Post-Translational Modification and Subcellular Distribution of Rac1: An Update
Abstract
1. Introduction
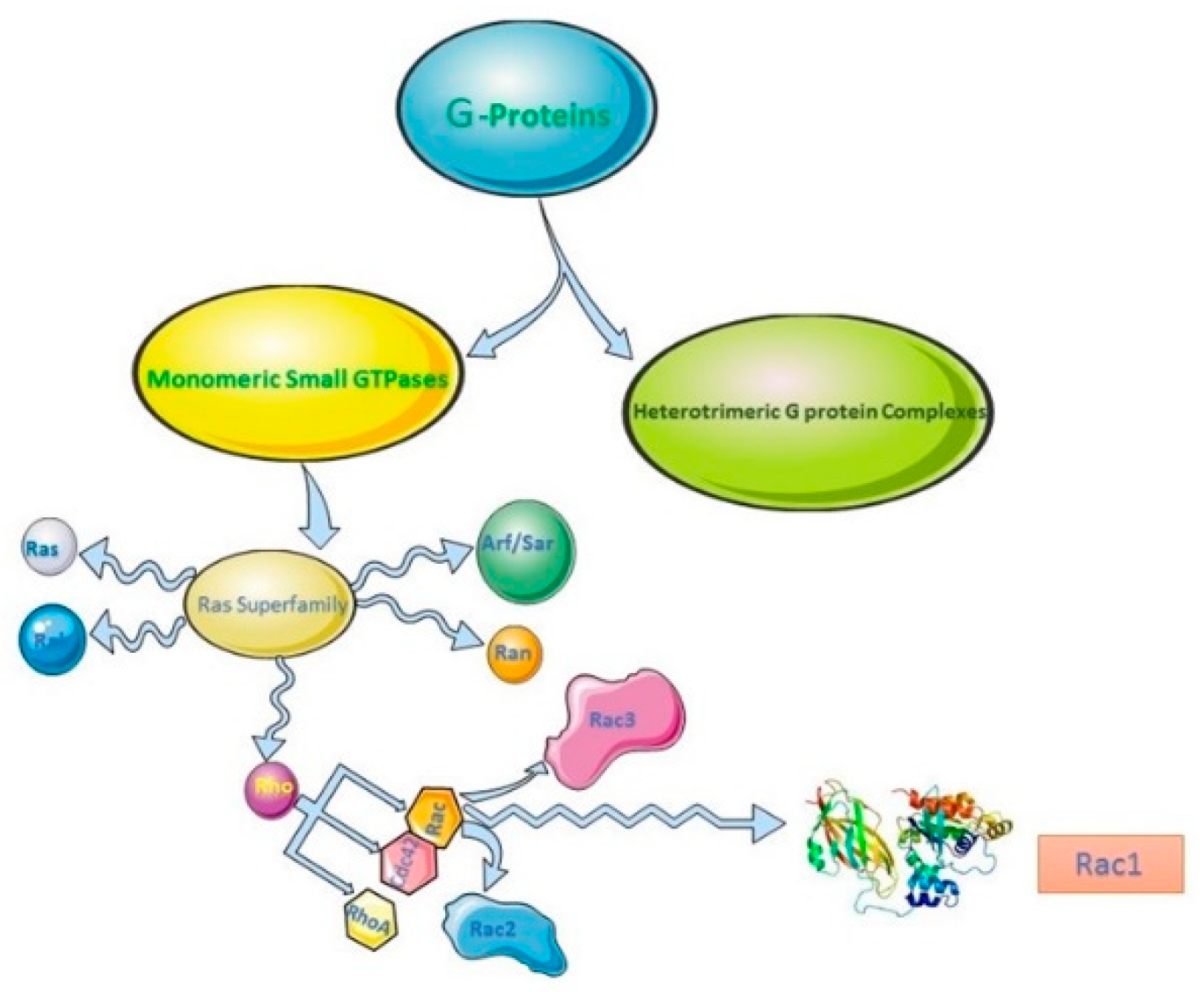
2. Rho Family of GTPases, Rac Subfamily and Rac1
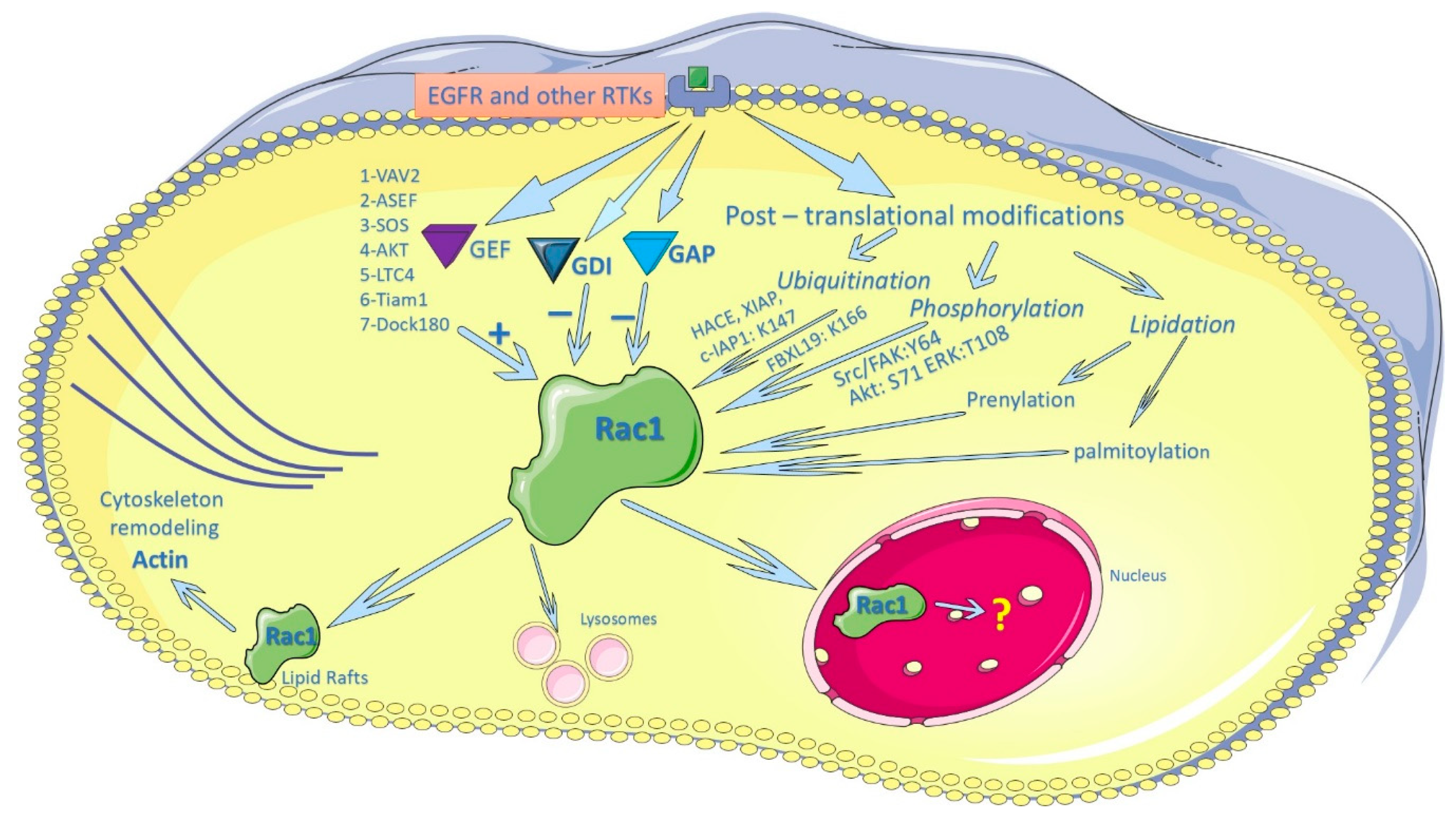
3. Regulation of Rac1 by EGFR and other Membrane Receptors
4. Regulation of Rac1 Activity by Post-Translational Modification
4.1. Regulation of Rac1 Activity by Lipidation
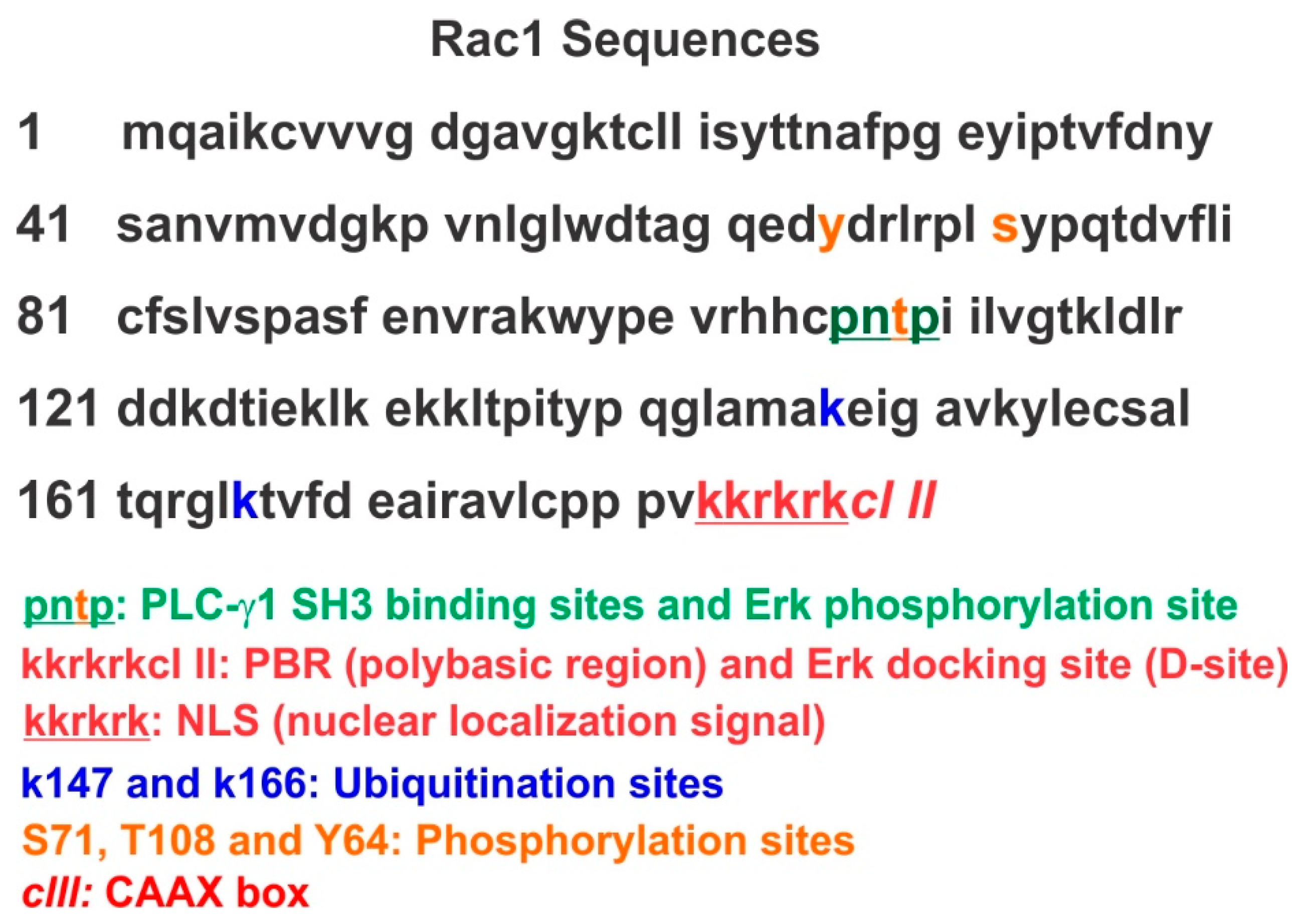
4.2. Rac1 Ubiquitination
4.3. Phosphorylation of Rac1 and other Rho GTPases
5. Regulation of Subcellular Localization of Rac1
5.1. Regulation of the PM Localization of Rac1
5.2. Regulation of the Nuclear Localization of Rac1
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Benitah, S.; Valeron, P.; van Aelst, L.; Marshall, C.; Lacal, J. Rho gtpases in human cancer: An unresolved link to upstream and downstream transcriptional regulation. Biochim. Biophys. Acta 2004, 1705, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Aznar, S.; Lacal, J. Rho signals to cell growth and apoptosis. Cancer Lett. 2001, 165, 1–10. [Google Scholar] [CrossRef]
- Van Aelst, L.; D’Souza-Schorey, C. Rho gtpases and signaling networks. Genes Dev. 1997, 11, 2295–2322. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Rho gtpases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef]
- Sugihara, K.; Nakatsuji, N.; Nakamura, K.; Nakao, K.; Hashimoto, R.; Otani, H.; Sakagami, H.; Kondo, H.; Nozawa, S.; Aiba, A.; et al. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene 1998, 17, 3427–3433. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Palmby, T.R.; Gavard, J.; Amornphimoltham, P.; Zheng, Y.; Gutkind, J.S. An essential role for Rac1 in endothelial cell function and vascular development. FASEB J. 2008, 22, 1829–1838. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Dickerson, J.B.; Guo, F.; Zheng, J.; Zheng, Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 2004, 101, 7618–7623. [Google Scholar] [CrossRef]
- Jaffe, A.B.; Hall, A. Rho GTPases: Biochemistry and biology. Ann. Rev. Cell Dev. Biol. 2005, 21, 247–269. [Google Scholar] [CrossRef]
- Parri, M.; Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun. Signal. 2010, 8, 23. [Google Scholar] [CrossRef]
- Zhou, W.; Li, X.; Premont, R.T. Expanding functions of git arf gtpase-activating proteins, Pix Rho guanine nucleotide exchange factors and git-pix complexes. J. Cell Sci. 2016, 129, 1963–1974. [Google Scholar] [CrossRef]
- Zou, T.; Mao, X.; Yin, J.; Li, X.; Chen, J.; Zhu, T.; Li, Q.; Zhou, H.; Liu, Z. Emerging roles of Rac1 in treating lung cancer patients. Clin. Genet. 2017, 91, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Chen, J.; Wang, Y.; Zhong, Y.; Dai, Q.; Wang, Q.; Tu, J. Mir-592 suppresses the development of glioma by regulating Rho-associated protein kinase. Neuroreport 2018, 29, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Hein, A.L.; Post, C.M.; Sheinin, Y.M.; Lakshmanan, I.; Natarajan, A.; Enke, C.A.; Batra, S.K.; Ouellette, M.M.; Yan, Y. Rac1 GTPase promotes the survival of breast cancer cells in response to hyper-fractionated radiation treatment. Oncogene 2016, 35, 6319–6329. [Google Scholar] [CrossRef] [PubMed]
- Sahai, E.; Marshall, C.J. Rho-GTPases and cancer. Nat. Rev. Cancer 2002, 2, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Orgaz, J.L.; Herraiz, C.; Sanz-Moreno, V. Rho GTPases modulate malignant transformation of tumor cells. Small GTPases 2014, 5, e29019. [Google Scholar] [CrossRef] [PubMed]
