.; Yuen, D.A.; Szászi, K.; Pedersen, S.F.; Kapus, A. Myocardin-related Transcription Factor Regulates Nox4 Protein Expression: Linking cytoskeletal organization to redox state. J. Biol. Chem. 2016, 291, 227–243. [Google Scholar] [CrossRef]
- Scharenberg, M.A.; Pippenger, B.E.; Sack, R.; Zingg, D.; Ferralli, J.; Schenk, S.; Martin, I.; Chiquet-Ehrismann, R. TGF- -induced differentiation into myofibroblasts involves specific regulation of two MKL1 isoforms. J. Cell Sci. 2014, 141, e707. [Google Scholar]
- Small, E.M.; Thatcher, J.E.; Sutherland, L.B.; Kinoshita, H.; Gerard, R.D.; Richardson, J.A.; DiMaio, J.M.; Sadek, H.; Kuwahara, K.; Olson, E.N. Myocardin-related transcription factor-A controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ. Res. 2010, 107, 294–304. [Google Scholar] [CrossRef]
- Foster, C.T.; Gualdrini, F.; Treisman, R. Mutual dependence of the MRTF–SRF and YAP–TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genome Res. 2017, 31, 2361–2375. [Google Scholar] [CrossRef]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef]
- Hao, H.; Starr, D.A. SUN/KASH interactions facilitate force transmission across the nuclear envelope. Nucleus 2019, 10, 73–80. [Google Scholar] [CrossRef]
- Olins, A.L.; Rhodes, G.; Welch, D.B.M.; Zwerger, M.; Olins, D.E. Lamin B receptor: Multi-tasking at the nuclear envelope. Nucleus 2010, 1, 53–70. [Google Scholar] [CrossRef]
- Pajerowski, J.D.; Dahl, K.N.; Zhong, F.L.; Sammak, P.J.; Discher, D.E. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 15619–15624. [Google Scholar] [CrossRef]
- Shivashankar, G. Mechanical regulation of genome architecture and cell-fate decisions. Curr. Opin. Cell Boil. 2019, 56, 115–121. [Google Scholar] [CrossRef]
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Boil. 2017, 216, 305–315. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Dekker, J.; Mirny, L. The 3D genome as moderator of chromosomal communication. Cell 2016, 164, 1110–1121. [Google Scholar] [CrossRef]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Graham, D.M.; Burridge, K. Mechanotransduction and nuclear function. Curr. Opin. Cell Boil. 2016, 40, 98–105. [Google Scholar] [CrossRef]
- Przybyla, L.; Muncie, J.M.; Weaver, V.M. Mechanical Control of Epithelial-to-Mesenchymal Transitions in Development and Cancer. Annu. Rev. Cell Dev. Boil. 2016, 32, 527–554. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G. Nuclear Mechanopathology and Cancer Diagnosis. Trends Cancer 2018, 4, 320–331. [Google Scholar] [CrossRef]
- Jain, N.; Iyer, K.V.; Kumar, A.; Shivashankar, G.V. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. USA 2013, 110, 11349–11354. [Google Scholar] [CrossRef]
- Wang, Y.; Nagarajan, M.; Uhler, C.; Shivashankar, G.V. Orientation and repositioning of chromosomes correlate with cell geometry–dependent gene expression. Mol. Boil. Cell 2017, 28, 1997–2009. [Google Scholar] [CrossRef]
- Khan, F.; Tanaka, M. Designing Smart Biomaterials for Tissue Engineering. Int. J. Mol. Sci. 2017, 19, 17. [Google Scholar] [CrossRef]
- Chen, F.-M.; Liu, X. Advancing biomaterials of human origin for tissue engineering. Prog. Polym. Sci. 2016, 53, 86–168. [Google Scholar] [CrossRef]
- Song, R.; Murphy, M.; Li, C.; Ting, K.; Soo, C.; Zheng, Z. Current development of biodegradable polymeric materials for biomedical applications. Drug Des. Dev. Ther. 2018, 12, 3117–3145. [Google Scholar] [CrossRef]
- Williams, D.F. Definitions in biomaterials: Proceedings of a consensus conference of the European Society for Biomaterials; Elsevier: Chester, UK, 1986. [Google Scholar]
- Hiew, V.V.; Simat, S.F.B.; Teoh, P.L. The Advancement of Biomaterials in Regulating Stem Cell Fate. Stem Cell Rev. Rep. 2018, 14, 43–57. [Google Scholar] [CrossRef]
