Centrosome Remodelling in Evolution
Abstract
1. Introduction
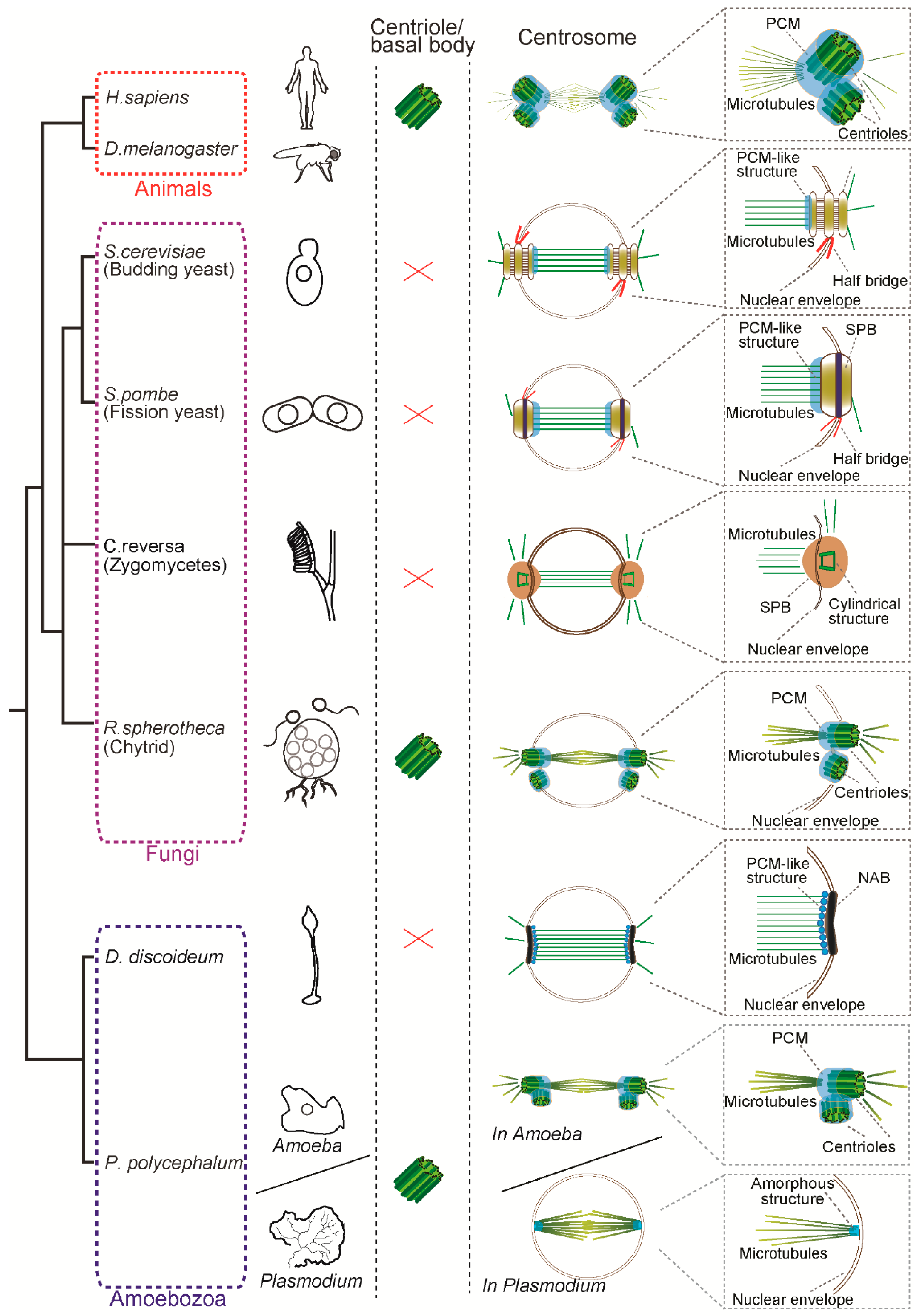
2. Canonical and Diverged-Centrosomes in Animal, Fungi and Amoebozoa
3. Common Components in Centrosome
4. Common Regulations in Centrosome Biogenesis
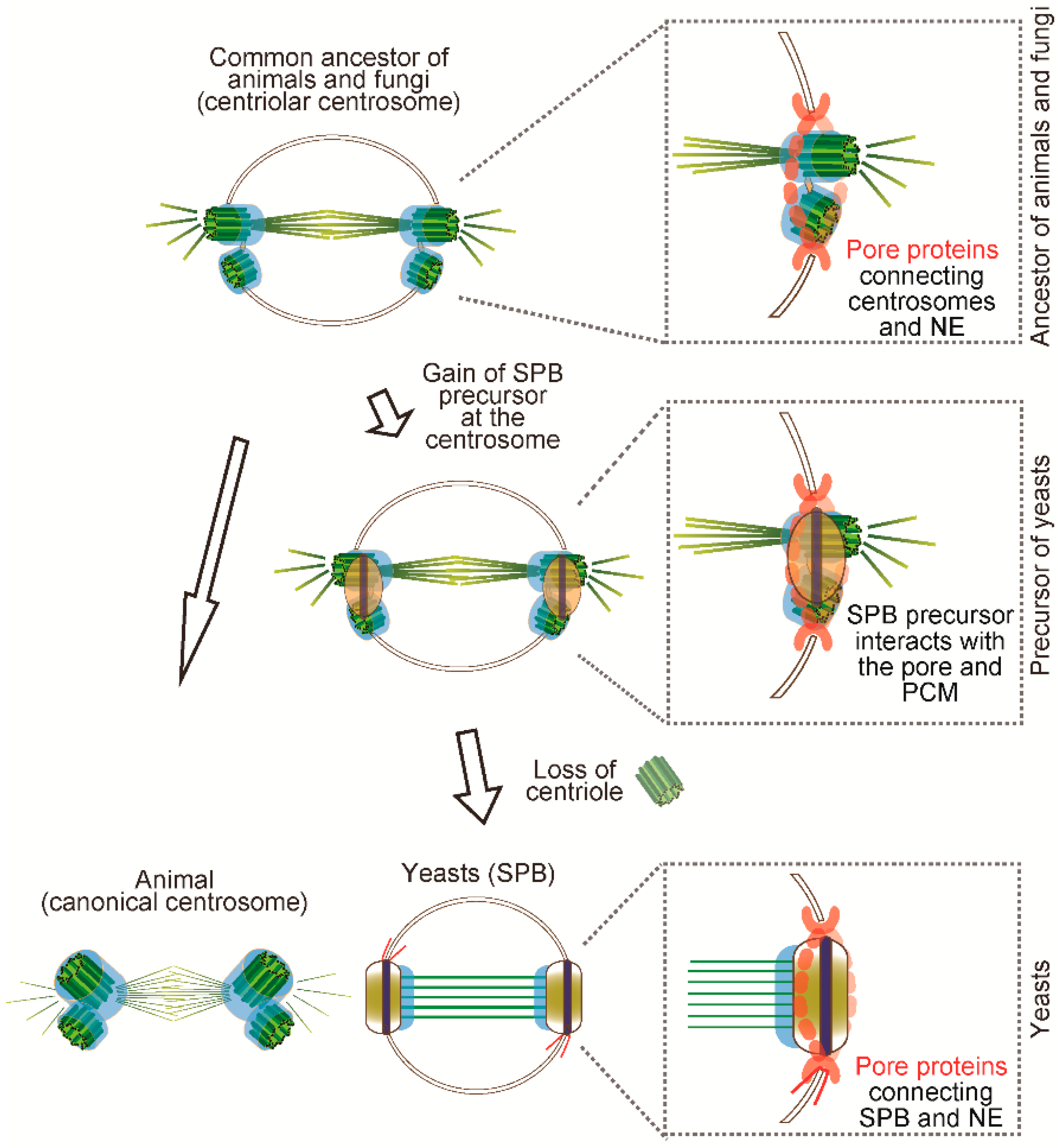
5. How Did the SPB Evolve?
6. Conclusions and Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scheer, U. Historical roots of centrosome research: Discovery of Boveri’s microscope slides in Wurzburg. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130469. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt-dias, M.; Glover, D.M. Centrosome biogenesis and function: Centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 2007, 8, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Santos, Z.; Azimzadeh, J.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Evolution: Tracing the origins of centrioles, cilia and flagella. J. Cell Biol. 2011, 194, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, J. Exploring the evolutionary history of centrosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130453. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, J.; Bornens, M. The Centrosome in Evolution. In Centrosomes in Development and Disease; Wiley-Blackwell: Hoboken, NJ, USA, 2005; pp. 93–122. ISBN 9783527603800. [Google Scholar]
- Bornens, M.; Azimzadeh, J. Origin and evolution of the centrosome. Adv. Exp. Med. Biol. 2007, 607, 119–129. [Google Scholar] [PubMed]
- Carvalho-Santos, Z.; Machado, P.; Branco, P.; Tavares-Cadete, F.; Rodrigues-Martins, A.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 2010, 123, 1414–1426. [Google Scholar] [CrossRef] [PubMed]
- Kilmartin, J.V. Lessons from yeast: The spindle pole body and the centrosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130456. [Google Scholar] [CrossRef] [PubMed]
- Graf, R.; Batsios, P.; Meyer, I. Evolution of centrosomes and the nuclear lamina: Amoebozoan assets. Eur. J. Cell Biol. 2015, 94, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Early evolution of eukaryote feeding modes, cell structural diversity and classification of the protozoan phyla Loukozoa, Sulcozoa and Choanozoa. Eur. J. Protistol. 2013, 49, 115–178. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Simpson, A.G.B.; Lane, C.E.; Lukes, J.; Bass, D.; Bowser, S.S.; Brown, M.W.; Burki, F.; Dunthorn, M.; Hampl, V.; et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012, 59, 429–493. [Google Scholar] [CrossRef] [PubMed]
- Jaspersen, S.L.; Winey, M. The Budding Yeast Spindle Pole Body: Structure, Duplication and Function. Annu. Rev. Cell Dev. Biol. 2004, 20, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, A.M.; Jaspersen, S.L. Big Lessons from Little Yeast: Budding and Fission Yeast Centrosome Structure, Duplication and Function. Annu. Rev. Genet. 2017, 51, 361–383. [Google Scholar] [CrossRef] [PubMed]
- Adams, I.R.; Kilmartin, J.V. Spindle pole body duplication: A model for centrosome duplication? Trends Cell Biol. 2000, 10, 329–335. [Google Scholar] [CrossRef]
