Buc Maintains Maternal RNA Stability and Embryogenesis in Zebrafish
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Maintenance
2.2. Generation of Mutant Lines
2.3. Overexpression Analysis
2.4. Western Blot
2.5. Quantitative RT-PCR
2.6. Immunofluorescence
2.7. RNA-seq and Data Processing of High-Throughput Sequencing
2.8. Protein Structure and Molecular Docking Analysis
2.9. Antibody Production
2.10. Statistics and Reproducibility
3. Results
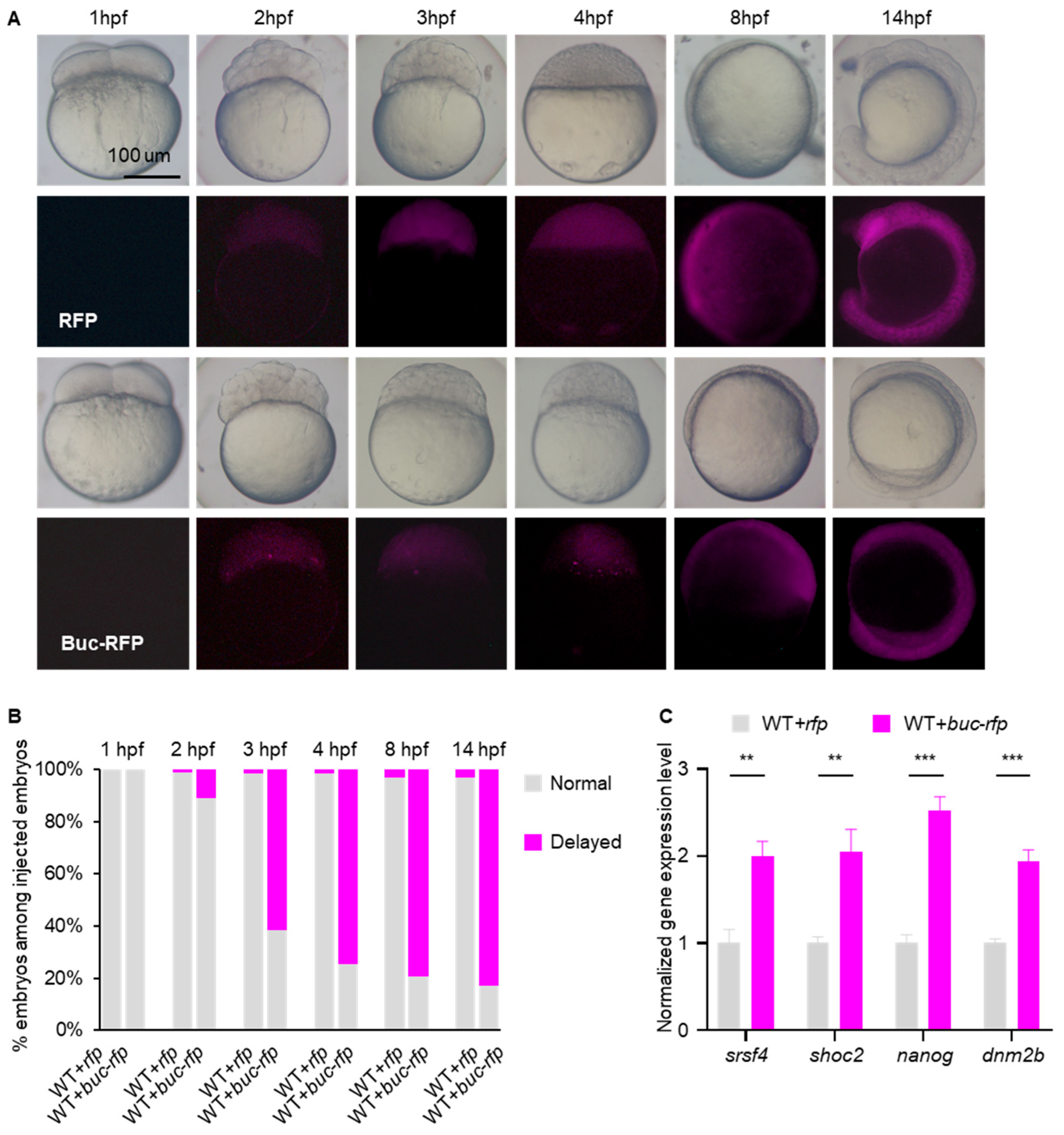
3.1. Overexpression of Buc Causes a Delay of Maternal mRNA Clearance and Embryo Development
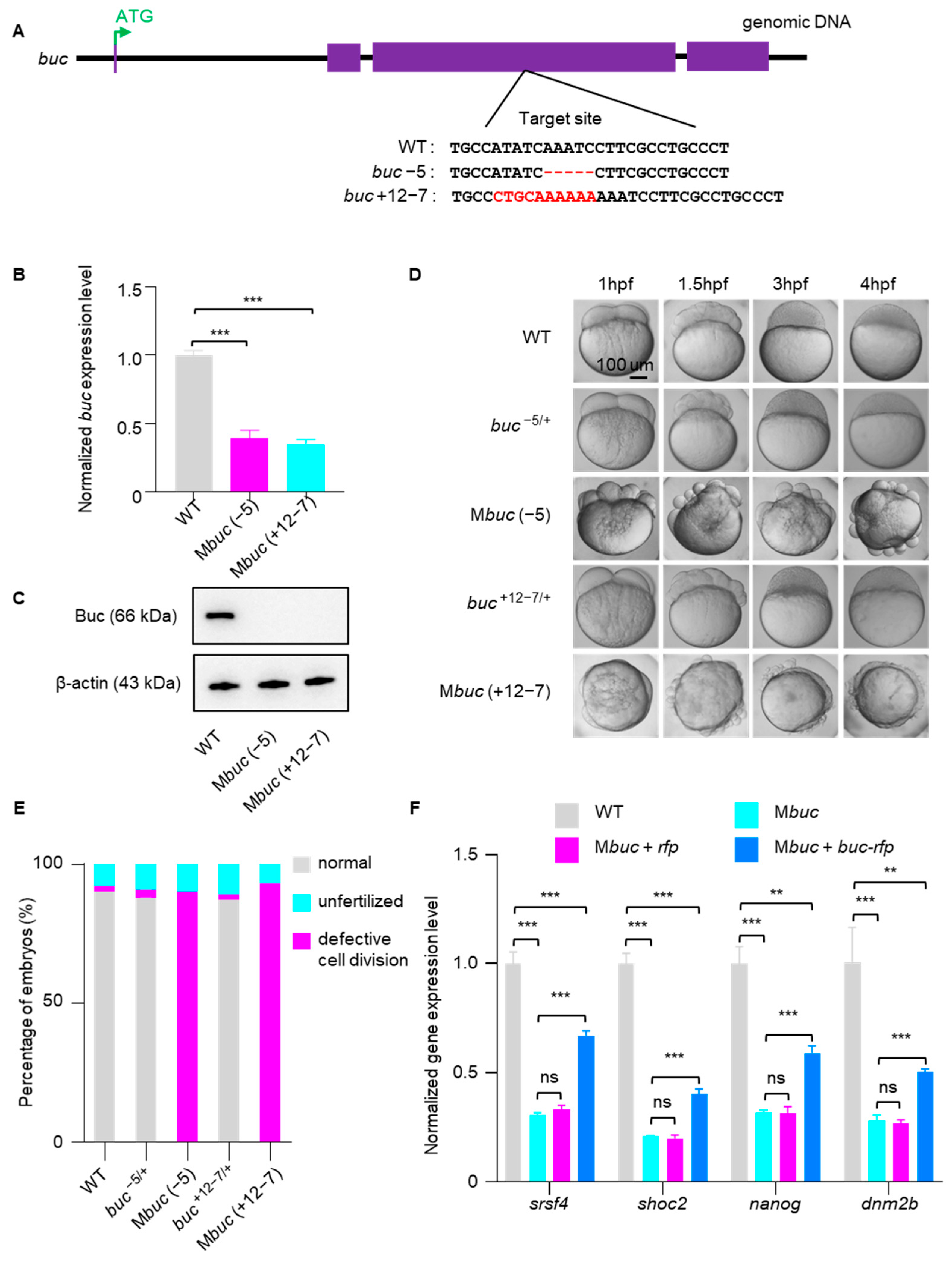
3.2. Mbuc Embryos Display Accelerated Decay of Maternal mRNA and Severe Defects in Embryo Development
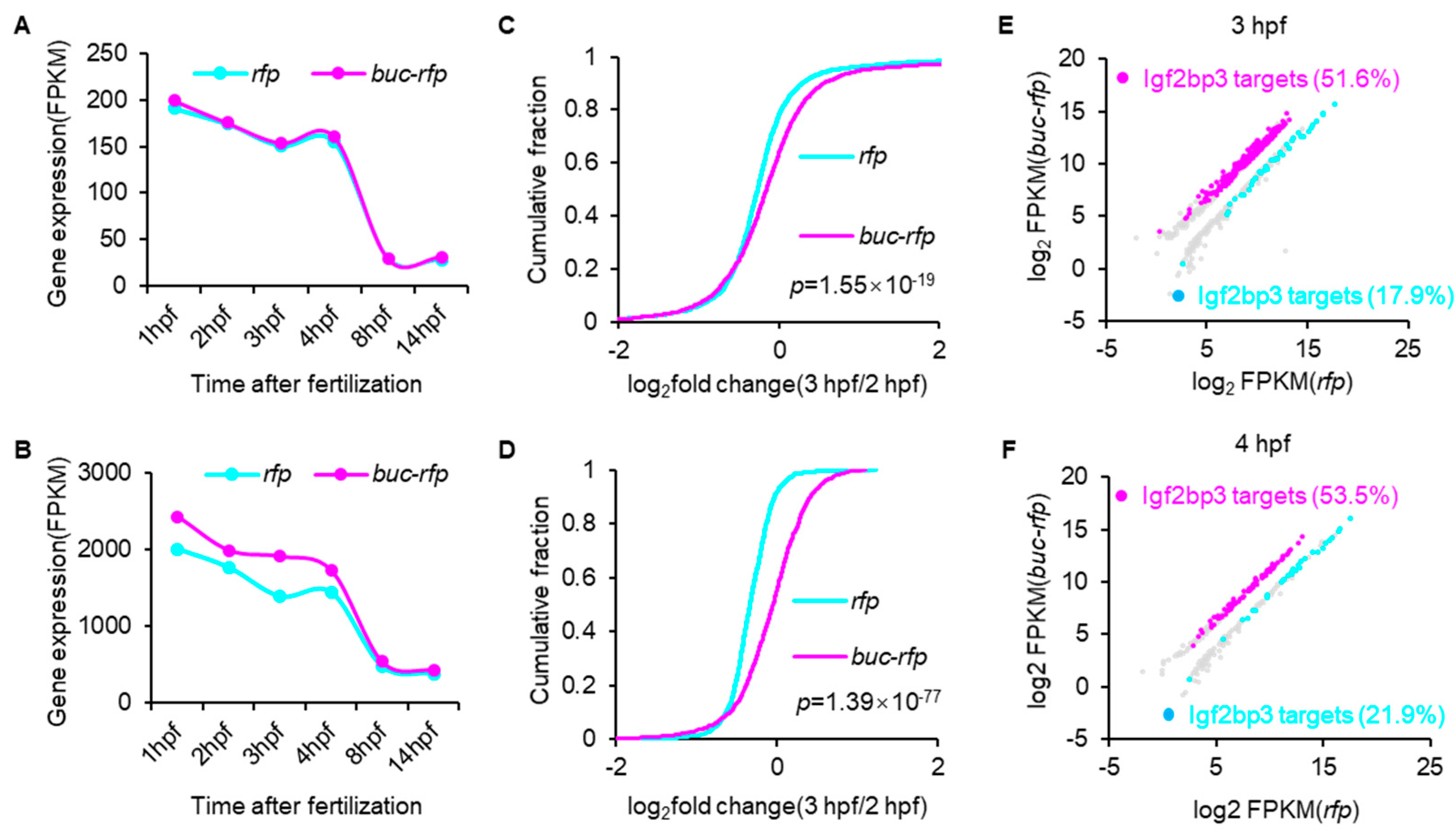
3.3. Buc Overexpression Decelerates the Decay of Bulk Maternal mRNA
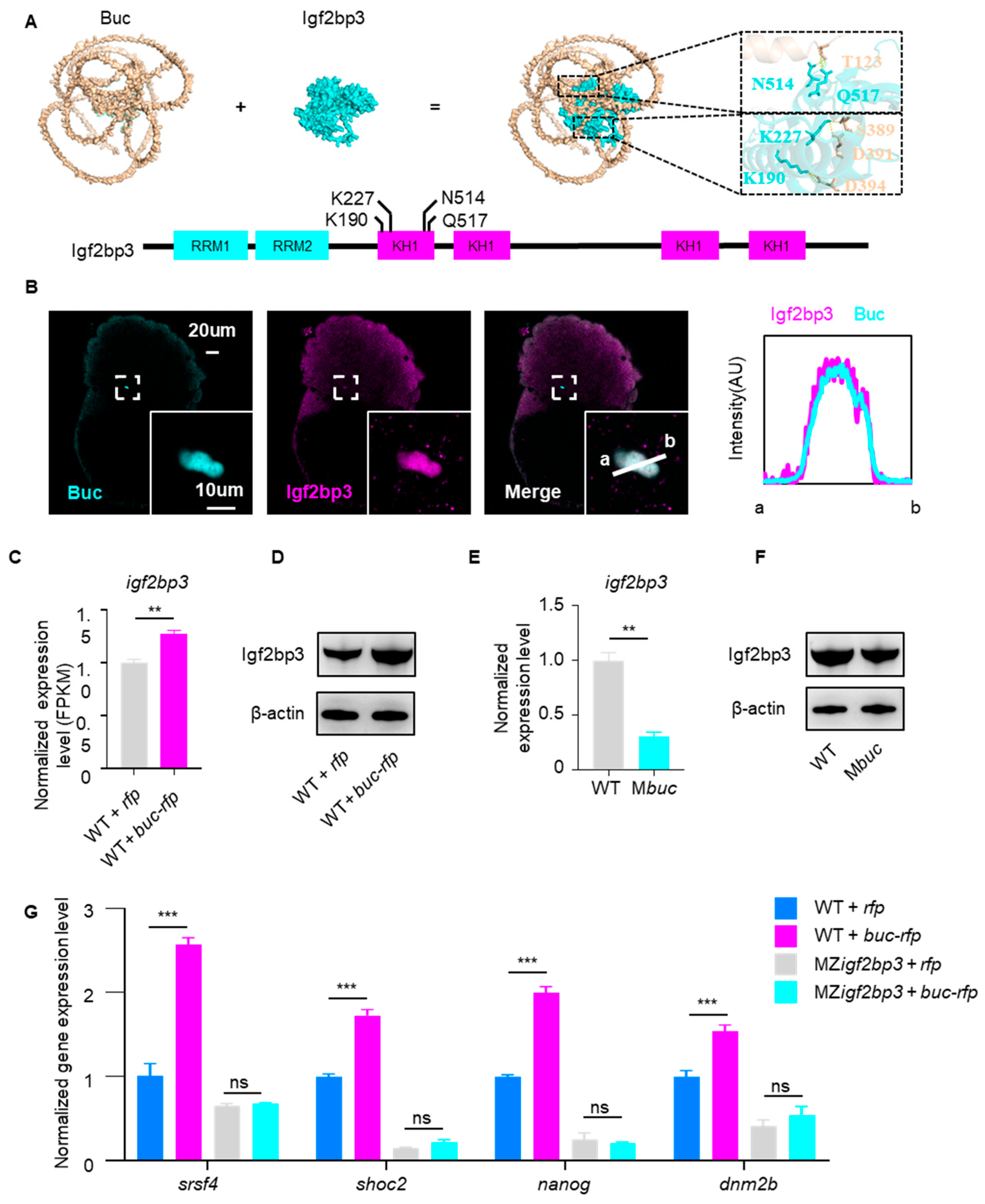
3.4. Buc Enhances the Stability of Igf2bp3-Targeted Maternal mRNAs
3.5. Buc Keeps Maternal mRNA Stability via Igf2bp3 Regulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farrell, J.A.; Wang, Y.; Riesenfeld, S.J.; Shekhar, K.; Regev, A.; Schier, A.F. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 2018, 360, eaar3131. [Google Scholar] [CrossRef]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146, dev161471. [Google Scholar] [CrossRef]
- Kojima, M.L.; Hoppe, C.; Giraldez, A.J. The maternal-to-zygotic transition: Reprogramming of the cytoplasm and nucleus. Nat. Rev. Genet. 2025, 26, 245–267. [Google Scholar] [CrossRef]
- Brantley, S.; Di Talia, S. The maternal-to-zygotic transition. Curr. Biol. 2024, 34, R519–R523. [Google Scholar] [CrossRef]
