2D- and 3D-Based Intestinal Stem Cell Cultures for Personalized Medicine
Abstract
:1. Introduction
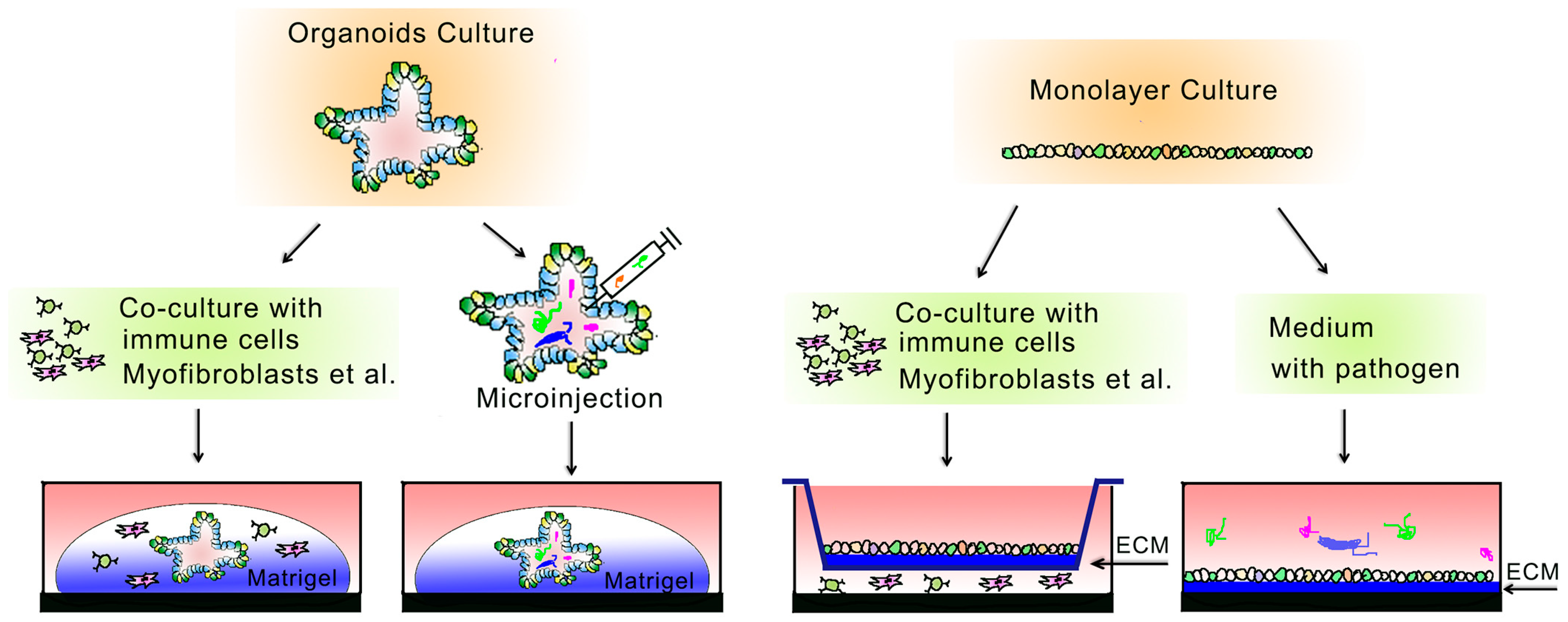
2. 3D-Based Organoid Culture System
3. 2D-Based Culture System
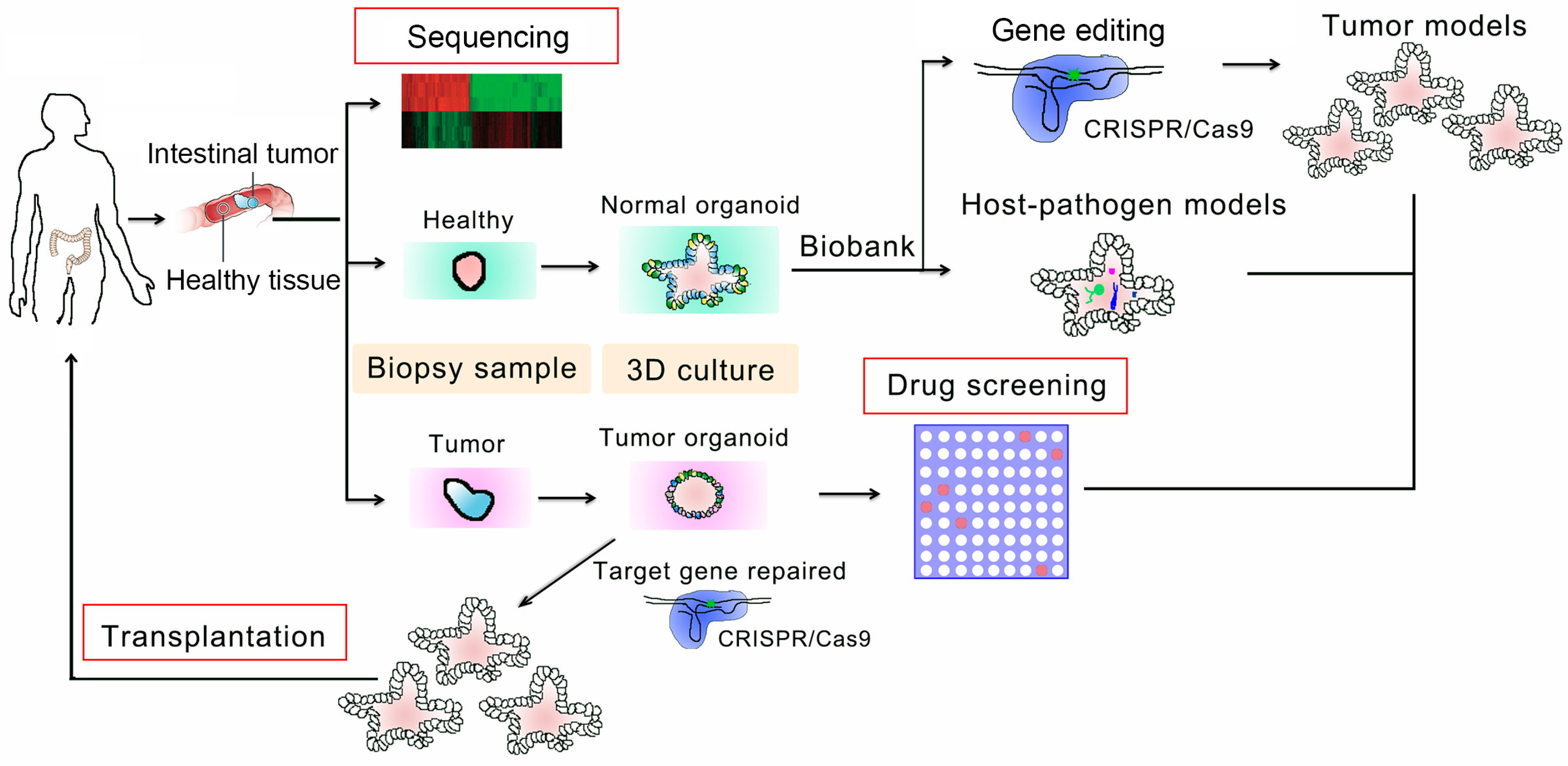
4. Applications in Personalized Therapy
5. Challenges, Limitations, and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Chen, Y.G. Regulation of intestinal stem cell fate specification. Sci. China Life Sci. 2015, 58, 570–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Nusse, Y.M.; Savage, A.K.; Marangoni, P.; Rosendahl-Huber, A.K.M.; Landman, T.A.; de Sauvage, F.J.; Locksley, R.M.; Klein, O.D. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 2018, 559, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; Snippert, H.J. Stem cell dynamics in homeostasis and cancer of the intestine. Nat. Rev. Cancer 2014, 14, 468–480. [Google Scholar] [CrossRef] [PubMed]
- van der Flier, L.G.; Clevers, H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009, 71, 241–260. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.C.; Bevins, C.L. Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 2013, 75, 289–311. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Colman, M.J.; Schewe, M.; Meerlo, M.; Stigter, E.; Gerrits, J.; Pras-Raves, M.; Sacchetti, A.; Hornsveld, M.; Oost, K.C.; Snippert, H.J.; et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Beumer, J.; Clevers, H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 2016, 143, 3639–3649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, J.P. Epithelial cell migration in the intestine. Cell. Biol. Int. 1996, 20, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.H.; Sato, T.; He, X.C.; Li, L. Current view: Intestinal stem cells and signaling. Gastroenterology 2008, 134, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Schuijers, J.; Clevers, H. Adult mammalian stem cells: The role of Wnt, Lgr5 and R-spondins. EMBO J. 2012, 31, 2685–2696. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Li, Y.; Zhao, B.; Xu, C.; Liu, Y.; Li, H.; Zhang, B.; Wang, X.; Yang, X.; Xie, W.; et al. BMP restricts stemness of intestinal Lgr5(+) stem cells by directly suppressing their signature genes. Nat. Commun. 2017, 8, 13824. [Google Scholar] [CrossRef] [PubMed]
- Beumer, J.; Artegiani, B.; Post, Y.; Reimann, F.; Gribble, F.; Nguyen, T.N.; Zeng, H.; Van den Born, M.; Van Es, J.H.; Clevers, H. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell. Biol. 2018, 20, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, Y.G. BMP signaling in homeostasis, transformation and inflammatory response of intestinal epithelium. Sci. China Life Sci. 2018, 61, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Tsuchiya, K.; Watanabe, M. Crosstalk between Wnt and Notch signaling in intestinal epithelial cell fate decision. J. Gastroenterol. 