- Mano, J.; Silva, G.; Azevedo, H.; Malafaya, P.; Sousa, R.; Silva, S.; Boesel, L.; Oliveira, J.M.; Santos, T.; Marques, A.; et al. Natural origin biodegradable systems in tissue engineering and regenerative medicine: present status and some moving trends. J. R. Soc. Interface 2007, 4, 999–1030. [Google Scholar] [CrossRef]
- Halper, J.; Kjaer, M. Basic Components of Connective Tissues and Extracellular Matrix: Elastin, Fibrillin, Fibulins, Fibrinogen, Fibronectin, Laminin, Tenascins and Thrombospondins. In Progress in Heritable Soft Connective Tissue Diseases; Halper, J., Ed.; Springer Netherlands: Heidelberg, Germany, 2014; Volume 802, pp. 31–47. ISBN 978-94-007-7892-4. [Google Scholar]
- Barnes, C.P.; Pemble, C.W.; Brand, D.D.; Simpson, D.G.; Bowlin, G.L. Cross-Linking Electrospun Type II Collagen Tissue Engineering Scaffolds with Carbodiimide in Ethanol. Tissue Eng. 2007, 13, 1593–1605. [Google Scholar] [CrossRef]
- Ravindran, S.; Song, Y.; George, A. Development of Three-Dimensional Biomimetic Scaffold to Study Epithelial–Mesenchymal Interactions. Tissue Eng. Part A 2010, 16, 327–342. [Google Scholar] [CrossRef]
- Liu, X.; Smith, L.A.; Hu, J.; Ma, P.X. Biomimetic Nanofibrous Gelatin/Apatite Composite Scaffolds for Bone Tissue Engineering. Biomaterials 2009, 30, 2252–2258. [Google Scholar] [CrossRef]
- Niknejad, H.; Peirovi, H.; Jorjani, M.; Ahmadiani, A.; Ghanavi, J.; Seifalian, A.M. Properties of the amniotic membrane for potential use in tissue engineering. Eur. Cell Mater 2008, 15, 88–99. [Google Scholar] [CrossRef]
- Dong, C.; Lv, Y. Application of Collagen Scaffold in Tissue Engineering: Recent Advances and New Perspectives. Polymers 2016, 8, 42. [Google Scholar] [CrossRef]
- Weisel, J.W.; Litvinov, R.I. Fibrin Formation, Structure and Properties. Sub-cellular Biochem. 2017, 82, 405–456. [Google Scholar]
- Brown, A.C.; Barker, T.H. Fibrin-based biomaterials: Modulation of macroscopic properties through rational design at the molecular level. Acta. Biomater. 2014, 10, 1502–1514. [Google Scholar] [CrossRef]
- Altman, G.H.; Diaz, F.; Jakuba, C.; Calabro, T.; Horan, R.L.; Chen, J.; Lu, H.; Richmond, J.; Kaplan, D.L. Silk-based biomaterials. Biomaterials 2003, 24, 401–416. [Google Scholar] [CrossRef]
- Brovold, M.; Almeida, J.I.; Pla-Palacín, I.; Sainz-Arnal, P.; Sánchez-Romero, N.; Rivas, J.J.; Almeida, H.; Dachary, P.R.; Serrano-Aulló, T.; Soker, S.; et al. Naturally-Derived Biomaterials for Tissue Engineering Applications. Single Mol. Single Cell Seq. 2018, 1077, 421–449. [Google Scholar]
- Cheng, G.; Davoudi, Z.; Xing, X.; Yu, X.; Cheng, X.; Li, Z.; Deng, H.; Wang, Q. Advanced Silk Fibroin Biomaterials for Cartilage Regeneration. ACS Biomater. Sci. Eng. 2018, 4, 2704–2715. [Google Scholar] [CrossRef]
- Rebouillat, S.; Pla, F. A Review: On Smart Materials Based on Some Polysaccharides; within the Contextual Bigger Data, Insiders, “Improvisation” and Said Artificial Intelligence Trends. J. Biomater. Nanobiotechnol. 2019, 10, 41–77. [Google Scholar] [CrossRef]
- Elieh-Ali-Komi, D.; Hamblin, M.R. Chitin and Chitosan: Production and Application of Versatile Biomedical Nanomaterials. Int. J. Adv. Res. 2016, 4, 411–427. [Google Scholar]
- Comblain, F.; Rocasalbas, G.; Gauthier, S.; Henrotin, Y. Chitosan: A promising polymer for cartilage repair and viscosupplementation. Bio-Med. Mater. Eng. 2017, 28, 209. [Google Scholar] [CrossRef]
- Johnson, N.R.; Wang, Y. Drug Delivery Systems for Wound Healing. Curr. Pharm. Biotechnol. 2015, 16, 621–629. [Google Scholar] [CrossRef]