- Lee, M.T.; Bonneau, A.R.; Giraldez, A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014, 30, 581–613. [Google Scholar] [CrossRef]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef]
- Walser, C.B.; Lipshitz, H.D. Transcript clearance during the maternal-to-zygotic transition. Curr. Opin. Genet. Dev. 2011, 21, 431–443. [Google Scholar] [CrossRef]
- Sakashita, A.; Kitano, T.; Ishizu, H.; Guo, Y.; Masuda, H.; Ariura, M.; Murano, K.; Siomi, H. Transcription of MERVL retrotransposons is required for preimplantation embryo development. Nat. Genet. 2023, 55, 484–495. [Google Scholar] [CrossRef]
- Yang, J.; Aguero, T.; King, M.L. The Xenopus maternal-to-zygotic transition from the perspective of the germline. Curr. Top. Dev. Biol. 2015, 113, 271–303. [Google Scholar] [CrossRef]
- Siddiqui, N.U.; Li, X.; Luo, H.; Karaiskakis, A.; Hou, H.; Kislinger, T.; Westwood, J.T.; Morris, Q.; Lipshitz, H.D. Genome-wide analysis of the maternal-to-zygotic transition in Drosophila primordial germ cells. Genome Biol. 2012, 13, R11. [Google Scholar] [CrossRef]
- D’Orazio, F.M.; Balwierz, P.J.; González, A.J.; Guo, Y.; Hernández-Rodríguez, B.; Wheatley, L.; Jasiulewicz, A.; Hadzhiev, Y.; Vaquerizas, J.M.; Cairns, B. Germ cell differentiation requires Tdrd7-dependent chromatin and transcriptome reprogramming marked by germ plasm relocalization. Dev. Cell 2021, 56, 641–656.e645. [Google Scholar] [CrossRef]
- Hanyu-Nakamura, K.; Sonobe-Nojima, H.; Tanigawa, A.; Lasko, P.; Nakamura, A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 2008, 451, 730–733. [Google Scholar] [CrossRef]
