Proteomic and Transcriptomic Profiling Identifies Early Developmentally Regulated Proteins in Dictyostelium Discoideum
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Protein Level Changes in Response to cAMP Pulsing Using Proteomics in Dictyostelium
2.2. Functional Annotation Clustering of Identified Proteins
2.3. Multiple Alignment of Proteins with Unknown Function
2.4. Transcriptomic Profiling Analysis in Response to cAMP in Dictyostelium
3. Results and Discussion
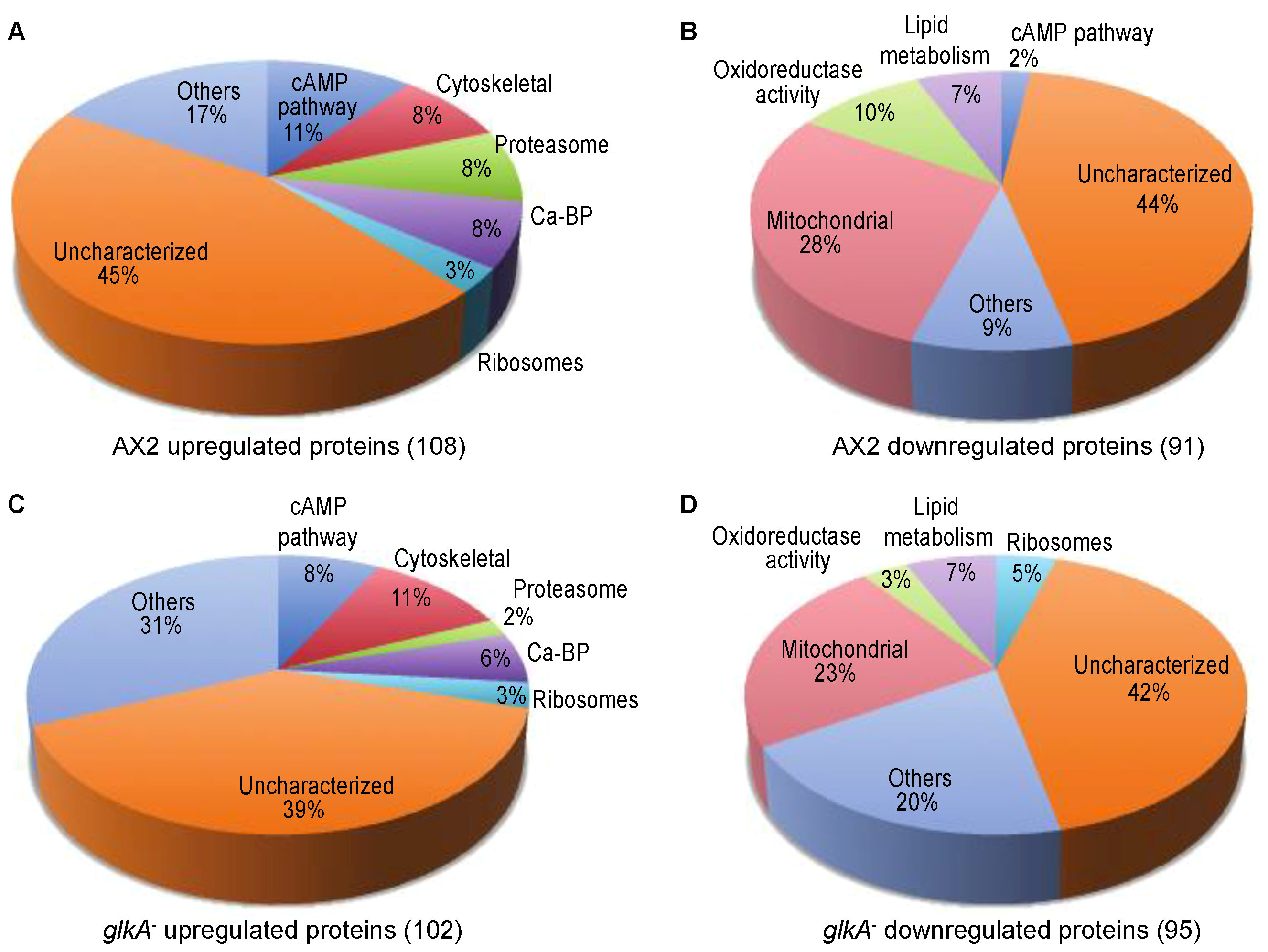
3.1. Identification of Protein Level Changes in Response to cAMP Pulsing in Dictyostelium Discoideum
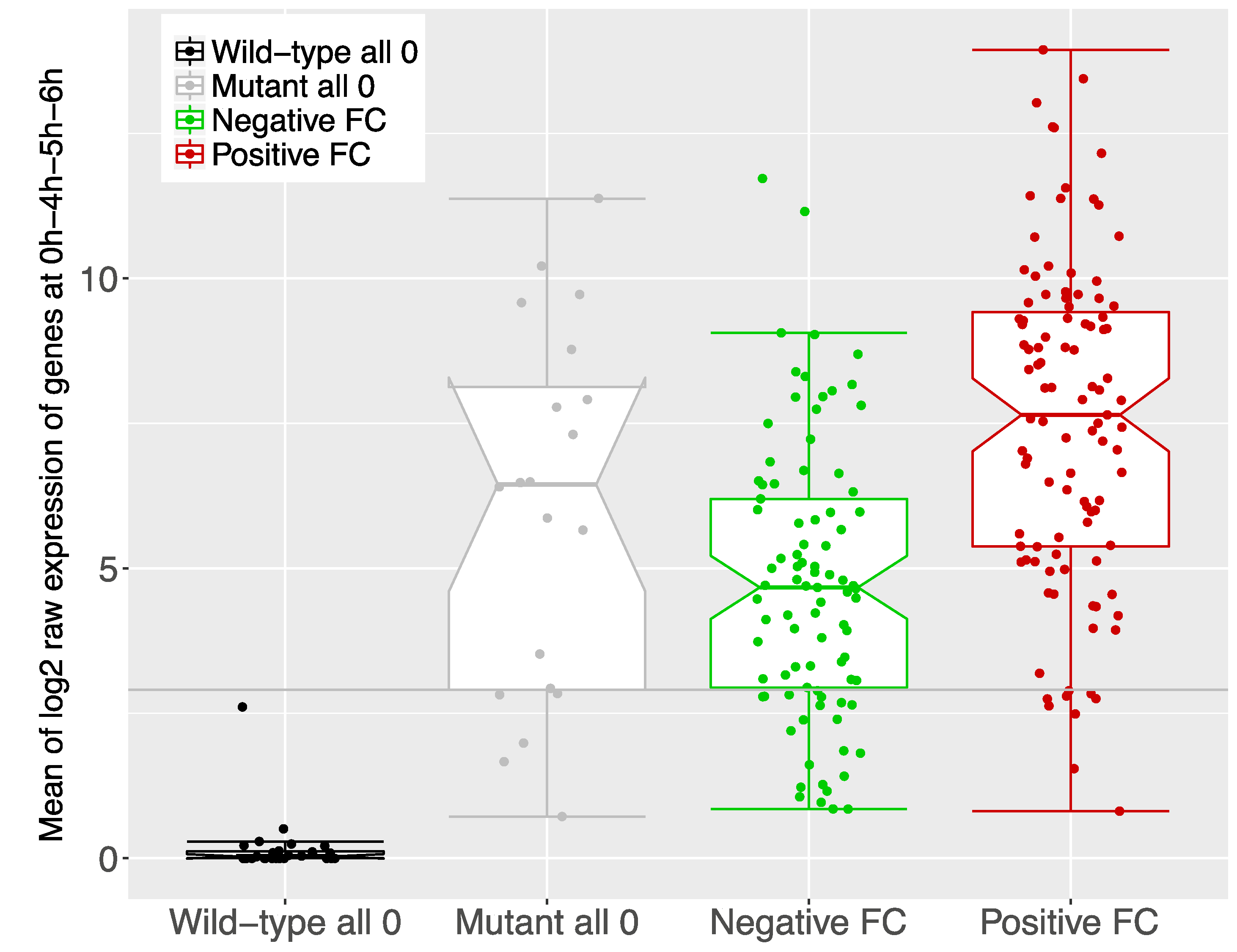
3.2. The Comparison of Proteomics and Transcriptomics Data Yielded a Significant Number of Concordant Genes
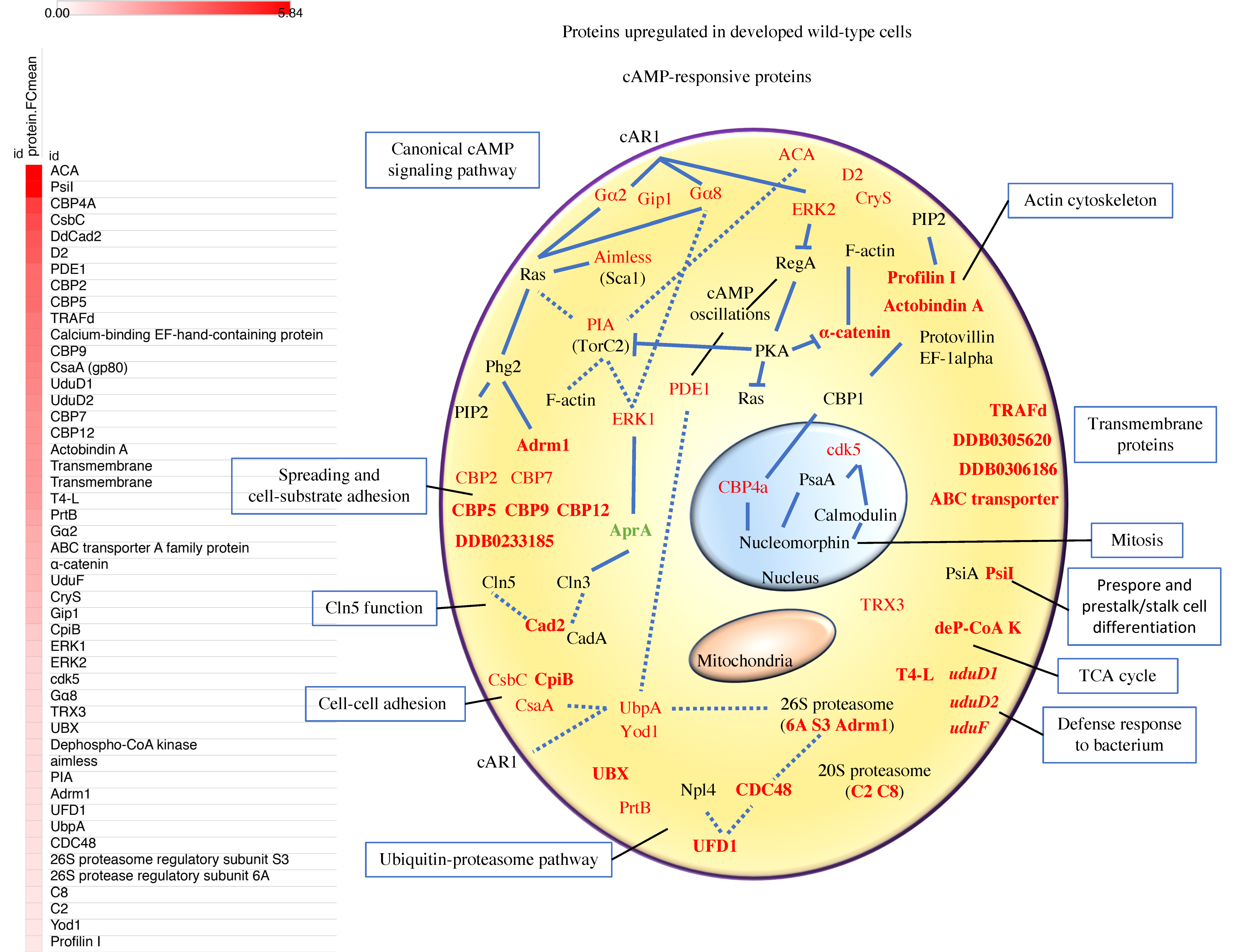
3.3. Proteins of the Canonical cAMP-Pathway, the Ubiquitin-Proteasome Pathway, Calcium Binding, and Cell Adhesion are Among the Most Abundant c-AMP-Responsive Proteins Upregulated in Early Development
3.3.1. cAMP-Signaling Pathway
3.3.2. Calcium-Binding Proteins (CBPs)
3.3.3. Adhesion Proteins and Cln5 Function
3.3.4. Actin Cytoskeleton
3.3.5. The Ubiquitin-Proteasome Pathway
3.3.6. Proteins with Unknown Functions
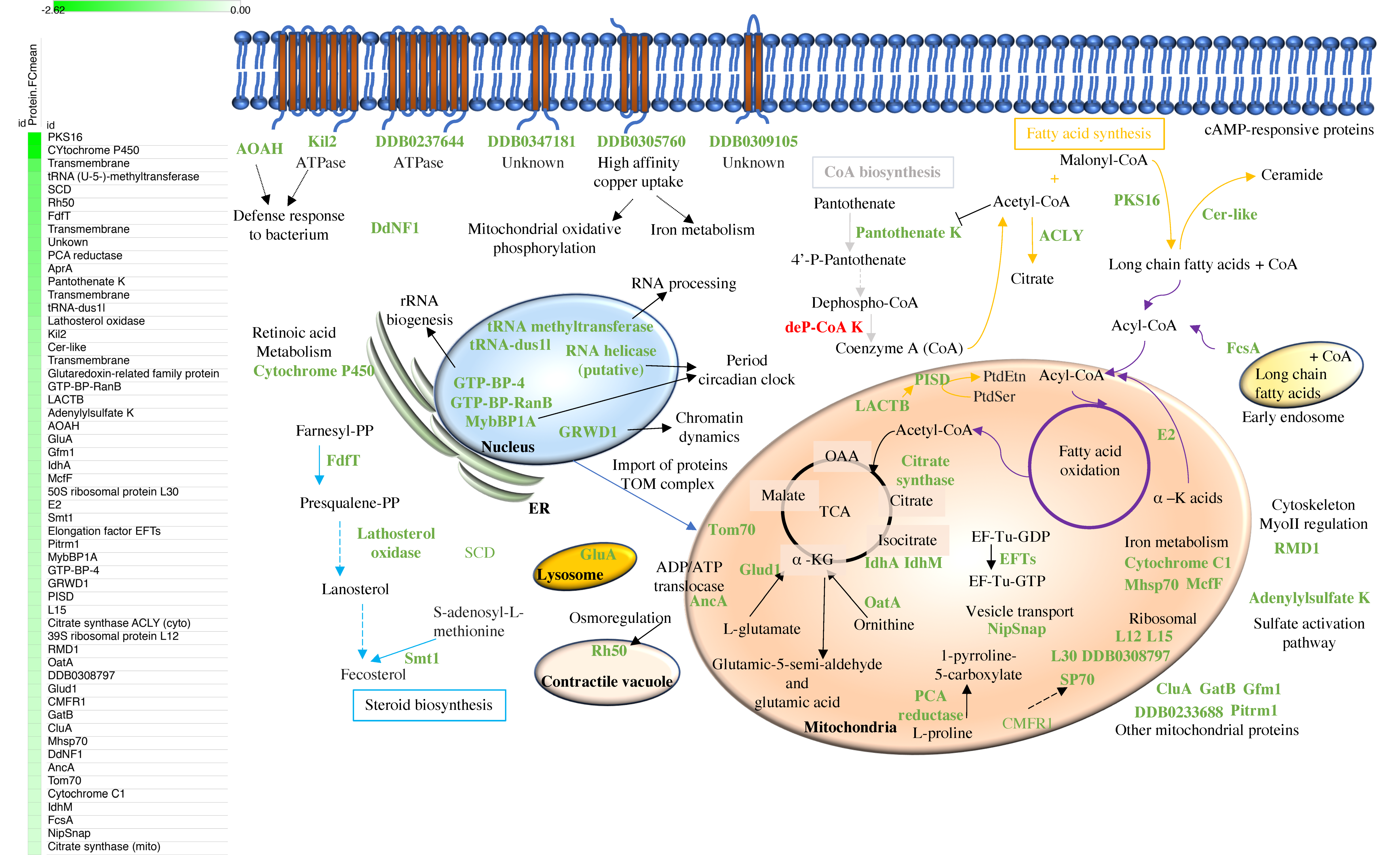
3.4. Mitochondrial Proteins Related to Metabolism, Proteins Involved in Fatty Acid, Coenzyme A, and Steroids Synthesis are Among the Most Abundant Proteins Downregulated in Early Development
3.4.1. Fatty Acid Metabolism
3.4.2. Coenzyme A (CoA) Biosynthesis
3.4.3. Steroid Biosynthesis
3.4.4. RNA Processing
3.4.5. Mitochondrial Proteins
3.4.6. Defense Response to Bacterium
3.4.7. Uncharacterized Proteins
3.4.8. Unknown Proteins
3.5. Proteomics in glkA-null Cells
3.6. Identification of GlkA-Dependent Candidates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Driessche, N.; Shaw, C.; Katoh, M.; Morio, T.; Sucgang, R.; Ibarra, M. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 2002, 129, 1543–1545. [Google Scholar]
- Garcia, G.L.; Parent, C.A. Signal relay during chemotaxis. J. Microsc. 2008, 231, 529–534. [Google Scholar]
- Cai, H.; Huang, C.H.; Devreotes, P.N.; Iijima, M. Analysis of chemotaxis in Dictyostelium. Methods Mol. Biol. 2012, 757, 451–468. [Google Scholar]
- Nichols, J.M.; Veltman, D.; Kay, R.R. Chemotaxis of a model organism: Progress with Dictyostelium. Curr. Opin. Cell Biol. 2015, 36, 7–12. [Google Scholar]
- Loomis, W.F. Dictyostelium Discoideum: A Developmental System; Academic Press: New York, NY, USA, 1975. [Google Scholar]
- Kessin, R.H. Dictyostelium—Evolution, Cell Biology and the Development of Multicellularity; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Trepat, X.; Chen, Z.; Jacobson, K. Cell migration. Compr. Physiol. 2012, 2, 2369–2392. [Google Scholar]
- Worbs, T.; Hammerschmidt, S.I.; Förster, R. Dendritic cell migration in health and disease. Nat. Rev. Immunol. 2017, 17, 30–48. [Google Scholar]
- Shaw, T.J.; Martin, P. Wound repair: A showcase for cell plasticity and migration. Curr. Opin. Cell Biol. 2016, 42, 29–37. [Google Scholar]
- Goichberg, P. Current Understanding of the Pathways Involved in Adult Stem and Progenitor Cell Migration for Tissue Homeostasis and Repair. Stem Cell Rev. 2016, 12, 421–437. [Google Scholar]
- Yumura, S.; Mori, H.; Fukui, Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol. 1984, 99, 894–899. [Google Scholar]
- Condeelis, J.; Bresnick, A.; Demma, M.; Dharmawardhane, S.; Eddy, R.; Hall, A.L.; Sauterer, R.; Warren, V. Mechanisms of amoeboid chemotaxis: An evaluation of the cortical expansion model. Dev. Genet. 1990, 11, 333–340. [Google Scholar]
- Cai, H.; Devreotes, P.N. Moving in the right direction: How eukaryotic cells migrate along chemical gradients. Semin. Cell Dev. Biol. 2011, 22, 834–841. [Google Scholar]
- McCann, C.P.; Parent, C.A. Eukaryotic chemotaxis. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 236–240. [Google Scholar]
- Tang, M.; Wang, M.; Shi, C.; Iglesias, P.A.; Devreotes, P.N.; Huang, C.H. Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis. Nat. Commun. 2014, 5, 5175. [Google Scholar] [CrossRef]
- Brock, D.A.; Gomer, R.H. A secreted factor represses cell proliferation in Dictyostelium. Development 2005, 132, 4553–4562. [Google Scholar] [CrossRef]