- Branski, L.K.; Herndon, D.N.; Celis, M.M.; Norbury, W.B.; Masters, O.E.; Jeschke, M.G. Amnion in the treatment of pediatric partial-thickness facial burns. Burn 2008, 34, 393–399. [Google Scholar] [CrossRef]
- Vaithilingam, V.; Tuch, B.E. Islet Transplantation and Encapsulation: An Update on Recent Developments. Rev. Diabet. Stud. 2011, 8, 51–67. [Google Scholar] [CrossRef]
- Coburn, J.; Pandit, A. Development of naturally-derived biomaterials and optimization of their biomechanical properties. Top. Tissue Eng. Country 2007, 3, 1–14. [Google Scholar]
- Lutolf, M.P.; A Hubbell, J. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005, 23, 47–55. [Google Scholar] [CrossRef]
- Turnbull, G.; Clarke, J.; Picard, F.; Riches, P.; Jia, L.; Han, F.; Li, B.; Shu, W. 3D bioactive composite scaffolds for bone tissue engineering. Bio. Mater. 2018, 3, 278–314. [Google Scholar] [CrossRef]
- Dorati, R.; DeTrizio, A.; Modena, T.; Conti, B.; Benazzo, F.; Gastaldi, G.; Genta, I. Biodegradable Scaffolds for Bone Regeneration Combined with Drug-Delivery Systems in Osteomyelitis Therapy. Pharmacology 2017, 10, 96. [Google Scholar] [CrossRef]
- Bose, S.; Vahabzadeh, S.; Bandyopadhyay, A. Bone tissue engineering using 3D printing. Mater. Today 2013, 16, 496–504. [Google Scholar] [CrossRef]
- Sisti, K.E.; de Andrés, M.C.; Johnston, D.; Almeida-Filho, E.; Guastaldi, A.C.; Oreffo, R.O.C. Skeletal stem cell and bone implant interactions are enhanced by LASER titanium modification. Biochem. Biophys. Res. Commun. 2016, 473, 719–725. [Google Scholar] [CrossRef]
- Hermawan, H.; Ramdan, D.; P. Djuansjah, J.R. Metals for Biomedical Applications. In Biomedical Engineering - From Theory to Applications; Fazel, R., Ed.; IntechOpen: London, UK, 2011. [Google Scholar]
- Eliaz, N. Corrosion of Metallic Biomaterials: A Review. Materials 2019, 12, 407. [Google Scholar] [CrossRef]
- Přikrylová, J.; Procházková, J.; Podzimek, Š. Side Effects of Dental Metal Implants: Impact on Human Health (Metal as a Risk Factor of Implantologic Treatment). BioMed. Res. Int. 2019, 2019, 1–5. [Google Scholar]
- Dong, H.; Qi, S.; Dong, H.; Qi, S. Realising the potential of graphene-based materials for biosurfaces—A future perspective. Biosurf. Biotribol. 2015, 1, 229–248. [Google Scholar] [CrossRef]
- Tadyszak, K.; Wychowaniec, J.K.; Litowczenko, J. Biomedical Applications of Graphene-Based Structures. Nanomaterial 2018, 8, 944. [Google Scholar] [CrossRef]
- Catanesi, M.; Panella, G.; Benedetti, E.; Fioravanti, G.; Perrozzi, F.; Ottaviano, L.; Di Leandro, L.; Ardini, M.; Giansanti, F.; D’Angelo, M.; et al. YAP/TAZ mechano-transduction as the underlying mechanism of neuronal differentiation induced by reduced graphene oxide. Nanomedicine 2018, 13, 3091–3106. [Google Scholar] [CrossRef]
- Holzwarth, J.M.; Ma, P.X. Biomimetic nanofibrous scaffolds for bone tissue engineering. Biomaterials 2011, 32, 9622–9629. [Google Scholar] [CrossRef]
- Green, D.W.; Goto, T.K.; Kim, K.-S.; Jung, H.-S. Calcifying tissue regeneration via biomimetic materials chemistry. J. R. Soc. Interface 2014, 11, 20140537. [Google Scholar] [CrossRef]
- Kabirian, F.; Mozafari, M. Decellularized ECM-derived bioinks: Prospects for the future. Methods 2019. [Google Scholar] [CrossRef]
- Adamski, M.; Fontana, G.; Gershlak, J.R.; Gaudette, G.R.; Le, H.D.; Murphy, W.L. Two Methods for Decellularization of Plant Tissues for Tissue Engineering Applications. J. Vis. Exp. 2018, e57586. [Google Scholar] [CrossRef]
- Fontana, G.; Gershlak, J.; Adamski, M.; Lee, J.; Matsumoto, S.; Le, H.D.; Binder, B.; Wirth, J.; Gaudette, G.; Murphy, W.L. Biofunctionalized plants as diverse biomaterials for human cell culture. Adv. Heal. Mater. 2017, 6, 1601225. [Google Scholar] [CrossRef]