- Hanyu-Nakamura, K.; Matsuda, K.; Cohen, S.M.; Nakamura, A. Pgc suppresses the zygotically acting RNA decay pathway to protect germ plasm RNAs in the Drosophila embryo. Development 2019, 146, dev167056. [Google Scholar] [CrossRef]
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Oude Vrielink, J.A.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Ørom, U.A. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [CrossRef]
- Mishima, Y.; Giraldez, A.J.; Takeda, Y.; Fujiwara, T.; Sakamoto, H.; Schier, A.F.; Inoue, K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006, 16, 2135–2142. [Google Scholar] [CrossRef]
- Oulhen, N.; Wessel, G.M. Differential Nanos 2 protein stability results in selective germ cell accumulation in the sea urchin. Dev. Biol. 2016, 418, 146–156. [Google Scholar] [CrossRef]
- Wessel, G.M.; Brayboy, L.; Fresques, T.; Gustafson, E.A.; Oulhen, N.; Ramos, I.; Reich, A.; Swartz, S.Z.; Yajima, M.; Zazueta, V. The biology of the germ line in echinoderms. Mol. Reprod. Dev. 2014, 81, 679–711. [Google Scholar] [CrossRef]
- Swartz, S.Z.; Reich, A.M.; Oulhen, N.; Raz, T.; Milos, P.M.; Campanale, J.P.; Hamdoun, A.; Wessel, G.M. Deadenylase depletion protects inherited mRNAs in primordial germ cells. Development 2014, 141, 3134–3142. [Google Scholar] [CrossRef]
- Vong, Y.H.; Sivashanmugam, L.; Leech, R.; Zaucker, A.; Jones, A.; Sampath, K. The RNA-binding protein Igf2bp3 is critical for embryonic and germline development in zebrafish. PLoS Genet. 2021, 17, e1009667. [Google Scholar] [CrossRef]