2007, 42, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Stanger, B.Z.; Datar, R.; Murtaugh, L.C.; Melton, D.A. Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. USA 2005, 102, 12443–12448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho, E.; Batlle, E.; Clevers, H. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell. Dev. Biol. 2004, 20, 695–723. [Google Scholar] [CrossRef] [PubMed]
- Le Bouteiller, M.; Jensen, K.B. Hippo signalling directs intestinal fate. Nat. Cell. Biol. 2015, 17, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Fodde, R.; Smits, R.; Clevers, H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer 2001, 1, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Kedrin, D.; Gala, M.K. Genetics of the serrated pathway to colorectal cancer. Clin. Transl. Gastroenterol. 2015, 6, e84. [Google Scholar] [CrossRef] [PubMed]
- Rutter, M.; Saunders, B.; Wilkinson, K.; Rumbles, S.; Schofield, G.; Kamm, M.; Williams, C.; Price, A.; Talbot, I.; Forbes, A. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology 2004, 126, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Jess, T.; Loftus, E.V., Jr.; Velayos, F.S.; Harmsen, W.S.; Zinsmeister, A.R.; Smyrk, T.C.; Schleck, C.D.; Tremaine, W.J.; Melton, L.J., 3rd; Munkholm, P.; et al. Risk of intestinal cancer in inflammatory bowel disease: A population-based study from olmsted county, Minnesota. Gastroenterology 2006, 130, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.B.; Harpaz, N.; Itzkowitz, S.; Hossain, S.; Matula, S.; Kornbluth, A.; Bodian, C.; Ullman, T. Histologic inflammation is a risk factor for progression to colorectal neoplasia in ulcerative colitis: A cohort study. Gastroenterology 2007, 133, 1099–1105; quiz 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Kiesler, P.; Fuss, I.J.; Strober, W. Experimental Models of Inflammatory Bowel Diseases. Cell. Mol. Gastroenterol. Hepatol. 2015, 1, 154–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Kohno, H.; Suzuki, R.; Yamada, Y.; Sugie, S.; Mori, H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003, 94, 965–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abba, K.; Sinfield, R.; Hart, C.A.; Garner, P. Pathogens associated with persistent diarrhoea in children in low and middle income countries: Systematic review. BMC Infect. Dis. 2009, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.M.; McLaws, M.L.; Ellis, J.T. Prevalence of gastrointestinal pathogens in developed and developing countries: Systematic review and meta-analysis. J. Public Health Res. 2013, 2, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, X.; Han, C.; Wan, G.; Huang, X.; Ivan, C.; Jiang, D.