- Carrizzo, A.; Forte, M.; Lembo, M.; Formisano, L.; Puca, A.A.; Vecchione, C. Rac-1 as a new therapeutic target in cerebro- and cardio-vascular diseases. Curr. Drug Targets 2014, 15, 1231–1246. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Rho proteins and cancer. Breast Cancer Res.Treat. 2004, 84, 13–19. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPase signalling in cell migration. Curr. Opin. Cell Biol. 2015, 36, 103–112. [Google Scholar] [CrossRef]
- Zuo, Y.; Oh, W.; Ulu, A.; Frost, J.A. Mouse models of Rho GTPase function in mammary gland development, tumorigenesis and metastasis. Mol. Endocrinol. 2016, 30, 278–289. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable ras: Mission possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zheng, Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin. Drug Discov. 2015, 10, 991–1010. [Google Scholar] [CrossRef] [PubMed]
- Symons, M.; Settleman, J. Rho family GTPases: More than simple switches. Trends Cell Biol. 2000, 10, 415–419. [Google Scholar] [CrossRef]
- Moon, S.; Zheng, Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003, 13, 13–22. [Google Scholar] [CrossRef]
- Goncalves, V.; Henriques, A.F.; Pereira, J.F.; Neves Costa, A.; Moyer, M.P.; Moita, L.F.; Gama-Carvalho, M.; Matos, P.; Jordan, P. Phosphorylation of srsf1 by srpk1 regulates alternative splicing of tumor-related rac1b in colorectal cells. RNA 2014, 20, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fu, X.; Chen, P.; Wu, P.; Fan, X.; Li, N.; Zhu, H.; Jia, T.T.; Ji, H.; Wang, Z.; et al. Spsb1-mediated hnrnp a1 ubiquitylation regulates alternative splicing and cell migration in egf signaling. Cell Res. 2017, 27, 540–558. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Bi, F.; Zhou, X.; Zheng, Y. Rho GTPase regulation by mirnas and covalent modifications. Trends Cell Biol. 2012, 22, 365–373. [Google Scholar] [CrossRef]
- Olson, M.F. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases 2018, 9, 203–215. [Google Scholar] [CrossRef]
- Worby, C.A.; Mattoo, S.; Kruger, R.P.; Corbeil, L.B.; Koller, A.; Mendez, J.C.; Zekarias, B.; Lazar, C.; Dixon, J.E. The Fic domain: Regulation of cell signaling by adenylylation. Mol. Cell 2009, 34, 93–103. [Google Scholar] [CrossRef]
- Matos, P.; Skaug, J.; Marques, B.; Beck, S.; Verissimo, F.; Gespach, C.; Boavida, M.G.; Scherer, S.W.; Jordan, P. Small GTPase Rac1: Structure, localization, and expression of the human gene. Biochem. Biophys. Res. Commun. 2000, 277, 741–751. [Google Scholar] [CrossRef]
- Brandwein, D.; Wang, Z. Interaction between Rho GTPases and 14-3-3 proteins. Int. J. Mol. Sci. 2017, 18, 2148. [Google Scholar] [CrossRef] [PubMed]
- Buchsbaum, R.J. Rho activation at a glance. J. Cell Sci. 2007, 120, 1149–1152. [Google Scholar] [CrossRef] [PubMed]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by gefs, gaps, and gdis. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef] [PubMed]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Boureux, A.; Vignal, E.; Faure, S.; Fort, P. Evolution of the Rho family of Ras-like GTPases in eukaryotes. Mol. Biol. Evol. 2007, 24, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Bishop, A.L.; Hall, A. Rho GTPases and their effector proteins. Biochem. J. 2000, 348, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Rho family proteins: Coordinating cell responses. Trends Cell Biol. 2001, 11, 471–477. [Google Scholar] [CrossRef]
- Boettner, B.; Van Aelst, L. The role of Rho GTPases in disease development. Gene 2002, 286, 155–174. [Google Scholar] [CrossRef]
- Burridge, K.; Wennerberg, K. Rho and Rac take center stage. Cell 2004, 116, 167–179. [Google Scholar] [CrossRef]