- McLaughlin, D.J.; Healy, R.A.; Celio, G.J.; Roberson, R.W.; Kumar, T.K.A. Evolution of zygomycetous spindle pole bodies: Evidence from Coemansia reversa mitosis. Am. J. Bot. 2015, 102, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Powell, M.J. Mitosis in the Aquatic Fungus Rhizophydium spherotheca (Chytridiales). Am. J. Bot. 1980, 67, 839–853. [Google Scholar] [CrossRef]
- Moens, P.B. Spindle and kinetochore morphology of Dictyostelium discoideum. J. Cell Biol. 1976, 68, 113–122. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.P.C.; Osmani, S.A. Mitosis, not just open or closed. Eukaryot. Cell 2007, 6, 1521–1527. [Google Scholar] [CrossRef] [PubMed]
- Gely, C.; Wright, M. The centriole cycle in the amoebae of the myxomycete Physarum polycephalum. Protoplasma 1986, 132, 23–31. [Google Scholar] [CrossRef]
- Aldrich, H.C. The ultrastructure of mitosis in myxamoebae and plasmodia of Physarum flavicomum. Am. J. Bot. 1969, 56, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Havercroft, J.C.; Quinlan, R.A.; Gull, K. Characterisation of a microtubule organising centre from Physarum polycephalum myxamoebae. J. Ultrastruct. Res. 1981, 74, 313–321. [Google Scholar] [CrossRef]
- Tanaka, K. Intranuclear microtubule organizing center in early prophase nuclei of the plasmodium of the slime mold, Physarum polycephalum. J. Cell Biol. 1973, 57, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.; Shigenaga, M. Electron microscopy of dividing cells. IV. Behaviour of spindle microtubules during nuclear division in the plasmodium of the myxomycete, Physarum polycephalum. Chromosoma 1972, 37, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Hinchee, A.A.; Haskins, E.F. Open spindle nuclear division in the amoebal phase of the acellular slime mold Echinostelium minutum with chromosomal movement related to the pronounced rearrangement of spindle microtubules. Protoplasma 1980, 102, 235–253. [Google Scholar] [CrossRef]
- Nigg, E.A.; Holland, A.J. Once and only once: Mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Gönczy, P. Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 2012, 13, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Van Breugel, M.; Hirono, M.; Andreeva, A.; Yanagisawa, H.; Yamaguchi, S.; Nakazawa, Y.; Morgner, N.; Petrovich, M.; Ebong, I.-O.; Robinson, C.V.; et al. Structures of SAS-6 suggest its organization in centrioles. Science 2011, 331, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, D.; Vakonakis, I.; Olieric, N.; Hilbert, M.; Keller, D.; Olieric, V.; Bortfeld, M.; Erat, M.C.; Flückiger, I.; Gönczy, P.; et al. Structural basis of the 9-fold symmetry of centrioles. Cell 2011, 144, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Chang, C.-W.; Hsu, W.-B.; Tang, C.-J.C.; Lin, Y.-N.; Chou, E.-J.; Wu, C.-T.; Tang, T.K. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP and is required for centriole assembly. EMBO J. 2013, 32, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-J.C.; Lin, S.-Y.; Hsu, W.-B.; Lin, Y.-N.; Wu, C.-T.; Lin, Y.-C.; Chang, C.-W.; Wu, K.-S.; Tang, T.K. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011, 30, 4790–4804. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Hiraki, M.; Kamiya, R.; Hirono, M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007, 17, 2169–2174. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-J.C.; Fu, R.-H.; Wu, K.-S.; Hsu, W.-B.; Tang, T.K. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009, 11, 825–831. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.I.; Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Lavoie, S.B.; Stierhof, Y.-D.; Nigg, E.A. Control of centriole length by CPAP and CP110. Curr. Biol. 2009, 19, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Kohlmaier, G.; Loncarek, J.; Meng, X.; McEwen, B.F.; Mogensen, M.M.; Spektor, A.; Dynlacht, B.D.; Khodjakov, A.; Gonczy, P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 2009, 19, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Glover, D.M. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012, 2, 120104. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Lipinszki, Z.; Rangone, H.; Min, M.; Mykura, C.; Chao-Chu, J.; Schneider, S.; Dzhindzhev, N.S.; Gottardo, M.; Riparbelli, M.G.; et al. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 2016, 18, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Mennella, V.; Keszthelyi, B.; McDonald, K.L.; Chhun, B.; Kan, F.; Rogers, G.C.; Huang, B.; Agard, D.A. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012, 14, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Lawo, S.; Hasegan, M.; Gupta, G.D.; Pelletier, L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 2012, 14, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- Delaval, B.; Doxsey, S.J. Pericentrin in cellular function and disease. J. Cell Biol. 2010, 188, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Megraw, T.L.; Sharkey, J.T.; Nowakowski, R.S. Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 2011, 21, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, J.B.; Ferreira Gomes, B.; Widlund, P.O.; Mahamid, J.; Honigmann, A.; Hyman, A.A. The Centrosome is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 2017, 169, 1066–1077. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Caballe, A.; Wainman, A.; Johnson, S.; Haensele, A.F.M.; Cottee, M.A.; Conduit, P.T.; Lea, S.M.; Raff, J.W. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell 2017, 169, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Kollman, J.M.; Merdes, A.; Mourey, L.; Agard, D.A. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 2011, 12, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Neuner, A.; Schiebel, E. Targeting of gamma-tubulin complexes to microtubule organizing centers: Conservation and divergence. Trends Cell Biol. 2015, 25, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Teixido-Travesa, N.; Roig, J.; Luders, J. The where, when and how of microtubule nucleation—One ring to rule them all. J. Cell Sci. 2012, 125, 4445–4456. [Google Scholar] [CrossRef] [PubMed]
- Gillingham, A.K.; Munro, S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000, 1, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Neuner, A.; Schlosser, Y.T.; Scharf, A.N.; Weber, L.; Schiebel, E. Cell-cycle dependent phosphorylation of yeast pericentrin regulates γ-TuSC-mediated microtubule nucleation. eLife 2014, 3, e02208. [Google Scholar] [CrossRef] [PubMed]