- Bakthavatsalam, D.; Brock, D.A.; Nikravan, N.N.; Houston, K.D.; Hatton, R.D.; Gomer, R.H. The secreted Dictyostelium protein CfaD is a chalone. J. Cell Sci. 2008, 121, 2473–2480. [Google Scholar] [CrossRef]
- Clarke, M.; Gomer, R.H. PSF and CMF, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia 1995, 51, 1124–1134. [Google Scholar]
- Tang, L.; Ammann, R.; Gao, T.; Gomer, R.H. A cell number-counting factor regulates group size in Dictyostelium by differentially modulating cAMP-induced cAMP and cGMP pulse sizes. J. Biol. Chem. 2001, 276, 27663–27669. [Google Scholar]
- Ray, S.; Chen, Y.; Ayoung, J.; Hanna, R.; Brazill, D. Phospholipase D controls Dictyostelium development by regulating G protein signaling. Cell Signal. 2011, 23, 335–343. [Google Scholar]
- Weijer, C.J. Dictyostelium morphogenesis. Curr. Opin. Genet. Dev. 2004, 14, 392–398. [Google Scholar]
- Maeda, Y. Regulation of Growth and Differentiation in Dictyostelium. Int. Rev. Cytol. 2005, 244, 287–332. [Google Scholar]
- Orlow, S.J.; Shapiro, R.I.; Franke, J.; Kessin, R.H. The extracellular cyclic nucleotide phosphodiesterase of Dictyostelium discoideum. Purification and characterization. J. Biol. Chem. 1981, 256, 7620–7627. [Google Scholar]
- Franke, J.; Kessin, R.H. The cyclic nucleotide phosphodiesterases of Dictyostelium discoideum: Molecular genetics and biochemistry. Cell Signal. 1992, 4, 471–478. [Google Scholar]
- Sucgang, R.; Weijer, C.J.; Siegert, F.; Franke, J.; Kessin, R.H. Null mutations of the Dictyostelium cyclic nucleotide phosphodiesterase gene block chemotactic cell movement in developing aggregates. Dev. Biol. 1997, 192, 181–192. [Google Scholar]
- Gerisch, G.; Wick, U. Intracellular oscillations and release of cyclic AMP from Dictyostelium cells. Biochem. Biophys. Res. Commun. 1975, 65, 364–370. [Google Scholar]
- Maeda, M.; Sakamoto, H.; Iranfar, N.; Fuller, D.T.; Ogihara, S.; Morio, T.; Urushihara, H.; Tanaka, Y.; Loomis, W.F. Changing patterns of gene expression in Dictyostelium prestalk cell subtypes recognized by in situ hybridization with genes from microarray analyses. Euk. Cell 2003, 2, 627–637. [Google Scholar]
- Iranfar, N.; Fuller, D.; Loomis, W.F. Genome-Wide Expression Analyses of Gene Regulation during Early Development of Dictyostelium discoideum. Eukaryot. Cell 2003, 2, 664–670. [Google Scholar]
- Iranfar, N.; Fuller, D.; Loomis, W.F. Transcriptional regulation of post-aggregation genes in Dictyostelium by a feed-forward loop involving GBF and LagC. Dev. Biol. 2006, 290, 460–469. [Google Scholar]
- Escalante, R.; Vicente, J.J. Dictyostelium discoideum: A model system for differentiation and patterning. Int. J. Dev. Biol. 2000, 44, 819–835. [Google Scholar]
- Kay, R.R. Chemotaxis and cell differentiation in Dictyostelium. Curr. Opin. Microbiol. 2002, 5, 575–579. [Google Scholar]
- Loomis, W.F. Genetic control of morphogenesis in Dictyostelium. Dev. Biol. 2015, 402, 146–161. [Google Scholar] [CrossRef]
- Anjard, C.; Loomis, W.F. Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. USA 2005, 102, 7607–7611. [Google Scholar]
- Anjard, C.; Loomis, W.F. GABA induces terminal differentiation of Dictyostelium through a GABAB receptor. Development 2006, 133, 2253–2261. [Google Scholar]
- Anjard, C.; Loomis, W.F. Cytokinins induce sporulation in Dictyostelium. Development 2008, 135, 819–827. [Google Scholar] [CrossRef]
- Eichinger, L.; Pachebat, J.A.; Glöckner, G.; Rajandream, M.A.; Sucgang, R.; Berriman, M. The genome of the social amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar]
- Morio, T.; Urushihara, H.; Saito, T.; Ugawa, Y.; Mizuno, H.; Yoshida, M. The Dictyostelium developmental cDNA project: Generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res. 1998, 5, 335–340. [Google Scholar]
- Stajdohar, M.; Rosengarten, R.D.; Kokosar, J.; Jeran, L.; Blenkus, D.; Shaulsky, G.; Zupan, B. dictyExpress: A web-based platform for sequence data management and analytics in Dictyostelium and beyond. BMC Bioinform. 2017, 18, 291. [Google Scholar]
- Saxe, L.; Johnson, R.; Devreotes, N.; Kimmel, R. Multiple genes for cell surface cAMP receptors in Dictyostelium discoideum. Dev. Genet. 1991, 12, 6–13. [Google Scholar]