- Williams, D.; Thayer, P.; Martinez, H.; Gatenholm, E.; Khademhosseini, A. A perspective on the physical, mechanical and biological specifications of bioinks and the development of functional tissues in 3D bioprinting. Bioprinting 2018, 9, 19–36. [Google Scholar] [CrossRef]
- Pal, S. Mechanical Properties of Biological Materials. In Design of Artificial Human Joints & Organs; Springer: Boston, MA, USA, 2014; pp. 23–40. ISBN 978-1-4614-6254-5. [Google Scholar]
- Basoli, F.; Giannitelli, S.M.; Gori, M.; Mozetic, P.; Bonfanti, A.; Trombetta, M.; Rainer, A. Biomechanical Characterization at the Cell Scale: Present and Prospects. Front. Physiol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Chameettachal, S.; Murab, S.; Vaid, R.; Midha, S.; Ghosh, S. Effect of visco-elastic silk-chitosan microcomposite scaffolds on matrix deposition and biomechanical functionality for cartilage tissue engineering: Viscoelastic silk-chitosan microcomposite scaffolds. J. Tissue Eng. Regener. Med. 2017, 11, 1212–1229. [Google Scholar] [CrossRef]
- Woods, T.; Gratzer, P.F. Effectiveness of three extraction techniques in the development of a decellularized bone–anterior cruciate ligament–bone graft. Biomaterials 2005, 26, 7339–7349. [Google Scholar] [CrossRef]
- Cartmell, J.S.; Dunn, M.G. Development of Cell-Seeded Patellar Tendon Allografts for Anterior Cruciate Ligament Reconstruction. Tissue Eng. 2004, 10, 1065–1075. [Google Scholar] [CrossRef]
- Harrison, R.D.; Gratzer, P.F. Effect of extraction protocols and epidermal growth factor on the cellular repopulation of decellularized anterior cruciate ligament allografts. J. Biomed. Mater. Res. Part A 2005, 75, 841–854. [Google Scholar] [CrossRef]
- Badylak, S.; Freytes, D.; Gilbert, T. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009, 5, 1–13. [Google Scholar] [CrossRef]
- Heydari, T.; Heidari, M.; Mashinchian, O.; Wojcik, M.; Xu, K.; Dalby, M.J.; Mahmoudi, M.; Ejtehadi, M.R. Development of a Virtual Cell Model to Predict Cell Response to Substrate Topography. ACS Nano 2017, 11, 9084–9092. [Google Scholar] [CrossRef]
- Versaevel, M.; Grevesse, T.; Gabriele, S. Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun. 2012, 3, 671. [Google Scholar] [CrossRef]
- Discher, D.E.; Mooney, D.J.; Zandstra, P.W. Growth factors, matrices, and forces combine and control stem cells. Science 2009, 324, 1673–1677. [Google Scholar] [CrossRef]
- Mente, P.L.; Lewis, J.L. Elastic modulus of calcified cartilage is an order of magnitude less than that of subchondral bone. J. Orthop. Res. 1994, 12, 637–647. [Google Scholar] [CrossRef]
- Vining, K.H.; Mooney, D.J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 2017, 18, 728–742. [Google Scholar] [CrossRef]
- Pelham, R.J.; Wang, Y. l Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA 1997, 94, 13661–13665. [Google Scholar] [CrossRef]
- Yeung, T.; Georges, P.C.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.; Janmey, P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton 2005, 60, 24–34. [Google Scholar] [CrossRef]
- Jenkins, T.L.; Little, D. Synthetic scaffolds for musculoskeletal tissue engineering: cellular responses to fiber parameters. NPJ Regen. Med. 2019, 4, 15. [Google Scholar] [CrossRef]
- Trappmann, B.; Gautrot, J.E.; Connelly, J.T.; Strange, D.G.T.; Li, Y.; Oyen, M.L.; Stuart, M.A.C.; Boehm, H.; Li, B.; Vogel, V.; et al. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater. 2012, 11, 642–649. [Google Scholar] [CrossRef]
- Wen, J.H.; Vincent, L.G.; Fuhrmann, A.; Choi, Y.S.; Hribar, K.C.; Taylor-Weiner, H.; Chen, S.; Engler, A.J. Interplay of Matrix Stiffness and Protein Tethering in Stem Cell Differentiation. Nat. Mater. 2014, 13, 979–987. [Google Scholar] [CrossRef]