- Knop, M.; Pereira, G.; Geissler, S.; Grein, K.; Schiebel, E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997, 16, 1550–1564. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Vinh, D.B.; Crawford, D.K.; Davis, T.N. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol. Biol. Cell 1998, 9, 2201–2216. [Google Scholar] [CrossRef] [PubMed]
- Soues, S.; Adams, I.R. SPC72: A spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J. Cell Sci. 1998, 111, 2809–2818. [Google Scholar] [PubMed]
- Knop, M.; Schiebel, E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998, 17, 3952–3967. [Google Scholar] [CrossRef] [PubMed]
- Bullitt, E.; Rout, M.P.; Kilmartin, J.V.; Akey, C.W. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell 1997, 89, 1077–1086. [Google Scholar] [CrossRef]
- Adams, I.R.; Kilmartin, J.V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999, 145, 809–823. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.G.D.; Snydsman, B.E.; Novik, I.; Hailey, D.W.; Gestaut, D.R.; Niemann, C.A.; O’Toole, E.T.; Giddings, T.H.J.; Sundin, B.A.; Davis, T.N. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell 2005, 16, 3341–3352. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.; Knop, M.; Schlenstedt, G.; Schiebel, E. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA 1999, 96, 6205–6210. [Google Scholar] [CrossRef] [PubMed]
- Kilmartin, J.V. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 2003, 162, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Spang, A.; Courtney, I.; Fackler, U.; Matzner, M.; Schiebel, E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 1993, 123, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sandercock, A.M.; Conduit, P.; Robinson, C.V.; Williams, R.L.; Kilmartin, J.V. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006, 173, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Flory, M.R.; Morphew, M.; Joseph, J.D.; Means, A.R.; Davis, T.N. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 2002, 13, 47–58. [Google Scholar] [PubMed]
- Samejima, I.; Miller, V.J.; Rincon, S.A.; Sawin, K.E. Fission yeast Mto1 regulates diversity of cytoplasmic microtubule organizing centers. Curr. Biol. 2010, 20, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Bestul, A.J.; Yu, Z.; Unruh, J.R.; Jaspersen, S.L. Molecular model of fission yeast centrosome assembly determined by superresolution imaging. J. Cell Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.S.; Sato, M.; Toda, T. Fission yeast Pcp1 links polo kinase-mediated mitotic entry to gamma-tubulin-dependent spindle formation. EMBO J. 2010, 29, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.A.; Tomlin, G.C.; McDonald, W.H.; Snydsman, B.E.; Muller, E.G.; Yates, J.R., 3rd; Gould, K.L. Ppc89 links multiple proteins, including the septation initiation network, to the core of the fission yeast spindle-pole body. Mol. Biol. Cell 2006, 17, 3793–3805. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Gould, K.L. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 5249–5254. [Google Scholar] [CrossRef] [PubMed]