- Pitt, S.; Milona, N.; Borleis, J.; Lin, C.; Reed, R.; Devreotes, N. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell 1992, 69, 305–315. [Google Scholar]
- Ginsburg, T.; Gollop, R.; Yu, Y.; Louis, M.; Saxe, L.; Kimmel, R. The regulation of Dictyostelium development by transmembrane signalling. J. Eukaryot. Microbiol. 1995, 42, 200–205. [Google Scholar]
- Shaulsky, G.; Escalante, R.; Loomis, F. Developmental signal transduction pathways uncovered by genetic suppressors. Proc. Natl. Acad. Sci. USA 1996, 93, 15260–15265. [Google Scholar]
- Galardi-Castilla, M.; Garciandia, A.; Suarez, T.; Sastre, L. The Dictyostelium discoideum acaA gene is transcribed from alternative promoters during aggregation and multicellular development. PLoS ONE 2010, 5, e13286. [Google Scholar]
- Booth, E.O.; Van Driessche, N.; Zhuchenko, O.; Kuspa, A.; Shaulsky, G. Microarray phenotyping in Dictyostelium reveals a regulon of chemotaxis genes. Bioinformatics 2005, 21, 4371–4377. [Google Scholar]
- Lacal, J.; Shen, Z.; Baumgardner, K.; Wei, J.; Briggs, S.P.; Firtel, R.A. The Dictyostelium GSK3 kinase GlkA coordinates signal relay and chemotaxis in response to growth conditions. Dev. Biol. 2018, 435, 56–72. [Google Scholar]
- Coulson, R.; Bowman, R.H. Excretion and degradation of exogenous adenosine 3′,5′–monophosphate by isolated perfused rat kidney. Life Sci. 1974, 14, 545–566. [Google Scholar]
- Constantin, J.; Suzuki-Kemmelmeier, F.; Yamamoto, N.S.; Bracht, A. Production, uptake, and metabolic effects of cyclic AMP in the bivascularly perfused rat liver. Biochem. Pharmacol. 1997, 54, 1115–1125. [Google Scholar]
- Strouch, M.B.; Jackson, E.K.; Mi, Z.; Metes, N.A.; Carey, G.B. Extracellular cyclic AMP-adenosine pathway in isolated adipocytes and adipose tissue. Obes. Res. 2005, 13, 974–981. [Google Scholar]
- Pilkis, S.J.; Claus, T.H.; Johnson, R.A.; Park, C.R. Hormonal control of cyclic 3′:5′-AMP levels and gluconeogenesis in isolated hepatocytes from fed rats. Biol. Chem. 1975, 250, 6328–6336. [Google Scholar]
- Ardaillou, N.; Placier, S.; Wahbe, F.; Ronco, P.; Ardaillou, R. Release of cyclic nucleotides from the apical and basolateral poles of cultured human glomerular epithelial cells. Exp. Nephrol. 1993, 1, 253–260. [Google Scholar]
- Boumendil-Podevin, E.F.; Podevin, R.A. Transport and metabolism of adenosine 3′:5′-monophosphate and N6, O2′–dibutyryl adenosine 3′:5′-monophosphate by isolated renal tubules. J. Biol. Chem. 1977, 252, 6675–6681. [Google Scholar]
- Butcher, R.W.; Baird, C.E. Effects of prostaglandins on adenosine 3′,5′-monophosphate levels in fat and other tissues. J. Biol. Chem. 1968, 243, 1713–1717. [Google Scholar]
- Exton, J.H.; Lewis, S.B.; Ho, R.J.; Robison, G.A.; Park, C.R. The role of cyclic AMP in the interaction of glucagon and insulin in the control of liver metabolism. Ann. N. Y. Acad. Sci. 1971, 185, 85–100. [Google Scholar]
- Godinho, R.O.; Duarte, T.; Pacini, E.S. New perspectives in signaling mediated by receptors coupled to stimulatory G protein: The emerging significance of cAMP efflux and extracellular cAMP-adenosine pathway. Front. Pharmacol. 2015, 6, 58. [Google Scholar] [CrossRef]
- Rosengarten, R.D.; Santhanam, B.; Fuller, D.; Katoh-Kurasawa, M.; Loomis, W.F.; Zupan, B.; Shaulsky, G. Leaps and lulls in the developmental transcriptome of Dictyostelium discoideum. BMC Genom. 2015, 16, 294. [Google Scholar]
- Smyth, G.K. Limma: Linear Models for Microarray Data; Springer: Berlin, Germany, 2005; pp. 397–420. [Google Scholar]
- Pupillo, M.; Insall, R.; Pitt, G.S.; Devreotes, P.N. Multiple cyclic AMP receptors are linked to adenylyl cyclase in Dictyostelium. Mol. Biol. Cell 1992, 3, 1229–1234. [Google Scholar]
- Loomis, W.F. Genetic analysis of the gene for N-acetylglucosaminidase in Dictyostelium discoideum. Genetics 1978, 88, 277–284. [Google Scholar]
- Kae, H.; Kortholt, A.; Rehmann, H.; Insall, R.H.; Van Haastert, P.J.; Spiegelman, G.B.; Weeks, G. Cyclic AMP signalling in Dictyostelium: G-proteins activate separate Ras pathways using specific RasGEFs. EMBO Rep. 2007, 8, 477–482. [Google Scholar]
- Lee, S.; Parent, C.A.; Insall, R.; Firtel, R.A. A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell 1999, 10, 2829–2845. [Google Scholar]