- Ren, F.; Lin, Q.; Gong, G.; Du, X.; Dan, H.; Qin, W.; Miao, R.; Xiong, Y.; Xiao, R.; Li, X. Igf2bp3 maintains maternal RNA stability and ensures early embryo development in zebrafish. Commun. Biol. 2020, 3, 94. [Google Scholar] [CrossRef]
- Ren, F.; Miao, R.; Xiao, R.; Mei, J. m6A reader Igf2bp3 enables germ plasm assembly by m6A-dependent regulation of gene expression in zebrafish. Sci. Bull. 2021, 66, 1119–1128. [Google Scholar] [CrossRef]
- Heim, A.E.; Hartung, O.; Rothhämel, S.; Ferreira, E.; Jenny, A.; Marlow, F.L. Oocyte polarity requires a Bucky ball-dependent feedback amplification loop. Development 2014, 141, 842–854. [Google Scholar] [CrossRef]
- Bontems, F.; Stein, A.; Marlow, F.; Lyautey, J.; Gupta, T.; Mullins, M.C.; Dosch, R. Bucky ball organizes germ plasm assembly in zebrafish. Curr. Biol. 2009, 19, 414–422. [Google Scholar] [CrossRef]
- Mickoleit, M.; Banisch, T.U.; Raz, E. Regulation of hub mRNA stability and translation by miR430 and the dead end protein promotes preferential expression in zebrafish primordial germ cells. Dev. Dyn. 2011, 240, 695–703. [Google Scholar] [CrossRef]
- Strome, S.; Updike, D. Specifying and protecting germ cell fate. Nat. Rev. Mol. Cell Biol. 2015, 16, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Wang, X.; Beadell, A.V.; Lu, Z.; Shi, H.; Kuuspalu, A.; Ho, R.K.; He, C. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 2017, 542, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.-L.; Zhang, M.; Ma, H.-L.; Sun, B.-F.; Li, A.; Xia, J.; Chen, J. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell 2019, 75, 1188–1202.e11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; Hou, W.-B.; Du, J.-W.; Zong, M.; Zheng, K.-L.; Wang, W.-J.; Wang, J.-Q.; Zhang, H.; Mu, Y.-S.; Yin, Z. Argonaute 2 is a key regulator of maternal mRNA degradation in mouse early embryos. Cell Death Discov. 2020, 6, 133. [Google Scholar] [CrossRef]
- Lykke-Andersen, K.; Gilchrist, M.J.; Grabarek, J.B.; Das, P.; Miska, E.; Zernicka-Goetz, M. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol. Biol. Cell 2008, 19, 4383–4392. [Google Scholar] [CrossRef]
- Pekovic, F.; Rammelt, C.; Kubíková, J.; Metz, J.; Jeske, M.; Wahle, E. RNA binding proteins Smaug and Cup induce CCR4–NOT-dependent deadenylation of the nanos mRNA in a reconstituted system. Nucleic Acids Res. 2023, 51, 3950–3970. [Google Scholar] [CrossRef]
- Niinuma, S.; Tomari, Y. ATP is dispensable for both miRNA-and Smaug-mediated deadenylation reactions. RNA 2017, 23, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Dumelie, J.G.; Li, X.; Cheng, M.H.; Yang, Z.; Laver, J.D.; Siddiqui, N.U.; Westwood, J.T.; Morris, Q.; Lipshitz, H.D. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014, 15, R4. [Google Scholar] [CrossRef] [PubMed]
- Jenny, A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 2006, 133, 2827. [Google Scholar] [CrossRef] [PubMed]
- Kim-Ha, J.; Smith, J.L.; Macdonald, P.M. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 1991, 66, 23–35. [Google Scholar] [CrossRef]
- Seth, M.; Shirayama, M.; Gu, W.; Ishidate, T.; Conte, D.; Mello, C.C. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 2013, 27, 656–663. [Google Scholar] [CrossRef]