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Rao, P.H.; et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature 2015, 520, 697–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563. [Google Scholar] [CrossRef] [PubMed]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, B.A.; Rietze, R.L. Neural stem cells and neurospheres—Re-evaluating the relationship. Nat. Methods 2005, 2, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Dontu, G.; Abdallah, W.M.; Foley, J.M.; Jackson, K.W.; Clarke, M.F.; Kawamura, M.J.; Wicha, M.S. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003, 17, 1253–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Dai, Y.; Shu, J.; Yu, R.; Guo, Y.; Chen, J. Spheroid cultures promote the stemness of corneal stromal cells. Tissue Cell 2015, 47, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Shevde, N.K.; Mael, A.A. Techniques in embryoid body formation from human pluripotent stem cells. Methods Mol. Biol. 2013, 946, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Aboulkheyr Es, H.; Montazeri, L.; Aref, A.R.; Vosough, M.; Baharvand, H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018, 36, 358–371. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Clevers, H. Growing self-organizing mini-guts from a single intestinal stem cell: Mechanism and applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Leushacke, M.; Barker, N. Ex vivo culture of the intestinal epithelium: Strategies and applications. Gut 2014, 63, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- de Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Degirmenci, B.; Valenta, T.; Dimitrieva, S.; Hausmann, G.; Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 2018, 558, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Shoshkes-Carmel, M.; Wang, Y.J.; Wangensteen, K.J.; Toth, B.; Kondo, A.; Massasa, E.E.; Itzkovitz, S.; Kaestner, K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 2018, 557, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; O’Leary, C.E.; von Moltke, J.; Liang, H.E.; Ang, Q.Y.; Turnbaugh, P.J.; Radhakrishnan, S.; Pellizzon, M.; Ma, A.; Locksley, R.M. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 2018, 174, 271–284.e14. [Google Scholar] [CrossRef] [PubMed]
- Basak, O.; Beumer, J.; Wiebrands, K.; Seno, H.; van Oudenaarden, A.; Clevers, H. Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells. Cell Stem Cell 2017, 20, 177–190.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kim, R.; Hinman, S.S.; Zwarycz, B.; Magness, S.T.; Allbritton, N.L. Bioengineered Systems and Designer Matrices That Recapitulate the Intestinal Stem Cell Niche. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 440–453.e1. [Google Scholar] [CrossRef] [PubMed]