- Xia, T.; Liu, W.; Yang, L.; Tingting, X.; Wanqian, L.; Li, Y. A review of gradient stiffness hydrogels used in tissue engineering and regenerative medicine. J. Biomed. Mater. Res. Part A 2017, 105, 1799–1812. [Google Scholar] [CrossRef]
- Caliari, S.R.; Perepelyuk, M.; Cosgrove, B.D.; Tsai, S.J.; Lee, G.Y.; Mauck, R.L.; Wells, R.G.; Burdick, J.A. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci. Rep. 2016, 6, 21387. [Google Scholar] [CrossRef]
- Chen, W.-L.; Cordero, R.; Tran, H.; Ober, C.K. 50th Anniversary Perspective: Polymer Brushes: Novel Surfaces for Future Materials. Macromolecules 2017, 50, 4089–4113. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Boil. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Guvendiren, M.; Burdick, J.A. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012, 3, 792. [Google Scholar] [CrossRef]
- Stowers, R.S.; Allen, S.C.; Suggs, L.J. Dynamic phototuning of 3D hydrogel stiffness. Proc. Natl. Acad. Sci. USA 2015, 112, 1953–1958. [Google Scholar] [CrossRef]
- Ventre, M.; Netti, P.A. Engineering Cell Instructive Materials to Control Cell Fate and Functions Through Material Cues and Surface Patterning. ACS Appl. Mater. Interfaces 2016, 8, 14896–14908. [Google Scholar] [CrossRef]
- Forti, F.L.; Goissis, G.; Plepis, A.M.G. Modifications on Collagen Structures Promoted by 1,4-Dioxane Improve Thermal and Biological Properties of Bovine Pericardium as a Biomaterial. J. Biomater. Appl. 2006, 20, 267–285. [Google Scholar] [CrossRef]
- Ganesan, V.V.; Dhanasekaran, M.; Thangavel, N.; Dhathathreyan, A. Elastic compliance of fibrillar assemblies in type I collagen. Biophys. Chem. 2018, 240, 15–24. [Google Scholar] [CrossRef]
- Kutys, M.L.; Doyle, A.D.; Yamada, K.M. Regulation of cell adhesion and migration by cell-derived matrices. Exp. Cell Res. 2013, 319, 2434–2439. [Google Scholar] [CrossRef]
- Petrie, R.J.; Koo, H.; Yamada, K.M. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 2014, 345, 1062–1065. [Google Scholar] [CrossRef]
- Munster, S.; Jawerth, L.M.; Leslie, B.A.; Weitz, J.I.; Fabry, B.; Weitz, D.A. Strain history dependence of the nonlinear stress response of fibrin and collagen networks. Proc. Natl. Acad. Sci. USA 2013, 110, 12197–12202. [Google Scholar] [CrossRef]
- Kubow, K.E.; VonCannon, S.K.; Horwitz, A.R. Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr. Biol. 2013, 23, 1607–1619. [Google Scholar] [CrossRef]
- Sun, M.; Chi, G.; Li, P.; Lv, S.; Xu, J.; Xu, Z.; Xia, Y.; Tan, Y.; Xu, J.; Li, L.; et al. Effects of Matrix Stiffness on the Morphology, Adhesion, Proliferation and Osteogenic Differentiation of Mesenchymal Stem Cells. Int. J. Med. Sci. 2018, 15, 257–268. [Google Scholar] [CrossRef]
- Anderson, H.J.; Sahoo, J.K.; Ulijn, R.V.; Dalby, M.J. Mesenchymal Stem Cell Fate: Applying Biomaterials for Control of Stem Cell Behavior. Front. Bioeng. Biotechnol. 2016, 4, 993. [Google Scholar] [CrossRef]
- Cimmino, C.; Rossano, L.; Netti, P.A.; Ventre, M. Spatio-Temporal Control of Cell Adhesion: Toward Programmable Platforms to Manipulate Cell Functions and Fate. Front. Bioeng. Biotechnol. 2018, 6, 6. [Google Scholar] [CrossRef]
- Ventre, M.; Natale, C.F.; Rianna, C.; Netti, P.A. Topographic cell instructive patterns to control cell adhesion, polarization and migration. J. R. Soc. Interface 2014, 11, 20140687. [Google Scholar] [CrossRef]
- Lamers, E.; Van Horssen, R.; Riet, J.T.; Van Delft, F.C.; Luttge, R.; Walboomers, X.F.; A Jansen, J. The influence of nanoscale topographical cues on initial osteoblast morphology and migration. Eur. Cell Mater. 2010, 20, 329–343. [Google Scholar] [CrossRef]