- Charest, P.G.; Firtel, R.A. “TORCing” neutrophil chemotaxis. Dev. Cell 2010, 19, 795–796. [Google Scholar] [CrossRef]
- Gaskins, C.; Maeda, M.; Firtel, R.A. Identification and functional analysis of a developmentally regulated extracellular signal-regulated kinase gene in Dictyostelium discoideum. Mol. Cell. Biol. 1994, 14, 6996–7012. [Google Scholar]
- Rijal, R.; Consalvo, K.M.; Lindsey, C.K.; Gomer, R.H. An endogenous chemorepellent directs cell movement by inhibiting pseudopods at one side of cells. Mol. Biol. Cell 2019, 30, 242–255. [Google Scholar] [CrossRef]
- Segall, J.E.; Kuspa, A.; Shaulsky, G.; Ecke, M.; Maeda, M.; Gaskins, C.; Firtel, R.A.; Loomis, W.F. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J. Cell Biol. 1995, 128, 405–413. [Google Scholar]
- Schwebs, D.J.; Pan, M.; Adhikari, N.; Kuburich, N.A.; Jin, T.; Hadwiger, J.A. Dictyostelium Erk2 is an atypical MAPK required for chemotaxis. Cell Signal. 2018, 46, 154–165. [Google Scholar]
- Maeda, M.; Firtel, R.A. Activation of the mitogen-activated protein kinase ERK2 by the chemoattractant folic acid in Dictyostelium. J. Biol. Chem. 1997, 272, 23690–23695. [Google Scholar]
- Laub, T.; Loomis, F. A molecular network that produces spontaneous oscillations in excitable cells of Dictyostelium. Mol. Biol. Cell 1998, 9, 3521–3532. [Google Scholar]
- Maeda, M.; Lu, S.; Shaulsky, G.; Miyazaki, Y.; Kuwayama, H.; Tanaka, Y.; Kuspa, A.; Loomis, F. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science 2004, 304, 875–878. [Google Scholar]
- Lacombe, L.; Podgorski, J.; Franke, J.; Kessin, H. Molecular cloning and developmental expression of the cyclic nucleotide phosphodiesterase gene of Dictyostelium discoideum. J. Biol. Chem. 1986, 261, 16811–16817. [Google Scholar]
- Masaki, N.; Fujimoto, K.; Honda-Kitahara, M.; Hada, E.; Sawai, S. Robustness of self-organizing chemoattractant field arising from precise pulse induction of its breakdown enzyme: A single-cell level analysis of PDE expression in Dictyostelium. Biophys. J. 2013, 104, 1191–1202. [Google Scholar] [CrossRef]
- Chen, Y.; Long, Y.; Devreotes, N. A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes. Dev. 1997, 11, 3218–3231. [Google Scholar]
- Kamimura, Y.; Miyanaga, Y.; Ueda, M. Heterotrimeric G-protein shuttling via Gip1 extends the dynamic range of eukaryotic chemotaxis. Proc. Natl. Acad. Sci. USA 2016, 113, 4356–4361. [Google Scholar] [CrossRef]
- Baumgardner, K.; Lin, C.; Firtel, R.A.; Lacal, J. Phosphodiesterase PdeD, dynacortin, and a Kelch repeat-containing protein are direct GSK3 substrates in Dictyostelium that contribute to chemotaxis towards cAMP. Environ. Microbiol. 2018, 20, 1888–1903. [Google Scholar] [CrossRef]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar]
- Lusche, D.F.; Wessels, D.; Soll, D.R. The effects of extracellular calcium on motility, pseudopod and uropod formation, chemotaxis, and the cortical localization of myosin II in Dictyostelium discoideum. Cell Motil. Cytoskelet. 2009, 66, 567–587. [Google Scholar] [CrossRef]
- Siu, C.H.; Sriskanthadevan, S.; Wang, J.; Hou, L.; Chen, G.; Xu, X.; Thomson, A.; Yang, C. Regulation of spatiotemporal expression of cell-cell adhesion molecules during development of Dictyostelium discoideum. Dev. Growth Differ. 2011, 53, 518–527. [Google Scholar] [CrossRef]
- O’Day, D.H.; Poloz, Y.; Myre, M.A. Differentiation inducing factor-1 (DIF-1) induces gene and protein expression of the Dictyostelium nuclear calmodulin-binding protein nucleomorphin. Cell Signal. 2009, 21, 317–323. [Google Scholar] [CrossRef]
- Dharamsi, A.; Tessarolo, D.; Coukell, B.; Pun, J. CBP1 associates with the Dictyostelium cytoskeleton and is important for normal cell aggregation under certain developmental conditions. Exp. Cell Res. 2000, 258, 298–309. [Google Scholar]
- Dorywalska, M.; Coukell, B.; Dharamsi, A. Characterization and hetereologous expression of cDNAs encoding two novel closely related Ca(2+)-binding proteins in Dictyostelium discoideum. Biochim. Biophys. Acta 2000, 1496, 356–361. [Google Scholar]
- Catalano, A.; O’Day, D.H. Rad53 homologue forkhead-associated kinase A (FhkA) and Ca2+-binding protein 4a (CBP4a) are nucleolar proteins that differentially redistribute during mitosis in Dictyostelium. Cell Div. 2013, 8, 4. [Google Scholar] [CrossRef]