- Beyaz, S.; Mana, M.D.; Roper, J.; Kedrin, D.; Saadatpour, A.; Hong, S.J.; Bauer-Rowe, K.E.; Xifaras, M.E.; Akkad, A.; Arias, E.; et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016, 531, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozuka, K.; He, Y.; Koo-McCoy, S.; Kumaraswamy, P.; Nie, B.; Shaw, K.; Chan, P.; Leadbetter, M.; He, L.; Lewis, J.G.; et al. Development and Characterization of a Human and Mouse Intestinal Epithelial Cell Monolayer Platform. Stem Cell Reports 2017, 9, 1976–1990. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Clevers, H. Organoid culture systems to study host-pathogen interactions. Curr. Opin. Immunol. 2017, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Jobin, C. Human Intestinal Microbiota and Colorectal Cancer: Moving Beyond Associative Studies. Gastroenterology 2017, 153, 1475–1478. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Qi, Z.; Li, X.; Du, Y.; Chen, Y.G. Monolayer culture of intestinal epithelium sustains Lgr5(+) intestinal stem cells. Cell Discov. 2018, 4, 32. [Google Scholar] [CrossRef] [PubMed]
- Puzan, M.; Hosic, S.; Ghio, C.; Koppes, A. Enteric Nervous System Regulation of Intestinal Stem Cell Differentiation and Epithelial Monolayer Function. Sci. Rep. 2018, 8, 6313. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Rouch, J.D.; Jabaji, Z.; Khalil, H.A.; Solorzano, S.; Lewis, M.; Martin, M.G.; Stelzner, M.G.; Dunn, J.C. Long-term renewable human intestinal epithelial stem cells as monolayers: A potential for clinical use. J. Pediatr. Surg. 2016, 51, 995–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorne, C.A.; Chen, I.W.; Sanman, L.E.; Cobb, M.H.; Wu, L.F.; Altschuler, S.J. Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev. Cell 2018, 44, 624–633.e4. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Martyn, K.; Yang, A.; Yin, X.; Mead, B.E.; Joshi, N.; Sherman, N.E.; Langer, R.S.; Karp, J.M. Towards a defined ECM and small molecule based monolayer culture system for the expansion of mouse and human intestinal stem cells. Biomaterials 2018, 154, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; DiSalvo, M.; Gunasekara, D.B.; Dutton, J.; Proctor, A.; Lebhar, M.S.; Williamson, I.A.; Speer, J.; Howard, R.L.; Smiddy, N.M.; et al. Self-renewing Monolayer of Primary Colonic or Rectal Epithelial Cells. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 165–182.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Qi, Z.; Li, Y.; Wang, C.; Fu, W.; Chen, Y.G. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 2015, 6, 7166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Yamamoto, Y.; Wilson, L.H.; Zhang, T.; Howitt, B.E.; Farrow, M.A.; Kern, F.; Ning, G.; Hong, Y.; Khor, C.C.; et al. Cloning and variation of ground state intestinal stem cells. Nature 2015, 522, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordonez-Moran, P.; Clevers, H.; Lutolf, M.P. Designer matrices for intestinal stem cell and organoid culture. Nature 2016, 539, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Braverman, J.; Yilmaz, O.H. From 3D Organoids back to 2D Enteroids. Dev. Cell 2018, 44, 533–534. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- Praharaj, P.P.; Bhutia, S.K.; Nagrath, S.; Bitting, R.L.; Deep, G. Circulating tumor cell-derived organoids: Current challenges and promises in medical research and precision medicine. Biochim. Biophys. Acta. Rev. Cancer 2018, 1869, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef] [PubMed]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Verissimo, C.S.; Overmeer, R.M.; Ponsioen, B.; Drost, J.; Mertens, S.; Verlaan-Klink, I.; Gerwen, B.V.; van der Ven, M.; Wetering, M.V.; Egan, D.A.; et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. Elife 2016, 5, e18489. [Google Scholar] [CrossRef] [PubMed]
- Schwank, G.; Koo, B.K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef] [PubMed]
- VanDussen, K.L.; Marinshaw, J.M.; Shaikh, N.; Miyoshi, H.; Moon, C.; Tarr, P.I.; Ciorba, M.A.; Stappenbeck, T.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 2015, 64, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model To Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Bijvelds, M.; Dang, W.; Xu, L.; van der Eijk, A.A.; Knipping, K.; Tuysuz, N.; Dekkers, J.F.; Wang, Y.; de Jonge, J.; et al. Modeling rotavirus infection and antiviral therapy using primary intestinal organoids. Antiviral Res. 2015, 123, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef] [PubMed]
- May, P.; Evans, S.; Parry, L. Organoids, organs-on-chips and other systems, and microbiota. Emerg. Top. Life Sci. 2017, 1, 385–400. [Google Scholar] [CrossRef] [Green Version]
- Voest, E.E.; Bernards, R. DNA-Guided Precision Medicine for Cancer: A Case of Irrational Exuberance? Cancer Discov. 2016, 6, 130–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayakawa, Y.; Sakitani, K.; Konishi, M.; Asfaha, S.; Niikura, R.; Tomita, H.; Renz, B.W.; Tailor, Y.; Macchini, M.; Middelhoff, M.; et al. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell 2017, 31, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yissachar, N.; Zhou, Y.; Ung, L.; Lai, N.Y.; Mohan, J.F.; Ehrlicher, A.; Weitz, D.A.; Kasper, D.L.; Chiu, I.M.; Mathis, D.; et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell 2017, 168, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Liu, B.; Wang, J.; Wei, S.; Qi, Z.; Wang, S.; Fu, W.; Chen, Y.G. A growth factor-free culture system underscores the coordination between Wnt and BMP signaling in Lgr5(+) intestinal stem cell maintenance. Cell Discov. 