- Ventre, M.; Coppola, V.; Iannone, M.; Netti, P.A.; Tekko, I.; Larraneta, E.; Rodgers, A.M.; Scott, C.J.; Kissenpfennig, A.; Donnelly, R.F.; et al. Nanotechnologies for tissue engineering and regeneration. In Nanotechnologies in Preventive and Regenerative Medicine; Elsevier: Chester, UK, 2018; pp. 93–206. [Google Scholar]
- Kim, E.; Tae, G. Direct reprogramming and biomaterials for controlling cell fate. Biomater. Res. 2016, 20, 2183. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Dickinson, L.E.; Kusuma, S.; Gerecht, S. Reconstructing the Differentiation Niche of Embryonic Stem Cells Using Biomaterials. Macromol. Biosci. 2011, 11, 36–49. [Google Scholar] [CrossRef]
- Battista, S.; Guarnieri, D.; Borselli, C.; Zeppetelli, S.; Borzacchiello, A.; Mayol, L.; Gerbasio, D.; Keene, D.R.; Ambrosio, L.; Netti, P.A. The effect of matrix composition of 3D constructs on embryonic stem cell differentiation. Biomaterials 2005, 26, 6194–6207. [Google Scholar] [CrossRef]
- Ji, H.; Atchison, L.; Chen, Z.; Chakraborty, S.; Jung, Y.; Truskey, G.A.; Christoforou, N.; Leong, K.W. Transdifferentiation of Human Endothelial Progenitors into Smooth Muscle Cells. Biomaterials 2016, 85, 180–194. [Google Scholar] [CrossRef]
- Willerth, S.M.; Arendas, K.J.; Gottlieb, D.I.; Sakiyama-Elbert, S.E. Optimization of Fibrin Scaffolds for Differentiation of Murine Embryonic Stem Cells into Neural Lineage Cells. Biomaterials 2006, 27, 5990–6003. [Google Scholar] [CrossRef]
- Lin, C.-C.; Anseth, K.S. PEG Hydrogels for the Controlled Release of Biomolecules in Regenerative Medicine. Pharm. Res. 2009, 26, 631–643. [Google Scholar] [CrossRef]
- Ingram, D.A. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood 2004, 104, 2752–2760. [Google Scholar] [CrossRef]
- Schuldiner, M.; Yanuka, O.; Itskovitz-Eldor, J.; Melton, D.A.; Benvenisty, N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2000, 97, 11307–11312. [Google Scholar] [CrossRef]
- Beckstead, B.L.; Santosa, D.M.; Giachelli, C.M. Mimicking cell–cell interactions at the biomaterial–cell interface for control of stem cell differentiation. J. Biomed. Mater. Res. Part A 2006, 79, 94–103. [Google Scholar] [CrossRef]
- Tung, J.C.; Paige, S.L.; Ratner, B.D.; Murry, C.E.; Giachelli, C.M. Engineered Biomaterials Control Differentiation and Proliferation of Human-Embryonic-Stem-Cell-Derived Cardiomyocytes via Timed Notch Activation. Stem Cell Rep. 2014, 2, 271–281. [Google Scholar] [CrossRef]
- Brusatin, G.; Panciera, T.; Gandin, A.; Citron, A.; Piccolo, S. Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat. Mater. 2018, 17, 1063–1075. [Google Scholar] [CrossRef]
- Zhang, H.; Deo, M.; Thompson, R.C.; Uhler, M.D.; Turner, D.L. Negative regulation of Yap during neuronal differentiation. Dev. Biol. 2012, 361, 103–115. [Google Scholar] [CrossRef]
- Hou, P.; Li, Y.; Zhang, X.; Liu, C.; Guan, J.; Li, H.; Zhao, T.; Ye, J.; Yang, W.; Liu, K.; et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef]
- Cao, N.; Huang, Y.; Zheng, J.; Spencer, C.I.; Zhang, Y.; Fu, J.-D.; Nie, B.; Xie, M.; Zhang, M.; Wang, H.; et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 2016, 352, 1216–1220. [Google Scholar] [CrossRef]
- Guilak, F.; Cohen, D.M.; Estes, B.T.; Gimble, J.M.; Liedtke, W.; Chen, C.S. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 2009, 5, 17–26. [Google Scholar] [CrossRef]
- Engler, A.J.; Sweeney, H.L.; E Discher, D.; E Schwarzbauer, J. Extracellular matrix elasticity directs stem cell differentiation. J. Musculoskelet. neuronal interactions 2007, 7, 335. [Google Scholar]