- Pertz, O. Spatio-temporal Rho GTPase signaling—where are we now? J. Cell Sci. 2010, 123, 1841–1850. [Google Scholar] [CrossRef]
- Schwartz, M. Rho signalling at a glance. J. Cell Sci. 2004, 117, 5457–5458. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; van Unen, J. Molecular perturbation strategies to examine spatiotemporal features of Rho gef and Rho GTPase activity in living cells. Small GTPases 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, Q.; Wang, Y.; Chen, X.; Wang, Z. Plc-gamma1 and Rac1 coregulate egf-induced cytoskeleton remodeling and cell migration. Mol. Endocrinol. 2009, 23, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation of Rac1 t108 by extracellular signal-regulated kinase in response to epidermal growth factor: A novel mechanism to regulate Rac1 function. Mol. Cell. Biol. 2013, 33, 4538–4551. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Li, L.; Ballermann, B.; Wang, Z. Phosphorylation and activation of RhoA by ERK in response to epidermal growth factor stimulation. PLoS ONE 2016, 11, e0147103. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, S.M.; Awadia, S.; Garcia-Mata, R. I’m coming to gef you: Regulation of Rhogefs during cell migration. Cell Adhes. Migr. 2014, 8, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Marei, H.; Malliri, A. Gefs: Dual regulation of Rac1 signaling. Small GTPases 2017, 8, 90–99. [Google Scholar] [CrossRef]
- Schmidt, A.; Hall, A. Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes Dev. 2002, 16, 1587–1609. [Google Scholar] [CrossRef]
- DerMardirossian, C.; Bokoch, G. Gdis: Central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005, 15, 356–363. [Google Scholar] [CrossRef]
- Dovas, A.; Couchman, J. Rhogdi: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 2005, 390, 1–9. [Google Scholar] [CrossRef]
- Eden, S.; Rohatgi, R.; Podtelejnikov, A.V.; Mann, M.; Kirschner, M.W. Mechanism of regulation of wave1-induced actin nucleation by Rac1 and nck. Nature 2002, 418, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Soon, L.L.; Yie, T.A.; Shvarts, A.; Levine, A.J.; Su, F.; Tchou-Wong, K.M. Overexpression of wisp-1 down-regulated motility and invasion of lung cancer cells through inhibition of Rac activation. J. Biol. Chem. 2003, 278, 11465–11470. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J.; Comoglio, P.M.; Hall, A. Regulation of scatter factor/hepatocyte growth factor responses by Ras, Rac, and Rho in mdck cells. Mol. Cell. Biol. 1995, 15, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Lamorte, L.; Rodrigues, S.; Naujokas, M.; Park, M. Crk synergizes with epidermal growth factor for epithelial invasion and morphogenesis and is required for the met morphogenic program. J. Biol. Chem. 2002, 277, 37904–37911. [Google Scholar] [CrossRef] [PubMed]
- Wertheimer, E.; Gutierrez-Uzquiza, A.; Rosemblit, C.; Lopez-Haber, C.; Sosa, M.S.; Kazanietz, M.G. Rac signaling in breast cancer: A tale of gefs and gaps. Cell. Signal. 2012, 24, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Yarden, Y.; Sliwkowski, M.X. Untangling the erbb signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Citri, A.; Yarden, Y. Egf-erbb signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef]
- Wang, Z. Erbb receptors and cancer. Methods Mol. Biol. 2017, 1652, 3–35. [Google Scholar]
- Scita, G.; Nordstrom, J.; Carbone, R.; Tenca, P.; Giardina, G.; Gutkind, S.; Bjarnegard, M.; Betsholtz, C.; Di Fiore, P.P. Eps8 and e3b1 transduce signals from Ras to Rac. Nature 1999, 401, 290–293. [Google Scholar] [CrossRef]