2018, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Takebe, T.; Wells, J.M.; Helmrath, M.A.; Zorn, A.M. Organoid Center Strategies for Accelerating Clinical Translation. Cell Stem Cell 2018, 22, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.F.; Wiegerinck, C.L.; de Jonge, H.R.; Bronsveld, I.; Janssens, H.M.; de Winter-de Groot, K.M.; Brandsma, A.M.; de Jong, N.W.; Bijvelds, M.J.; Scholte, B.J.; et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013, 19, 939–945. [Google Scholar] [CrossRef] [PubMed]
| 3D-Based Culture System | 2D-Based Culture System | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Spheroids | PDX | Organoids | Monolayer | Air-Liquid Interface Culture | |||||
| Culture Technique | Low-melting Agrose | Transplanted into immune-deficient mouse | Embended in Matrigel | Cell lines | Plate coated with Collagen I | Transwell coated with Collagen I | Plate coated with Matrigel | Seeded onto 3T3-J2 cells which cultured on transwell coated with 20% Matrigel | |
| Origin | Cell lines | Patient-derived tumors | mIEC; hIEC; Patient-derived tumors | Cell lines | mIEC; hIEC | mIEC; hIEC | mIEC; hIEC; Tumors | hIEC; Tumors | |
| Key Features | In vivo-like complexity | ✗ | ✓ | ✓ | ✗ | Lack of mature secretory lineage | Lack of mature secretory lineage | ✓ | ✓ |
| Stemness and multipotency | - | ✓ | ✓ | - | ✓ | ✓ | ✓ | ✓ | |
| Easily passaged | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ | |
| Applications | Cancer subtype modeling (gene manipulation) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Host-pathogen interaction | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Co-culture | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| High-throughput screening | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | |
| High-throughput imaging | ✓ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ | |
| Time consumption for modeling | Medium | Long | Medium | Short | Medium | Medium | Medium | Medium | |
| Cost benefits | Low | High | High | Low | Medium | Medium | Medium | High | |
| Reference | [34] | [34] | [34,35,36,39,47,58,59,63] | [31,32,34] | [47,55,59] | [47,54,59] | [47,50,53,59] | [57] | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, Y.-G. 2D- and 3D-Based Intestinal Stem Cell Cultures for Personalized Medicine. Cells 2018, 7, 225. https://doi.org/10.3390/cells7120225
Liu Y, Chen Y-G. 2D- and 3D-Based Intestinal Stem Cell Cultures for Personalized Medicine. Cells. 2018; 7(12):225. https://doi.org/10.3390/cells7120225
Chicago/Turabian StyleLiu, Yuan, and Ye-Guang Chen. 2018. "2D- and 3D-Based Intestinal Stem Cell Cultures for Personalized Medicine" Cells 7, no. 12: 225. https://doi.org/10.3390/cells7120225
APA StyleLiu, Y., & Chen, Y.-G. (2018). 2D- and 3D-Based Intestinal Stem Cell Cultures for Personalized Medicine. Cells, 7(12), 225. https://doi.org/10.3390/cells7120225