- Choi, B.; Park, K.-S.; Kim, J.-H.; Ko, K.-W.; Kim, J.-S.; Han, D.K.; Lee, S.-H. Stiffness of Hydrogels Regulates Cellular Reprogramming Efficiency Through Mesenchymal-to-Epithelial Transition and Stemness Markers: Stiffness of Hydrogels Regulates Cellular Reprogramming Efficiency. Macromol. Biosci. 2016, 16, 199–206. [Google Scholar] [CrossRef]
- Christopherson, G.T.; Song, H.; Mao, H.-Q. The influence of fiber diameter of electrospun substrates on neural stem cell differentiation and proliferation. Biomaterials 2009, 30, 556–564. [Google Scholar] [CrossRef]
- Lim, S.H.; Mao, H.-Q. Electrospun scaffolds for stem cell engineering. Adv. Drug Deliv. Rev. 2009, 61, 1084–1096. [Google Scholar] [CrossRef]
- Levenberg, S.; Huang, N.F.; Lavik, E.; Rogers, A.B.; Itskovitz-Eldor, J.; Langer, R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc. Natl. Acad. Sci. USA 2003, 100, 12741–12746. [Google Scholar] [CrossRef]
- Kulangara, K.; Adler, A.F.; Wang, H.; Chellappan, M.; Hammett, E.; Yasuda, R.; Leong, K.W. The Effect of Substrate Topography on Direct Reprogramming of Fibroblasts to Induced Neurons. Biomaterials 2014, 35, 5327–5336. [Google Scholar] [CrossRef]
- Frey, M.T.; Tsai, I.Y.; Russell, T.P.; Hanks, S.K.; Wang, Y.-L. Cellular Responses to Substrate Topography: Role of Myosin II and Focal Adhesion Kinase. Biophys. J. 2006, 90, 3774–3782. [Google Scholar] [CrossRef]
- Fu, K.; Griebenow, K.; Hsieh, L.; Klibanov, A.M.; Langer, R. FTIR characterization of the secondary structure of proteins encapsulated within PLGA microspheres. J. Control Release 1999, 58, 357–366. [Google Scholar] [CrossRef]
- Moribe, K.; Nomizu, N.; Izukura, S.; Yamamoto, K.; Tozuka, Y.; Sakurai, M.; Ishida, A.; Nishida, H.; Miyazaki, M. Physicochemical, Morphological and Therapeutic Evaluation of Agarose Hydrogel Particles as a Reservoir for Basic Fibroblast Growth Factor. Pharm. Dev. Technol. 2008, 13, 541–547. [Google Scholar] [CrossRef]
- Bratt-Leal, A.M.; Carpenedo, R.L.; Ungrin, M.D.; Zandstra, P.W.; McDevitt, T.C. Incorporation of biomaterials in multicellular aggregates modulates pluripotent stem cell differentiation. Biomaterials 2011, 32, 48–56. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Medvedev, S.P.; Shevchenko, A.I.; Zakian, S.M. Induced Pluripotent Stem Cells: Problems and Advantages when Applying them in Regenerative Medicine. Acta Nat. 2010, 2, 18–28. [Google Scholar] [CrossRef]
- Kanninen, L.K.; Harjumäki, R.; Peltoniemi, P.; Bogacheva, M.S.; Salmi, T.; Porola, P.; Niklander, J.; Smutný, T.; Urtti, A.; Yliperttula, M.L.; et al. Laminin-511 and laminin-521-based matrices for efficient hepatic specification of human pluripotent stem cells. Biomaterials 2016, 103, 86–100. [Google Scholar] [CrossRef]
- Nagy, Á.G.; Kámán, J.; Horváth, R.; Bonyár, A. Spring constant and sensitivity calibration of FluidFM micropipette cantilevers for force spectroscopy measurements. Sci. Rep. 2019, 9, 10287. [Google Scholar] [CrossRef]
- Brouwer, T.B.; Kaczmarczyk, A.; Pham, C.; Van Noort, J. Unraveling DNA Organization with Single-Molecule Force Spectroscopy Using Magnetic Tweezers. Methods Mol. Biol. 2018, 1837, 317–349. [Google Scholar]
- Sumbul, F.; Rico, F. Single-Molecule Force Spectroscopy: Experiments, Analysis, and Simulations. Methods Mol. Biol. 2019, 1886, 163–189. [Google Scholar]
- Li, F.; Redick, S.D.; Erickson, H.P.; Moy, V.T. Force Measurements of the α5β1 Integrin–Fibronectin Interaction. Biophys. J. 2003, 84, 1252–1262. [Google Scholar] [CrossRef]
- Taubenberger, A.V.; Quent, V.M.; Thibaudeau, L.; Clements, J.A.; Hutmacher, D.W. Delineating breast cancer cell interactions with engineered bone microenvironments: Delineating breast cancer cell-bone interactions. J. Bone Miner. Res. 2013, 28, 1399–1411. [Google Scholar] [CrossRef]