- Wedeles, C.J.; Wu, M.Z.; Claycomb, J.M. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 2013, 27, 664–671. [Google Scholar] [CrossRef]
- Ashe, A.; Sapetschnig, A.; Weick, E.-M.; Mitchell, J.; Bagijn, M.P.; Cording, A.C.; Doebley, A.-L.; Goldstein, L.D.; Lehrbach, N.J.; Le Pen, J. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012, 150, 88–99. [Google Scholar] [CrossRef]
- Riemer, S.; Bontems, F.; Krishnakumar, P.; Gömann, J.; Dosch, R. A functional Bucky ball-GFP transgene visualizes germ plasm in living zebrafish. Gene Expr. Patterns 2015, 18, 44–52. [Google Scholar] [CrossRef]
- Westerich, K.J.; Tarbashevich, K.; Schick, J.; Gupta, A.; Zhu, M.; Hull, K.; Romo, D.; Zeuschner, D.; Goudarzi, M.; Gross-Thebing, T. Spatial organization and function of RNA molecules within phase-separated condensates in zebrafish are controlled by Dnd1. Dev. Cell 2023, 58, 1578–1592.e1575. [Google Scholar] [CrossRef]
- Roovers, E.F.; Kaaij, L.J.; Redl, S.; Bronkhorst, A.W.; Wiebrands, K.; de Jesus Domingues, A.M.; Huang, H.-Y.; Han, C.-T.; Riemer, S.; Dosch, R. Tdrd6a regulates the aggregation of Buc into functional subcellular compartments that drive germ cell specification. Dev. Cell 2018, 46, 285–301.e289. [Google Scholar] [CrossRef]
- Boke, E.; Ruer, M.; Wühr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Hyman, A.A.; Mitchison, T.J. Amyloid-like self-assembly of a cellular compartment. Cell 2016, 166, 637–650. [Google Scholar] [CrossRef]