- Myre MAO’Day, D.H. Dictyostelium calcium-binding protein 4a interacts with nucleomorphin, a BRCT-domain protein that regulates nuclear number. Biochem. Biophys. Res. Commun. 2004, 322, 665–671. [Google Scholar]
- O’Day, D.H. Proteins of the Nucleolus of Dictyostelium discoideum: Nucleolar Compartmentalization, Targeting Sequences, Protein Translocations and Binding Partners. Cells 2019, 8, 167. [Google Scholar] [CrossRef]
- Bhutani, N.; Venkatraman, P.; Goldberg, A.L. Puromycin-sensitive aminopeptidase is the major peptidaseresponsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 2007, 26, 1385–1396. [Google Scholar]
- Finck BNKelly, D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Investig. 2006, 116, 615–622. [Google Scholar]
- Uldry, M.; Yang, W.; St-Pierre, J.; Lin, J.; Seale, P.; Spiegelman, B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006, 3, 333–341. [Google Scholar]
- Miura, S.; Kawanaka, K.; Kai, Y.; Tamura, M.; Goto, M.; Shiuchi, T.; Minokoshi, Y.; Ezaki, O. An increase in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to exercise is mediated by beta-adrenergic receptor activation. Endocrinology 2007, 148, 3441–3448. [Google Scholar]
- Hu, Y.; Pan, S.; Zhang, H.T. Interaction of Cdk5 and cAMP/PKA Signaling in the Mediation of Neuropsychiatric and Neurodegenerative Diseases. Adv. Neurobiol. 2017, 17, 45–61. [Google Scholar]
- Sakamoto, H.; Nishio, K.; Tomisako, M.; Kuwayama, H.; Tanaka, Y.; Suetake, I.; Tajima, S.; Ogihara, S.; Coukell, B.; Maeda, M. Identification and characterization of novel calcium-binding proteins of Dictyostelium and their spatial expression patterns during development. Dev. Growth Differ. 2003, 45, 507–514. [Google Scholar]
- Huber, R.J.; Myre, M.A.; Cotman, S.L. Aberrant adhesion impacts early development in a Dictyostelium model for juvenile neuronal ceroid lipofuscinosis. Cell Adhes. Migr. 2017, 11, 399–418. [Google Scholar] [CrossRef]
- André, B.; Noegel, A.A.; Schleicher, M. Dictyostelium discoideum contains a family of calmodulin-related EF-hand proteins that are developmentally regulated. FEBS Lett. 1996, 382, 198–202. [Google Scholar]
- Chisholm, R.L.; Firtel, R.A. Insights into morphogenesis from a simple developmental system. Nat. Rev. Mol. Cell Biol. 2004, 5, 531–541. [Google Scholar] [CrossRef]
- Malchow, D.; Mutzel, R.; Schlatterer, C. On the role of calcium during chemotactic signalling and differentiation of the cellular slime mould Dictyostelium discoideum. Int. J. Dev. Biol. 1996, 40, 135–139. [Google Scholar]
- Yumura, S.; Furuya, K.; Takeuchi, I. Intracellular free calcium responses during chemotaxis of Dictyostelium cells. J. Cell Sci. 1996, 109 Pt 11, 2673–2678. [Google Scholar]
- McLaren, M.D.; Mathavarajah, S.; Huber, R.J. Recent Insights into NCL Protein Function Using the Model Organism Dictyostelium discoideum. Cells 2019, 8, 115. [Google Scholar] [CrossRef]
- Huber, R.J.; Mathavarajah, S. Cln5 is secreted and functions as a glycoside hydrolase in Dictyostelium. Cell Signal. 2018, 42, 236–248. [Google Scholar] [CrossRef]
- Cherix, N.; Froquet, R.; Charette, S.J.; Blanc, C.; Letourneur, F.; Cosson, P. A Phg2-Adrm1 pathway participates in the nutrient-controlled developmental response in Dictyostelium. Mol. Biol. Cell 2006, 17, 4982–4987. [Google Scholar]
- Qiu, X.B.; Ouyang, S.Y.; Li, C.J.; Miao, S.; Wang, L.; Goldberg, A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006, 25, 5742–5753. [Google Scholar]
- Jiang, R.T.; Yemelyanova, A.; Xing, D.; Anchoori, R.K.; Hamazaki, J.; Murata, S.; Seidman, J.D.; Wang, T.L.; Roden, R.B. Early and consistent overexpression of ADRM1 in ovarian high-grade serous carcinoma. J. Ovarian Res. 2017, 10, 53. [Google Scholar] [CrossRef]
- Chadwick, C.M.; Ellison, J.E.; Garrod, D.R. Dual role for Dictyostelium contact site B in phagocytosis and developmental size regulation. Nature 1984, 307, 646–647. [Google Scholar]
- Noegel, A.; Gerisch, G.; Stadler, J.; Westphal, M. Complete sequence and transcript regulation of a cell adhesion protein from aggregating Dictyostelium cells. EMBO J. 1986, 5, 1473–1476. [Google Scholar]