- Krapp, A.; Schmidt, S.; Cano, E.; Simanis, V.S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 2001, 11, 1559–1568. [Google Scholar] [CrossRef]
- Bouhlel, I.B.; Ohta, M.; Mayeux, A.; Bordes, N.; Dingli, F.; Boulanger, J.; Velve Casquillas, G.; Loew, D.; Tran, P.T.; Sato, M.; et al. Cell cycle control of spindle pole body duplication and splitting by Sfi1 and Cdc31 in fission yeast. J. Cell Sci. 2015, 128, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Wang, N.; Hu, W.; Schott, K.; Bähler, J.; Giddings, T.H.; Pringle, J.R.; Du, L.-L.; Wu, J.-Q. Regulation of spindle-pole body assembly and cytokinesis by the centrin-binding protein Sfi1 in fission yeast. Mol. Biol. Cell 2014, 25, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sanz, J.; Yang, A.; Blouquit, Y.; Duchambon, P.; Assairi, L.; Craescu, C.T. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006, 273, 4504–4515. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, A.; Moudjou, M.; Paintrand, M.; Salisbury, J.L.; Bornens, M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996, 109, 3089–3102. [Google Scholar] [PubMed]
- Dantas, T.J.; Wang, Y.; Lalor, P.; Dockery, P.; Morrison, C.G. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J. Cell Biol. 2011, 193, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Kleylein-Sohn, J.; Westendorf, J.; Le Clech, M.; Habedanck, R.; Stierhof, Y.-D.; Nigg, E.A. Plk4-induced centriole biogenesis in human cells. Dev. Cell 2007, 13, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Byers, B.; Goetsch, L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 1975, 124, 511–523. [Google Scholar] [PubMed]
- McCully, E.K.; Robinow, C.F. Mitosis in the fission yeast Schizosaccharomyces pombe: A comparative study with light and electron microscopy. J. Cell Sci. 1971, 9, 475–507. [Google Scholar] [PubMed]
- Ding, R.; West, R.R.; Morphew, D.M.; Oakley, B.R.; McIntosh, J.R. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell 1997, 8, 1461–1479. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, E.T.; Winey, M.; McIntosh, J.R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 2017–2031. [Google Scholar] [CrossRef] [PubMed]
- Chial, H.J.; Rout, M.P.; Giddings, T.H.; Winey, M. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 1998, 143, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- West, R.R.; Vaisberg, E.V.; Ding, R.; Nurse, P.; McIntosh, J.R. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol. Biol. Cell 1998, 9, 2839–2855. [Google Scholar] [CrossRef] [PubMed]
- Ruthnick, D.; Neuner, A.; Dietrich, F.; Kirrmaier, D.; Engel, U.; Knop, M.; Schiebel, E. Characterization of spindle pole body duplication reveals a regulatory role for nuclear pore complexes. J. Cell Biol. 2017, 216, 2425–2442. [Google Scholar] [CrossRef] [PubMed]