- Campbell, P.D.; Heim, A.E.; Smith, M.Z.; Marlow, F.L. Kinesin-1 interacts with Bucky ball to form germ cells and is required to pattern the zebrafish body axis. Development 2015, 142, 2996–3008. [Google Scholar] [CrossRef] [PubMed]
- Pamula, M.C.; Lehmann, R. How germ granules promote germ cell fate. Nat. Rev. Genet. 2024, 25, 803–821. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Chen, S.; Ma, M.; Fan, H.-C.; Borwick, E.; Böke, E.; Mei, W.; Yang, J. Solubility phase transition of maternal RNAs during vertebrate oocyte-to-embryo transition. Dev. Cell 2023, 58, 2776–2788.e2775. [Google Scholar] [CrossRef] [PubMed]
- Eichler, C.E.; Hakes, A.C.; Hull, B.; Gavis, E.R. Compartmentalized oskar degradation in the germ plasm safeguards germline development. eLife 2020, 9, e49988. [Google Scholar] [CrossRef]
- Dodson, A.E.; Kennedy, S. Phase separation in germ cells and development. Dev. Cell 2020, 55, 4–17. [Google Scholar] [CrossRef]
- Dufourt, J.; Bontonou, G.; Chartier, A.; Jahan, C.; Meunier, A.-C.; Pierson, S.; Harrison, P.F.; Papin, C.; Beilharz, T.H.; Simonelig, M. piRNAs and Aubergine cooperate with Wispy poly (A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun. 2017, 8, 1305. [Google Scholar] [CrossRef]
- Noble, S.L.; Allen, B.L.; Goh, L.K.; Nordick, K.; Evans, T.C. Maternal mRNAs are regulated by diverse P body–related mRNP granules during early Caenorhabditis elegans development. J. Cell Biol. 2008, 182, 559–572. [Google Scholar] [CrossRef]
- Boag, P.R.; Atalay, A.; Robida, S.; Reinke, V.; Blackwell, T.K. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J. Cell Biol. 2008, 182, 543–557. [Google Scholar] [CrossRef]