- Marcoux, N.; Vuori, K. Egf receptor mediates adhesion-dependent activation of the Rac GTPase: A role for phosphatidylinositol 3-kinase and Vav2. Oncogene 2003, 22, 6100–6106. [Google Scholar] [CrossRef]
- Ray, R.M.; Vaidya, R.J.; Johnson, L.R. Mek/erk regulates adherens junctions and migration through Rac1. Cell Motil. Cytoskelet. 2007, 64, 143–156. [Google Scholar] [CrossRef]
- Itoh, R.E.; Kiyokawa, E.; Aoki, K.; Nishioka, T.; Akiyama, T.; Matsuda, M. Phosphorylation and activation of the Rac1 and Cdc42 gef asef in a431 cells stimulated by egf. J. Cell Sci. 2008, 121, 2635–2642. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Tsuji, S.; Sagara, M.; Echizen, K.; Shibata, Y.; Akiyama, T. Adenomatous polyposis coli and asef function downstream of hepatocyte growth factor and phosphatidylinositol 3-kinase. J. Biol. Chem. 2009, 284, 22436–22443. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.A.; Govek, E.E.; Bottner, B.; Van, A.L. Rho GTPases: Signaling, migration, and invasion. Exp. Cell Res. 2000, 261, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Ogura, Y.; Sawada, M.; Nakayama, R.; Takano, K.; Minato, Y.; Takemoto, Y.; Tashiro, E.; Watanabe, H.; Imoto, M. Involvement of 14-3-3 proteins in the second epidermal growth factor-induced wave of Rac1 activation in the process of cell migration. J. Biol. Chem. 2011, 286, 39259–39268. [Google Scholar] [CrossRef] [PubMed]
- Magi, S.; Takemoto, Y.; Kobayashi, H.; Kasamatsu, M.; Akita, T.; Tanaka, A.; Takano, K.; Tashiro, E.; Igarashi, Y.; Imoto, M. 5-Lipoxygenase and cysteinyl leukotriene receptor 1 regulate epidermal growth factor-induced cell migration through tiam1 upregulation and Rac1 activation. Cancer Sci. 2014, 105, 290–296. [Google Scholar] [CrossRef]
- Liu, B.P.; Burridge, K. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol. Cell. Biol. 2000, 20, 7160–7169. [Google Scholar] [CrossRef]
- Feng, H.; Hu, B.; Jarzynka, M.J.; Li, Y.; Keezer, S.; Johns, T.G.; Tang, C.K.; Hamilton, R.L.; Vuori, K.; Nishikawa, R.; et al. Phosphorylation of dedicator of cytokinesis 1 (dock180) at tyrosine residue y722 by src family kinases mediates egfrviii-driven glioblastoma tumorigenesis. Proceed. Natl. Acad. Sci. USA 2012, 109, 3018–3023. [Google Scholar] [CrossRef]
- Feng, H.; Hu, B.; Vuori, K.; Sarkaria, J.N.; Furnari, F.B.; Cavenee, W.K.; Cheng, S.Y. Egfrviii stimulates glioma growth and invasion through pka-dependent serine phosphorylation of dock180. Oncogene 2014, 33, 2504–2512. [Google Scholar] [CrossRef]
- Zhu, G.; Fan, Z.; Ding, M.; Zhang, H.; Mu, L.; Ding, Y.; Zhang, Y.; Jia, B.; Chen, L.; Chang, Z.; et al. An egfr/pi3k/akt axis promotes accumulation of the Rac1-gef tiam1 that is critical in egfr-driven tumorigenesis. Oncogene 2015, 34, 5971–5982. [Google Scholar] [CrossRef]
- Trenkle, T.; Hakim, S.G.; Jacobsen, H.C.; Sieg, P. Differential gene expression of the proto-oncogene vav3 and the transcript variant vav3.1 in oral squamous cell carcinoma. Anticancer Res. 2015, 35, 2593–2600. [Google Scholar] [PubMed]
- Duan, L.; Raja, S.M.; Chen, G.; Virmani, S.; Williams, S.H.; Clubb, R.J.; Mukhopadhyay, C.; Rainey, M.A.; Ying, G.; Dimri, M.; et al. Negative regulation of egfr-vav2 signaling axis by cbl ubiquitin ligase controls egf receptor-mediated epithelial cell adherens junction dynamics and cell migration. J. Biol. Chem. 2011, 286, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Makino, Y.; Tsuda, M.; Ichihara, S.; Watanabe, T.; Sakai, M.; Sawa, H.; Nagashima, K.; Hatakeyama, S.; Tanaka, S. Elmo1 inhibits ubiquitylation of dock180. J. Cell Sci. 2006, 119, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Singh, S.; Georgescu, M.M.; Birge, R.B. A role for