- Ruthnick, D.; Schiebel, E. Duplication and Nuclear Envelope Insertion of the Yeast Microtubule Organizing Centre, the Spindle Pole Body. Cells 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Burns, S.; Avena, J.S.; Unruh, J.R.; Yu, Z.; Smith, S.E.; Slaughter, B.D.; Winey, M.; Jaspersen, S.L. Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Hagan, I.; Yanagida, M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 1995, 129, 1033–1047. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Alvarez, A.; Bez, C.; O’Toole, E.T.; Morphew, M.; Cooper, J.P. Mitotic Nuclear Envelope Breakdown and Spindle Nucleation are Controlled by Interphase Contacts between Centromeres and the Nuclear Envelope. Dev. Cell 2016, 39, 544–559. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Smoyer, C.J.; Slaughter, B.D.; Unruh, J.R.; Jaspersen, S.L. The SUN protein Mps3 controls Ndc1 distribution and function on the nuclear membrane. J. Cell Biol. 2014, 204, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Jaspersen, S.L.; Martin, A.E.; Glazko, G.; Giddings, T.H.J.; Morgan, G.; Mushegian, A.; Winey, M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 2006, 174, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, A.; Bordes, N.; Haddad, R.; Schwartz, C.L.; Chang, F.; Bornens, M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell 2003, 14, 2793–2808. [Google Scholar] [CrossRef] [PubMed]
- Hodges, M.E.; Scheumann, N.; Wickstead, B.; Langdale, J.A.; Gull, K. Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 2010, 123, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- Ito, D.; Zitouni, S.; Jana, S.C.; Duarte, P.; Surkont, J.; Carvalho-Santos, Z.; Pereira-Leal, J.B.; Godinho Ferreira, M.; Bettencourt-Dias, M. An ancestral role of pericentrin in centriole formation through SAS-6 recruitment. bioRxiv 2018. [Google Scholar] [CrossRef]
- Nigg, E.A.; Stearns, T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011, 13, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Ruthnick, D.; Schiebel, E. Duplication of the Yeast Spindle Pole Body Once per Cell Cycle. Mol. Cell. Biol. 2016, 36, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Hagan, I.M.; Glover, D.M. The Centrosome and Its Duplication Cycle. Cold Spring Harb. Perspect. Biol. 2015, 7, a015800. [Google Scholar] [CrossRef] [PubMed]
- Boveri, T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008, 121, 1–84. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A.; Raff, J.W. Centrioles, centrosomes and cilia in health and disease. Cell 2009, 139, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt-Dias, M.; Hildebrandt, F.; Pellman, D.; Woods, G.; Godinho, S.A. Centrosomes and cilia in human disease. Trends Genet. 2011, 27, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Gonczy, P. Centrosomes and cancer: Revisiting a long-standing relationship. Nat. Rev. Cancer 2015, 15, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Elserafy, M.; Saric, M.; Neuner, A.; Lin, T.; Zhang, W.; Seybold, C.; Sivashanmugam, L.; Schiebel, E. Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr. Biol. 2014, 24, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Avena, J.S.; Burns, S.; Yu, Z.; Ebmeier, C.C.; Old, W.M.; Jaspersen, S.L.; Winey, M. Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1. PLoS Genet. 2014, 10, e1004666. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt-Dias, M.; Rodrigues-Martins, A.; Carpenter, L.; Riparbelli, M.; Lehmann, L.; Gatt, M.K.; Carmo, N.; Balloux, F.; Callaini, G.; Glover, D.M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005, 15, 2199–2207. [Google Scholar] [CrossRef] [PubMed]