- Schaap-Oziemlak, A.M.; Kühn, P.T.; Van Kooten, T.G.; Van Rijn, P. Biomaterial–stem cell interactions and their impact on stem cell response. RSC Adv. 2014, 4, 53307–53320. [Google Scholar] [CrossRef]
- Kenry; Lee, W.C.; Loh, K.P.; Lim, C.T. When stem cells meet graphene: Opportunities and challenges in regenerative medicine. Biomaterials 2018, 155, 236–250. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, M.; Zhao, J.; Chai, R.; Kang, J. Looking into the Future: Toward Advanced 3D Biomaterials for Stem-Cell-Based Regenerative Medicine. Adv. Mater. 2018, 30, 1705388. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Mol. Cell Boil. 2017, 18, 717–727. [Google Scholar] [CrossRef]
- Dalby, M.J.; Biggs, M.J.; Gadegaard, N.; Kalna, G.; Wilkinson, C.D.; Curtis, A.S. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J. Cell. Biochem. 2007, 100, 326–338. [Google Scholar] [CrossRef]
- Murphy, W.L.; McDevitt, T.C.; Engler, A.J. Materials as stem cell regulators. Nat. Mater. 2014, 13, 547–557. [Google Scholar] [CrossRef]
- Roberts, J.N.; Sahoo, J.K.; McNamara, L.E.; Burgess, K.V.; Yang, J.; Alakpa, E.V.; Anderson, H.J.; Hay, J.; Turner, L.-A.; Yarwood, S.J.; et al. Dynamic Surfaces for the Study of Mesenchymal Stem Cell Growth through Adhesion Regulation. ACS Nano 2016, 10, 6667–6679. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Y.; Ni, X.; Li, N.; Zhang, B.; Fang, X. Enhancement of the nonamyloidogenic pathway by exogenous NGF in an Alzheimer transgenic mouse model. Neuropeptides 2014, 48, 233–238. [Google Scholar] [CrossRef]
- Young, J.L.; Engler, A.J. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials 2011, 32, 1002–1009. [Google Scholar] [CrossRef]
- Hay, J.J.; Rodrigo-Navarro, A.; Petaroudi, M.; Bryksin, A.V.; García, A.J.; Barker, T.H.; Dalby, M.J.; Salmeron-Sanchez, M. Bacteria-Based Materials for Stem Cell Engineering. Adv. Mater. 2018, 30, 1804310. [Google Scholar] [CrossRef]
- Applegate, M.B.; Coburn, J.; Partlow, B.P.; Moreau, J.E.; Mondia, J.P.; Marelli, B.; Kaplan, D.L.; Omenetto, F.G. Laser-based three-dimensional multiscale micropatterning of biocompatible hydrogels for customized tissue engineering scaffolds. Proc. Natl. Acad. Sci. USA 2015, 112, 12052–12057. [Google Scholar] [CrossRef]
- Tong, M.H.; Huang, N.; Zhang, W.; Zhou, Z.L.; Ngan, A.H.W.; Du, Y.; Chan, B.P. Multiphoton photochemical crosslinking-based fabrication of protein micropatterns with controllable mechanical properties for single cell traction force measurements. Sci. Rep. 2016, 6, 20063. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
d’Angelo, M.; Benedetti, E.; Tupone, M.G.; Catanesi, M.; Castelli, V.; Antonosante, A.; Cimini, A. The Role of Stiffness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration. Cells 2019, 8, 1036. https://doi.org/10.3390/cells8091036
d’Angelo M, Benedetti E, Tupone MG, Catanesi M, Castelli V, Antonosante A, Cimini A. The Role of Stiffness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration. Cells. 2019; 8(9):1036. https://doi.org/10.3390/cells8091036
Chicago/Turabian Styled’Angelo, Michele, Elisabetta Benedetti, Maria Grazia Tupone, Mariano Catanesi, Vanessa Castelli, Andrea Antonosante, and Annamaria Cimini. 2019. "The Role of Stiffness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration" Cells 8, no. 9: 1036. https://doi.org/10.3390/cells8091036
APA Styled’Angelo, M., Benedetti, E., Tupone, M. G., Catanesi, M., Castelli, V., Antonosante, A., & Cimini, A. (2019). The Role of Stiffness in Cell Reprogramming: A Potential Role for Biomaterials in Inducing Tissue Regeneration. Cells, 8(9), 1036. https://doi.org/10.3390/cells8091036