mer tyrosine kinase in alphavbeta5 integrin-mediated phagocytosis of apoptotic cells. J. Cell Sci. 2005, 118, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Misek, S.A.; Chen, J.; Schroeder, L.; Rattanasinchai, C.; Sample, A.; Sarkaria, J.N.; Gallo, K.A. Egfr signals through a dock180-mlk3 axis to drive glioblastoma cell invasion. Mol. Cancer Res. 2017, 15, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Caloca, M.J.; Wang, H.; Kazanietz, M.G. Characterization of the Rac-gap (Rac-GTPase-activating protein) activity of beta2-chimaerin, a ‘non-protein kinase c’ phorbol ester receptor. Biochem. J. 2003, 375, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, C.; Leskow, F.C.; Sun, J.; Canagarajah, B.; Hurley, J.H.; Kazanietz, M.G. Phospholipase cgamma/diacylglycerol-dependent activation of beta2-chimaerin restricts egf-induced Rac signaling. EMBO J. 2006, 25, 2062–2074. [Google Scholar] [CrossRef]
- Wang, H.; Kazanietz, M.G. P23/tmp21 differentially targets the Rac-gap beta2-chimaerin and protein kinase c via their c1 domains. Mol. Biol. Cell 2010, 21, 1398–1408. [Google Scholar] [CrossRef]
- Griner, E.M.; Caino, M.C.; Sosa, M.S.; Colon-Gonzalez, F.; Chalmers, M.J.; Mischak, H.; Kazanietz, M.G. A novel cross-talk in diacylglycerol signaling: The Rac-gap beta2-chimaerin is negatively regulated by protein kinase cdelta-mediated phosphorylation. J. Biol. Chem. 2010, 285, 16931–16941. [Google Scholar] [CrossRef]
- Yamada, H.; Tsutsumi, K.; Nakazawa, Y.; Shibagaki, Y.; Hattori, S.; Ohta, Y. Src family tyrosine kinase signaling regulates filgap through association with rbm10. PLoS ONE 2016, 11, e0146593. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Chang, J.Y.; Chang, K.Y.; Chang, W.C.; Chen, B.K. Epidermal growth factor-induced pyruvate dehydrogenase kinase 1 expression enhances head and neck squamous cell carcinoma metastasis via up-regulation of fibronectin. FASEB 2017, 31, 4265–4276. [Google Scholar] [CrossRef] [PubMed]
- Caggia, S.; Chunduri, H.; Millena, A.C.; Perkins, J.N.; Venugopal, S.V.; Vo, B.T.; Li, C.; Tu, Y.; Khan, S.A. Novel role of gialpha2 in cell migration: Downstream of pi3-kinase-akt and Rac1 in prostate cancer cells. J. Cell. Physiol. 2018, 234, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Shen, N.; Jiang, X.; Sun, H.; Xu, N.; Zhou, D.; Nong, L.; Ren, K. Periodic mechanical stress activates egfr-dependent Rac1 mitogenic signals in rat nucleus pulpous cells via ERK1/2. Biochem. Biophys. Res. Commun. 2016, 469, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Lof-Ohlin, Z.M.; Nyeng, P.; Bechard, M.E.; Hess, K.; Bankaitis, E.; Greiner, T.U.; Ameri, J.; Wright, C.V.; Semb, H. Egfr signalling controls cellular fate and pancreatic organogenesis by regulating apicobasal polarity. Nat. Cell Biol. 2017, 19, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Kyriakakis, E.; Maslova, K.; Frachet, A.; Ferri, N.; Contini, A.; Pfaff, D.; Erne, P.; Resink, T.J.; Philippova, M. Cross-talk between egfr and t-cadherin: Egfr activation promotes t-cadherin localization to intercellular contacts. Cell. Signal. 2013, 25, 1044–1053. [Google Scholar] [CrossRef] [PubMed]
- Morrison Joly, M.; Williams, M.M.; Hicks, D.J.; Jones, B.; Sanchez, V.; Young, C.D.; Sarbassov, D.D.; Muller, W.J.; Brantley-Sieders, D.; Cook, R.S. Two distinct mtorc2-dependent pathways converge on Rac1 to drive breast cancer metastasis. Breast Cancer Res. 2017, 19, 74. [Google Scholar] [CrossRef]
- Samuel, F.; Hynds, D.L. Rho GTPase signaling for axon extension: Is prenylation important? Mol. Neurobiol. 2010, 42, 133–142. [Google Scholar] [CrossRef]