- Habedanck, R.; Stierhof, Y.-D.; Wilkinson, C.J.; Nigg, E.A. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005, 7, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Arquint, C.; Sonnen, K.F.; Stierhof, Y.-D.; Nigg, E.A. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 2012, 125, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Arquint, C.; Gabryjonczyk, A.-M.; Imseng, S.; Bohm, R.; Sauer, E.; Hiller, S.; Nigg, E.A.; Maier, T. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife 2015, 4, e07888. [Google Scholar] [CrossRef] [PubMed]
- Moyer, T.C.; Clutario, K.M.; Lambrus, B.G.; Daggubati, V.; Holland, A.J. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 2015, 209, 863–878. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Ashikawa, T.; Nozaki, Y.; Kozuka-Hata, H.; Goto, H.; Inagaki, M.; Oyama, M.; Kitagawa, D. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014, 5, 5267. [Google Scholar] [CrossRef] [PubMed]
- Steere, N.; Wagner, M.; Beishir, S.; Smith, E.; Breslin, L.; Morrison, C.G.; Hochegger, H.; Kuriyama, R. Centrosome amplification in CHO and DT40 cells by inactivation of cyclin-dependent kinases. Cytoskeleton 2011, 68, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Vidwans, S.J.; Wong, M.L.; O’Farrell, P.H. Anomalous centriole configurations are detected in Drosophila wing disc cells upon Cdk1 inactivation. J. Cell Sci. 2003, 116, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Arquint, C.; Nigg, E.A. STIL Microcephaly Mutations Interfere with APC/C-Mediated Degradation and Cause Centriole Amplification. Curr. Biol. 2014, 24, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Zitouni, S.; Francia, M.E.; Leal, F.; Lorca, T.; Lince-faria, M.; Zitouni, S.; Francia, M.E.; Leal, F.; Gouveia, S.M.; Nabais, C.; et al. CDK1 Prevents Unscheduled PLK4-STIL Complex Assembly in Centriole Biogenesis Article CDK1 Prevents Unscheduled PLK4-STIL Complex Assembly in Centriole Biogenesis. Curr. Biol. 2016, 26, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Saunders, W.S.; Hoyt, M.A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 1992, 70, 451–458. [Google Scholar] [CrossRef]
- Crasta, K.; Lim, H.H.; Giddings, T.H.; Winey, M.; Surana, U. Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle. Nat. Cell Biol. 2008, 10, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Huang, P.; Morgan, G.; Winey, M.; Surana, U. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 2006, 25, 2551–2563. [Google Scholar] [CrossRef] [PubMed]
- Chee, M.K.; Haase, S.B. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010, 6, e1000935. [Google Scholar] [CrossRef] [PubMed]
- Hagan, I.; Yanagida, M. Novel potential mitotic motor protein encoded by the fission yeast cut7+ gene. Nature 1990, 347, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Hagan, I.; Yanagida, M. Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 1992, 356, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Blangy, A.; Lane, H.A.; d’Herin, P.; Harper, M.; Kress, M.; Nigg, E.A. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 1995, 83, 1159–1169. [Google Scholar] [PubMed]