- Wong, L.M.; Siu, C.H. Cloning of cDNA for the contact site A glycoprotein of Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 1986, 83, 4248–4252. [Google Scholar]
- Vandekerckhove, J.; Van Damme, J.; Vancompernolle, K.; Bubb, M.R.; Lambooy, P.K.; Korn, E.D. The covalent structure of Acanthamoeba actobindin. J. Biol. Chem. 1990, 265, 12801–12805. [Google Scholar]
- Arasada, R.; Gloss, A.; Tunggal, B.; Joseph, J.M.; Rieger, D.; Mondal, S.; Faix, J.; Schleicher, M.; Noegel, A.A. Profilin isoforms in Dictyostelium discoideum. Biochim. Biophys. Acta 2007, 1773, 631–641. [Google Scholar]
- Dickinson, D.J.; Robinson, D.N.; Nelson, W.J.; Weis, W.I. α-catenin and IQGAP regulate myosin localization to control epithelial tube morphogenesis in Dictyostelium. Dev. Cell 2012, 23, 533–546. [Google Scholar] [CrossRef]
- Nagasaki, A.; Kanada, M.; Uyeda, T.Q. Cell adhesion molecules regulate contractile ring-independent cytokinesis in Dictyostelium discoideum. Cell Res. 2009, 19, 236–246. [Google Scholar] [CrossRef]
- Daugherty, R.L.; Serebryannyy, L.; Yemelyanov, A.; Flozak, A.S.; Yu, H.J.; Kosak, S.T.; deLanerolle, P.; Gottardi, C.J. α-Catenin is an inhibitor of transcription. Proc. Natl. Acad. Sci. USA 2014, 111, 5260–5265. [Google Scholar] [CrossRef]
- Le Clainche, C.; Dwivedi, S.P.; Didry, D.; Carlier, M.F. Vinculin is a dually regulated actin filament barbed end-capping and side-binding protein. J. Biol. Chem. 2010, 285, 23420–23432. [Google Scholar] [CrossRef]
- Pergolizzi, B.; Bozzaro, S.; Bracco, E. G-Protein Dependent Signal Transduction and Ubiquitination in Dictyostelium. Int. J. Mol. Sci. 2017, 18, 2180. [Google Scholar] [CrossRef]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 1999, 96, 635–644. [Google Scholar]
- VerPlank, J.J.S.; Lokireddy, S.; Zhao, J.; Goldberg, A.L. 26S Proteasomes are rapidly activated by diverse hormones and physiological states that raise cAMP and cause Rpn6 phosphorylation. Proc. Natl. Acad. Sci. USA 2019, 116, 201809254. [Google Scholar] [CrossRef]
- Sugden, C.; Ross, S.; Bloomfield, G.; Ivens, A.; Skelton, J.; Mueller-Taubenberger, A.; Williams, J.G. Two novel Src homology 2 domain proteins interact to regulate Dictyostelium gene expression during growth and early development. J. Biol. Chem. 2010, 285, 22927–22935. [Google Scholar] [CrossRef]
- Bandau, S.; Knebel, A.; Gage, Z.O.; Wood, N.T.; Alexandru, G. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation. BMC Biol. 2012, 10, 36. [Google Scholar] [CrossRef]
- Linsey, D.F.; Amerik, A.; Deery, W.J.; Bishop, J.D.; Hochstrasser, M.; Gomer, R.H. A deubiquitinating enzyme that disassembles free polyubiquitin chains is required for development but not growth in Dictyostelium. J. Biol. Chem. 1998, 273, 29178–29187. [Google Scholar]
- Sawai, S.; Thomason, P.A.; Cox, E.C. An autoregulatory circuit for long-range self-organization in Dictyostelium cell populations. Nature 2005, 433, 323–326. [Google Scholar]
- El-Halawany, S.; Ohkouchi, S.; Shibata, H.; Hitomi, K.; Maki, M. Identification of cysteine protease inhibitors that belong to cystatin family 1 in the cellular slime mold Dictyostelium discoideum. Biol. Chem. 2004, 385, 547–550. [Google Scholar]
- Li, Z.; Dugan, A.S.; Bloomfield, G.; Skelton, J.; Ivens, A.; Losick, V.; Isberg, R.R. DupA: A key regulator of the amoebal MAP kinase response to Legionella pneumophila. Cell Host Microbe 2009, 6, 253–267. [Google Scholar]
- Kawata, T.; Nakagawa, M.; Shimada, N.; Fujii, S.; Oohata, A.A. A gene encoding, prespore-cell-inducing factor in Dictyostelium discoideum. Dev. Growth Differ. 2004, 46, 383–392. [Google Scholar]
- Yamada, Y.; Minamisawa, H.; Fukuzawa, M.; Kawata, T.; Oohata, A.A. Prespore cell inducing factor, psi factor, controls both prestalk and prespore gene expression in Dictyostelium development. Dev. Growth Differ. 2010, 52, 377–383. [Google Scholar] [CrossRef]
- Zucko, J.; Skunca, N.; Curk, T.; Zupan, B.; Long, P.F.; Cullum, J.; Kessin, R.H.; Hranueli, D. Polyketide synthase genes and the natural products potential of Dictyostelium discoideum. Bioinformatics 2007, 23, 2543–2549. [Google Scholar]
- Narita, T.B.; Chen, Z.H.; Schaap, P.; Saito, T. The hybrid type polyketide synthase SteelyA is required for cAMP signalling in early Dictyostelium development. PLoS ONE 2014, 9, e106634. [Google Scholar] [CrossRef]