- Kinsella, B.; Erdman, R.; Maltese, W. Carboxyl-terminal isoprenylation of Ras-related GTP-binding proteins encoded by Rac1, Rac2, and rala. J. Biol. Chem. 1991, 266, 9786–9794. [Google Scholar]
- Berg, T.J.; Gastonguay, A.J.; Lorimer, E.L.; Kuhnmuench, J.R.; Li, R.; Fields, A.P.; Williams, C.L. Splice variants of smggds control small GTPase prenylation and membrane localization. J. Biol. Chem. 2010, 285, 35255–35266. [Google Scholar] [CrossRef]
- Sharrocks, A.; Yang, S.; Galanis, A. Docking domains and substrate-specificity determination for map kinases. Trends Biochem. Sci. 2000, 25, 448–453. [Google Scholar] [CrossRef]
- Enslen, H.; Davis, R. Regulation of map kinases by docking domains. Biol. Cell 2001, 93, 5–14. [Google Scholar] [CrossRef]
- Navarro-Lerida, I.; Sanchez-Perales, S.; Calvo, M.; Rentero, C.; Zheng, Y.; Enrich, C.; Del Pozo, M.A. A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 2012, 31, 534–551. [Google Scholar] [CrossRef] [PubMed]
- Rocks, O.; Gerauer, M.; Vartak, N.; Koch, S.; Huang, Z.P.; Pechlivanis, M.; Kuhlmann, J.; Brunsveld, L.; Chandra, A.; Ellinger, B.; et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 2010, 141, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Torrino, S.; Visvikis, O.; Doye, A.; Boyer, L.; Stefani, C.; Munro, P.; Bertoglio, J.; Gacon, G.; Mettouchi, A.; Lemichez, E. The e3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev. Cell 2011, 21, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Lluva, S.; Tan, C.T.; Daugaard, M.; Sorensen, P.H.; Malliri, A. The tumour suppressor HACE1 controls cell migration by regulating Rac1 degradation. Oncogene 2013, 32, 1735–1742. [Google Scholar] [CrossRef] [PubMed]
- Oberoi, T.K.; Dogan, T.; Hocking, J.C.; Scholz, R.P.; Mooz, J.; Anderson, C.L.; Karreman, C.; Meyer zu Heringdorf, D.; Schmidt, G.; Ruonala, M.; et al. Iaps regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012, 31, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, J.; Wei, J.; Bowser, R.K.; Khoo, A.; Liu, Z.; Luketich, J.D.; Pennathur, A.; Ma, H.; Zhao, Y. F-box protein complex fbxl19 regulates tgfbeta1-induced e-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol. Cancer 2014, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- Goka, E.T.; Lippman, M.E. Loss of the e3 ubiquitin ligase HACE1 results in enhanced Rac1 signaling contributing to breast cancer progression. Oncogene 2015, 34, 5395–5405. [Google Scholar] [CrossRef] [PubMed]
- Loirand, G.; Guilluy, C.; Pacaud, P. Regulation of Rho proteins by phosphorylation in the cardiovascular system. Trends Cariovasc. Med. 2006, 16, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Kwon, D.; Chun, J.; Kim, J.; Kang, S. Akt protein kinase inhibits Rac1-GTP binding through phosphorylation at serine 71 of Rac1. J. Biol. Chem. 2000, 275, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Schoentaube, J.; Olling, A.; Tatge, H.; Just, I.; Gerhard, R. Serine-71 phosphorylation of Rac1/Cdc42 diminishes the pathogenic effect of clostridium difficile toxin a. Cell. Microbiol. 2009, 11, 1816–1826. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.; Proff, J.; Havemeier, A.; Ladwein, M.; Rottner, K.; Barlag, B.; Pich, A.; Tatge, H.; Just, I.; Gerhard, R. Serine-71 phosphorylation of Rac1 modulates downstream signaling. PLoS ONE 2012, 7, e44358. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.; Lemmon, C.; Lietha, D.; Eck, M.; Romer, L. Tyrosine phosphorylation of Rac1: A role in regulation of cell spreading. PLoS ONE 2011, 6, e28587. [Google Scholar] [CrossRef] [PubMed]