- Sawin, K.E.; Mitchison, T.J. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA 1995, 92, 4289–4293. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Hegarat, N.; Vesely, C.; Roseboom, I.; Larch, C.; Streicher, H.; Straatman, K.; Flynn, H.; Skehel, M.; Hirota, T.; et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J. 2011, 30, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- Agircan, F.G.; Schiebel, E.; Mardin, B.R. Separate to operate: Control of centrosome positioning and separation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130461. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jiang, Q.; Zhang, C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J. Cell Sci. 2014, 127, 4111–4122. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.M.; Bayliss, R.; Roig, J. Mitotic Regulation by NEK Kinase Networks. Front. Cell Dev. Biol. 2017, 5, 102. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Shibata, T. Role of gamma-tubulin in mitosis-specific microtubule nucleation from the Schizosaccharomyces pombe spindle pole body. J. Cell Sci. 1996, 109, 165–177. [Google Scholar] [PubMed]
- Woodruff, J.B.; Wueseke, O.; Hyman, A.A. Pericentriolar material structure and dynamics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130459. [Google Scholar] [CrossRef] [PubMed]
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Wälde, S.; King, M.C. The KASH protein Kms2 coordinates mitotic remodeling of the spindle pole body. J. Cell Sci. 2014, 127, 3625–3640. [Google Scholar] [CrossRef] [PubMed]
- Lane, H.A.; Nigg, E.A. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996, 135, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Steegmaier, M.; Hoffmann, M.; Baum, A.; Lenart, P.; Petronczki, M.; Krssak, M.; Gurtler, U.; Garin-Chesa, P.; Lieb, S.; Quant, J.; et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007, 17, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Lénárt, P.; Petronczki, M.; Steegmaier, M.; di Fiore, B.; Lipp, J.J.; Hoffmann, M.; Rettig, W.J.; Kraut, N.; Peters, J. The Small-Molecule Inhibitor BI 2536 Reveals Novel Insights into Mitotic Roles of Polo-like Kinase 1. Curr. Biol. 2007, 17, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Rhee, K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 2011, 195, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J.; et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ito, D.; Bettencourt-Dias, M. Centrosome Remodelling in Evolution. Cells 2018, 7, 71. https://doi.org/10.3390/cells7070071
Ito D, Bettencourt-Dias M. Centrosome Remodelling in Evolution. Cells. 2018; 7(7):71. https://doi.org/10.3390/cells7070071
Chicago/Turabian StyleIto, Daisuke, and Mónica Bettencourt-Dias. 2018. "Centrosome Remodelling in Evolution" Cells 7, no. 7: 71. https://doi.org/10.3390/cells7070071
APA StyleIto, D., & Bettencourt-Dias, M. (2018). Centrosome Remodelling in Evolution. Cells, 7(7), 71. https://doi.org/10.3390/cells7070071