- Paschke, P.; Pawolleck, N.; Haenel, F.; Otto, H.; Rühling, H.; Maniak, M. The isoform B of the Dictyostelium long-chain fatty-acyl-coenzyme A synthetase is initially inserted into the ER and subsequently provides peroxisomes with an activity important for efficient phagocytosis. Eur. J. Cell Biol. 2012, 91, 717–727. [Google Scholar]
- Sampath, H.; Ntambi, J.M. The role of stearoyl-CoA desaturase in obesity, insulin resistance, and inflammation. Ann. N. Y. Acad. Sci. 2011, 1243, 47–53. [Google Scholar] [CrossRef]
- Casimir, D.A.; Ntambi, J.M. cAMP activates the expression of stearoyl-CoA desaturase gene 1 during early preadipocyte differentiation. J. Biol. Chem. 1996, 271, 29847–29853. [Google Scholar]
- Martinez, D.L.; Tsuchiya, Y.; Gout, I. Coenzyme A biosynthetic machinery in mammalian cells. Biochem. Soc. Trans. 2014, 42, 1112–1117. [Google Scholar]
- Han, X.; Shen, L.; Wang, Q.; Cen, X.; Wang, J.; Wu, M.; Li, P.; Zhao, W.; Zhang, Y.; Zhao, G. Cyclic AMP Inhibits the Activity and Promotes the Acetylation of Acetyl-CoA Synthetase through Competitive Binding to the ATP/AMP Pocket. J. Biol. Chem. 2017, 292, 1374–1384. [Google Scholar] [CrossRef]
- Manna, P.R.; Slominski, A.T.; King, S.R.; Stetson, C.L.; Stocco, D.M. Synergistic activation of steroidogenic acute regulatory protein expression and steroid biosynthesis by retinoids: Involvement of cAMP/PKA signaling. Endocrinology 2014, 155, 576–591. [Google Scholar] [CrossRef][Green Version]
- Anjard, C.; Su, Y.; Loomis, W.F. Steroids initiate a signaling cascade that triggers rapid sporulation in Dictyostelium. Development 2009, 136, 803–812. [Google Scholar] [CrossRef]
- Johansson, M.J.; Byström, A.S. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA 2002, 8, 324–335. [Google Scholar]
- Cloutlier, P.; Lavallée-Adam, M.; Faubert, D.; Blanchette, M.; Coulombe, B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013, 9, e1003210. [Google Scholar] [CrossRef]
- Sugimoto, N.; Maehara, K.; Yoshida, K.; Yasukouchi, S.; Osano, S.; Watanabe, S.; Aizawa, M.; Yugawa, T.; Kiyono, T.; Kurumizaka, H.; et al. Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res. 2015, 43, 5898–5911. [Google Scholar] [CrossRef]
- Steggerda, S.M.; Paschal, B.M. Regulation of nuclear import and export by the GTPase Ran. Int. Rev. Cytol. 2002, 217, 41–91. [Google Scholar]
- Keckesova, Z.; Donaher, J.L.; De Cock, J.; Freinkman, E.; Lingrell, S.; Bachovchin, D.A.; Bierie, B.; Tischler, V.; Noske, A.; Okondo, M.C.; et al. LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature 2017, 543, 681–686. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar]
- Shan, Y.; Cortopassi, G. Mitochondrial Hspa9/Mortalin regulates erythroid differentiation via iron-sulfur cluster assembly. Mitochondrion 2016, 26, 94–103. [Google Scholar] [CrossRef]
- Ren, M.; Phoon, C.K.; Schlame, M. Metabolism and function of mitochondrial cardiolipin. Prog. Lipid. Res. 2014, 55, 1–16. [Google Scholar] [CrossRef]
- Deery, W.J.; Gomer, R.H. A putative receptor mediating cell-density sensing in Dictyostelium. J. Biol. Chem. 1999, 274, 34476–34482. [Google Scholar]
- Lelong, E.; Marchetti, A.; Guého, A.; Lima, W.C.; Sattler, N.; Molmeret, M.; Hagedorn, M.; Soldati, T.; Cosson, P. Role of magnesium and a phagosomal P-type ATPase in intracellular bacterial killing. Cell Microbiol. 2011, 13, 246–258. [Google Scholar] [CrossRef]
- Gorelik, A.; Illes, K.; Nagar, B. Crystal structure of the mammalian lipopolysaccharide detoxifier. Proc. Natl. Acad. Sci. USA 2018, 115, E896–E905. [Google Scholar]
- Coston, B.; Loomis, F. Isozymes of beta-glucosidase in Dictyostelium discoideum. J. Bacteriol. 1969, 100, 1208–1217. [Google Scholar]
- Chen, Z.H.; Singh, R.; Cole, C.; Lawal, H.M.; Schilde, C.; Febrer, M.; Barton, G.J.; Schaap, P. Adenylate cyclase A acting on PKA mediates induction of stalk formation by cyclic diguanylate at the Dictyostelium organizer. Proc. Natl. Acad. Sci. USA 2017, 114, 516–521. [Google Scholar]
- Benghezal, M.; Gotthardt, D.; Cornillon, S.; Cosson, P. Localization of the Rh50-like protein to the contractile vacuole in Dictyostelium. Immunogenetics 2001, 52, 284–288. [Google Scholar]
- Hallows and Tempel. J. Neurosci. 1998, 15, 5682–5691.