- Moissoglu, K.; Schwartz, M.A. Spatial and temporal control of Rho GTPase functions. Cellular Logist. 2014, 4, e943618. [Google Scholar] [CrossRef] [PubMed]
- Palamidessi, A.; Frittoli, E.; Garre, M.; Faretta, M.; Mione, M.; Testa, I.; Diaspro, A.; Lanzetti, L.; Scita, G.; Di Fiore, P.P. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell 2008, 134, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Kraynov, V.S.; Chamberlain, C.; Bokoch, G.M.; Schwartz, M.A.; Slabaugh, S.; Hahn, K.M. Localized Rac activation dynamics visualized in living cells. Science 2000, 290, 333–337. [Google Scholar] [CrossRef]
- Ando, S.; Kaibuchi, K.; Sasaki, T.; Hiraoka, K.; Nishiyama, T.; Mizuno, T.; Asada, M.; Nunoi, H.; Matsuda, I.; Matsuura, Y.; et al. Post-translational processing of Rac p21s is important both for their interaction with the GDP/GTP exchange proteins and for their activation of nadph oxidase. J. Biol. Chem. 1992, 267, 25709–25713. [Google Scholar]
- Afshordel, S.; Wood, W.G.; Igbavboa, U.; Muller, W.E.; Eckert, G.P. Impaired geranylgeranyltransferase-i regulation reduces membrane-associated Rho protein levels in aged mouse brain. J. Neurochem. 2014, 129, 732–742. [Google Scholar] [CrossRef]
- Williams, C. He polybasic region of Ras and Rho family small GTPases: A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003, 15, 1071–1080. [Google Scholar] [CrossRef]
- Michaely, P.A.; Mineo, C.; Ying, Y.S.; Anderson, R.G. Polarized distribution of endogenous Rac1 and RhoA at the cell surface. J. Biol. Chem. 1999, 274, 21430–21436. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Eng, C.H.; Schlaepfer, D.D.; Marcantonio, E.E.; Gundersen, G.G. Localized stabilization of microtubules by integrin- and fak-facilitated Rho signaling. Science 2004, 303, 836–839. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, M.; Alderson, N.; Kiosses, W.; Chiang, H.; Anderson, R.; Schwartz, M. Integrins regulate Rac targeting by internalization of membrane domains. Science 2004, 303, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Prior, I.A.; Muncke, C.; Parton, R.G.; Hancock, J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003, 160, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.D.; Philips, M.R. Rac1 gets fattier. EMBO J. 2012, 31, 517–518. [Google Scholar] [CrossRef] [PubMed]
- Gungor, B.; Gombos, I.; Crul, T.; Ayaydin, F.; Szabo, L.; Torok, Z.; Mates, L.; Vigh, L.; Horvath, I. Rac1 participates in thermally induced alterations of the cytoskeleton, cell morphology and lipid rafts, and regulates the expression of heat shock proteins in b16f10 melanoma cells. PLoS ONE 2014, 9, e89136. [Google Scholar] [CrossRef]
- Lanning, C.C.; Daddona, J.L.; Ruiz-Velasco, R.; Shafer, S.H.; Williams, C.L. The Rac1 F-terminal polybasic region regulates the nuclear localization and protein degradation of Rac1. J. Biol. Chem. 2004, 279, 44197–44210. [Google Scholar] [CrossRef]
- Michaelson, D.; Abidi, W.; Guardavaccaro, D.; Zhou, M.; Ahearn, I.; Pagano, M.; Philips, M.R. Rac1 accumulates in the nucleus during the g2 phase of the cell cycle and promotes cell division. J. Cell Biol. 2008, 181, 485–496. [Google Scholar] [CrossRef]
- Dubash, A.D.; Guilluy, C.; Srougi, M.C.; Boulter, E.; Burridge, K.; Garcia-Mata, R. The small GTPase RhoA localizes to the nucleus and is activated by net1 and DNA damage signals. PLoS ONE 2011, 6, e17380. [Google Scholar] [CrossRef]
- Lanning, C.; Ruiz-Velasco, R.; Williams, C. Novel mechanism of the co-regulation of nuclear transport of smggds and Rac1. J. Biol. Chem. 2003, 278, 12495–12506. [Google Scholar] [CrossRef]