| Gene Name | Primer Sequence (5′-3′) | Purpose |
|---|---|---|
| srsf-F | CCGAGATGGTGGCAACAG | qRT-PCR |
| srsf-R | CCTGTAATCTGTGCGTGTCG | qRT-PCR |
| shoc-F | TCCATCTGTTGCCCTCGTC | qRT-PCR |
| shoc-R | TGGTGATGCGGTTGAAGC | qRT-PCR |
| nanog-F | GGCGTCCCGAATCTGAG | qRT-PCR |
| nanog-R | CCGTTCTGCGAGTGTCCC | qRT-PCR |
| dnm2b-F | TTCCCTCCAGACCCACT | qRT-PCR |
| dnm2b-R | TCGGACGGATGATTGTG | qRT-PCR |
| igf2bp3-F | AGCGAGTGGAGGGATTTCA | qRT-PCR |
| igf2bp3-R | ATTGACGCACCAGCGAAGC | qRT-PCR |
| buc-F | CCACAAGTGACCCAAGAGCG | qRT-PCR |
| buc-R | CCTACCACCACCAACATAAACA | qRT-PCR |
| β-actin-F | CGAGCAGGAGATGGGAACC | qRT-PCR |
| β-actin-R | CAACGGAAACGCTCATTGC | qRT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, R.; Xie, Y.; Han, Q.; Meng, Y.; Tang, Q.; Mei, J.; Ren, F. Buc Maintains Maternal RNA Stability and Embryogenesis in Zebrafish. Cells 2025, 14, 1879. https://doi.org/10.3390/cells14231879
Miao R, Xie Y, Han Q, Meng Y, Tang Q, Mei J, Ren F. Buc Maintains Maternal RNA Stability and Embryogenesis in Zebrafish. Cells. 2025; 14(23):1879. https://doi.org/10.3390/cells14231879
Chicago/Turabian StyleMiao, Ran, Yan Xie, Qingqing Han, Yinglu Meng, Qin Tang, Jie Mei, and Fan Ren. 2025. "Buc Maintains Maternal RNA Stability and Embryogenesis in Zebrafish" Cells 14, no. 23: 1879. https://doi.org/10.3390/cells14231879
APA StyleMiao, R., Xie, Y., Han, Q., Meng, Y., Tang, Q., Mei, J., & Ren, F. (2025). Buc Maintains Maternal RNA Stability and Embryogenesis in Zebrafish. Cells, 14(23), 1879. https://doi.org/10.3390/cells14231879