- Guo, R.; Rowe, P.S.; Liu, S.; Simpson, L.G.; Xiao, Z.S.; Quarles, L.D. Inhibition of MEPE cleavage by Phex. Biochem. Biophys. Res. Commun. 2002, 297, 38–45. [Google Scholar]
- Mijanovic, O.; Branković, A.; Panin, A.N.; Savchuk, S.; Timashev, P.; Ulasov, I.; Lesniak, M.S. Cathepsin B: A sellsword of cancer progression. Cancer Lett. 2019, 449, 207–214. [Google Scholar]
- Chung, C.Y.; Lee, S.; Briscoe, C.; Ellsworth, C.; Firtel, R.A. Role of Rac in controlling the actin cytoskeleton and chemotaxis in motile cells. Proc. Natl. Acad. Sci. USA 2000, 97, 5225–5230. [Google Scholar]
- Chung, C.Y.; Firtel, R.A. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J. Cell Biol. 1999, 147, 559–576. [Google Scholar]
- Chung, C.Y.; Potikyan, G.; Firtel, R.A. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 2001, 7, 937–947. [Google Scholar]
- Miranda, E.R.; Zhuchenko, O.; Toplak, M.; Santhanam, B.; Zupan, B.; Kuspa, A.; Shaulsky, G. ABC Transporters in Dictyostelium discoideum Development. PLoS ONE 2013, 8, e70040. [Google Scholar]
- Konijn, T.M.; Van De Meene, J.G.; Bonner, J.T.; Barkley, D.S. The acrasin activity of adenosine-3′,5′-cyclic phosphate. Proc. Natl. Acad. Sci. USA 1967, 58, 1152–1154. [Google Scholar]
- Klein, P.; Vaughan, R.; Borleis, J.; Devreotes, P. The surface cyclic AMP receptor in Dictyostelium. Levels of ligand-induced phosphorylation, solubilization, identification of primary transcript, and developmental regulation of expression. J. Biol. Chem. 1987, 262, 358–364. [Google Scholar]
- Pupillo, M.; Klein, P.; Vaughan, R.; Pitt, G.; Lilly, P.; Sun, T.; Devreotes, P.; Kumagai, A.; Firtel, R. cAMP receptor and G-protein interactions control development in Dictyostelium. Cold Spring Harb. Symp. Quant. Biol. 1988, 53 Pt 2, 657–665. [Google Scholar]
- Insall, R.; Kuspa, A.; Lilly, P.J.; Shaulsky, G.; Levin, L.R.; Loomis, W.F.; Devreotes, P. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J. Cell Biol. 1994, 126, 1537–1545. [Google Scholar]
- Maeda, M.; Aubry, L.; Insall, R.; Gaskins, C.; Devreotes, P.N.; Firtel, R.A. Seven helix chemoattractant receptors transiently stimulate mitogen-activated protein kinase in Dictyostelium. Role of heterotrimeric G proteins. J. Biol. Chem. 1996, 271, 3351–3354. [Google Scholar]
- Swaney, K.F.; Huang, C.H.; Devreotes, P.N. Eukaryotic chemotaxis: A network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 2010, 39, 265–289. [Google Scholar] [CrossRef]
- Scherer, A.; Kuhl, S.; Wessels, D.; Lusche, D.F.; Raisley, B.; Soll, D.R. Ca2+ chemotaxis in Dictyostelium discoideum. J. Cell Sci. 2010, 123 Pt 21, 3756–3767. [Google Scholar] [CrossRef]
| Proteins Upregulated in Wild-Type and glkA Null Cells | Proteins Downregulated in Wild-Type and glkA Null Cells | ||
|---|---|---|---|
| UniProt | Protein | UniProt | Protein |
| P13773 | Cyclic AMP receptor 1 | Q55DR6 | Fatty acyl-CoA synthetase A |
| Q54QB1 | ERK2 | Q54N49 | Inositol-3-phosphate synthase |
| P34117 | Cyclin-dependent kinase 5 homolog | Q54YA0 | Probable ATP-citrate synthase |
| P16051 | G alpha-2 | Q54I98 | Cycloartenol-C-24-methyltransferase |
| O60952 | LIM domain-containing protein E | Q54DR1 | Squalene synthase (SQS) |
| Q55DU3 | Actobindin-A | Q553V1 | Citrate synthase |
| Q65YR7 | Cystatin-A2 | Q54KB7 | Glutamate dehydrogenase |
| Q1ZXH5 | Calcium-binding protein | Q55BI2 | Isocitrate dehydrogenase subunit A |
| P54653 | Calcium-binding protein 2 | O97470 | Substrate carrier family protein ancA |
| Q54RF4 | Calcium-binding protein 4a | Q54JP5 | Probable ornithine aminotransferase |
| P54679 | Probable membrane ATPase | Q8MP58 | Uncharacterized protein |
| Q86AA1 | Probable T4-type lysozyme 2 | Q54IS1 | Uncharacterized protein |
| Q54FS0 | Uncharacterized protein | Q55EK2 | Probable cytochrome P450 524A1 |
| Q54FV6 | Uncharacterized protein | Q869W9 | Probable polyketide synthase 16 |
| Q54G71 | Uncharacterized protein | Q556T4 | Uncharacterized protein |
| Q54GR0 | Uncharacterized protein | Q1ZXN5 | Uncharacterized protein |
| Q54I40 | Uncharacterized protein | Q54CD7 | Uncharacterized protein |
| Q54IK3 | Uncharacterized protein | Q54J99 | Uncharacterized protein |
| Q54Q34 | Uncharacterized protein | Q54R89 | Uncharacterized protein |
| Q54UX5 | Uncharacterized protein | Q54T87 | Uncharacterized protein |
| Q54WT5 | Uncharacterized protein | Q54WK0 | Uncharacterized protein |
| Q556W6 | Uncharacterized protein | Q86KA1 | Uncharacterized protein |
| Q55BQ2 | Uncharacterized protein | Q54NS9 | Apoptosis-inducing factor homolog A |
| Q55E22 | Uncharacterized protein | Q5XM24 | Autocrine proliferation repressor protein A |
| Q86AC9 | Uncharacterized protein | Q9GPS1 | Complex III assembly factor LYRM7 |
| Q55DE7 | Ataxin-2 homolog | Q55G75 | PH domain-containing protein |
| P90532 | Cell division cycle protein 48 | Q55BZ5 | Protein dcd1A |
| Q54ST6 | Membrane protein subunit | Q54F74 | Sulfate adenylyltransferase |
| Q54I92 | Protein psiI | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Velasco, Ó.; De Las Rivas, J.; Lacal, J. Proteomic and Transcriptomic Profiling Identifies Early Developmentally Regulated Proteins in Dictyostelium Discoideum. Cells 2019, 8, 1187. https://doi.org/10.3390/cells8101187
González-Velasco Ó, De Las Rivas J, Lacal J. Proteomic and Transcriptomic Profiling Identifies Early Developmentally Regulated Proteins in Dictyostelium Discoideum. Cells. 2019; 8(10):1187. https://doi.org/10.3390/cells8101187
Chicago/Turabian StyleGonzález-Velasco, Óscar, Javier De Las Rivas, and Jesus Lacal. 2019. "Proteomic and Transcriptomic Profiling Identifies Early Developmentally Regulated Proteins in Dictyostelium Discoideum" Cells 8, no. 10: 1187. https://doi.org/10.3390/cells8101187
APA StyleGonzález-Velasco, Ó., De Las Rivas, J., & Lacal, J. (2019). Proteomic and Transcriptomic Profiling Identifies Early Developmentally Regulated Proteins in Dictyostelium Discoideum. Cells, 8(10), 1187. https://doi.org/10.3390/cells8101187