- Sandrock, K.; Bielek, H.; Schradi, K.; Schmidt, G.; Klugbauer, N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic 2010, 11, 198–209. [Google Scholar] [CrossRef]
- Baldassare, J.J.; Jarpe, M.B.; Alferes, L.; Raben, D.M. Nuclear translocation of RhoA mediates the mitogen-induced activation of phospholipase d involved in nuclear envelope signal transduction. J. Biol. Chem. 1997, 272, 4911–4914. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Chen, Y.C.; Lan, T.; Qian, H.; Wang, Y.; Jiang, L. Lps-induced nuclear translocation of RhoA is dependent on nf-kappab in the human lung cancer cell line a549. Oncol. Lett. 2012, 3, 1283–1287. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Yang, X.; Chen, Y.; Chen, M. Nuclear translocation of small g protein RhoA via active transportation in gastric cancer cells. Oncol. Rep. 2013, 30, 1878–1882. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Chen, Y.C.; Li, Y.Y.; Yang, S.Q.; Xu, W.R. Localization and translocation of RhoA protein in the human gastric cancer cell line sgc-7901. World J. Gastroenterol. 2008, 14, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y.; Xu, J. Factors influencing RhoA protein distribution in the nucleus. Mol. Med. Rep. 2011, 4, 1115–1119. [Google Scholar] [PubMed]
- Habets, G.G.; Scholtes, E.H.; Zuydgeest, D.; van der Kammen, R.A.; Stam, J.C.; Berns, A.; Collard, J.G. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell 1994, 77, 537–549. [Google Scholar] [CrossRef]
- Bustelo, X.R. The Vav family of signal transduction molecules. Crit. Rev. Oncog. 1996, 7, 65–88. [Google Scholar] [CrossRef]
- Bustelo, X.R. Regulatory and signaling properties of the vav family. Mol. Cell. Biol. 2000, 20, 1461–1477. [Google Scholar] [CrossRef]
- Mertens, A.E.; Roovers, R.C.; Collard, J.G. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003, 546, 11–16. [Google Scholar] [CrossRef]
- Hinz, M.; Krappmann, D.; Eichten, A.; Heder, A.; Scheidereit, C.; Strauss, M. Nf-kappab function in growth control: Regulation of cyclin d1 expression and g0/g1-to-s-phase transition. Mol. Cell. Biol. 1999, 19, 2690–2698. [Google Scholar] [CrossRef]
- Simon, A.R.; Vikis, H.G.; Stewart, S.; Fanburg, B.L.; Cochran, B.H.; Guan, K.L. Regulation of stat3 by direct binding to the Rac1 GTPase. Science 2000, 290, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Bao, Y.C.; Nomura, Y.; Moon, Y.; Tonozuka, Y.; Minoshima, Y.; Hatori, T.; Tsuchiya, A.; Kiyono, M.; Nosaka, T.; et al. Rac1 and a GTPase-activating protein, mgcracgap, are required for nuclear translocation of stat transcription factors. J. Cell Biol. 2006, 175, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Lerida, I.; Pellinen, T.; Sanchez, S.A.; Guadamillas, M.C.; Wang, Y.; Mirtti, T.; Calvo, E.; Del Pozo, M.A. Rac1 nucleocytoplasmic shuttling drives nuclear shape changes and tumor invasion. Dev. Cell 2015, 32, 318–334. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdrabou, A.; Wang, Z. Post-Translational Modification and Subcellular Distribution of Rac1: An Update. Cells 2018, 7, 263. https://doi.org/10.3390/cells7120263
Abdrabou A, Wang Z. Post-Translational Modification and Subcellular Distribution of Rac1: An Update. Cells. 2018; 7(12):263. https://doi.org/10.3390/cells7120263
Chicago/Turabian StyleAbdrabou, Abdalla, and Zhixiang Wang. 2018. "Post-Translational Modification and Subcellular Distribution of Rac1: An Update" Cells 7, no. 12: 263. https://doi.org/10.3390/cells7120263
APA StyleAbdrabou, A., & Wang, Z. (2018). Post-Translational Modification and Subcellular Distribution of Rac1: An Update. Cells, 7(12), 263. https://doi.org/10.3390/cells7120263




