Chronic Inflammation and Altered Immune Responses in LongCOVID Associate with Neurological Manifestations and Accelerated Aging
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants and Blood Collection
2.2. Plasma Collection
2.3. Peripheral Blood Mononuclear Cell (PBMC) Collection
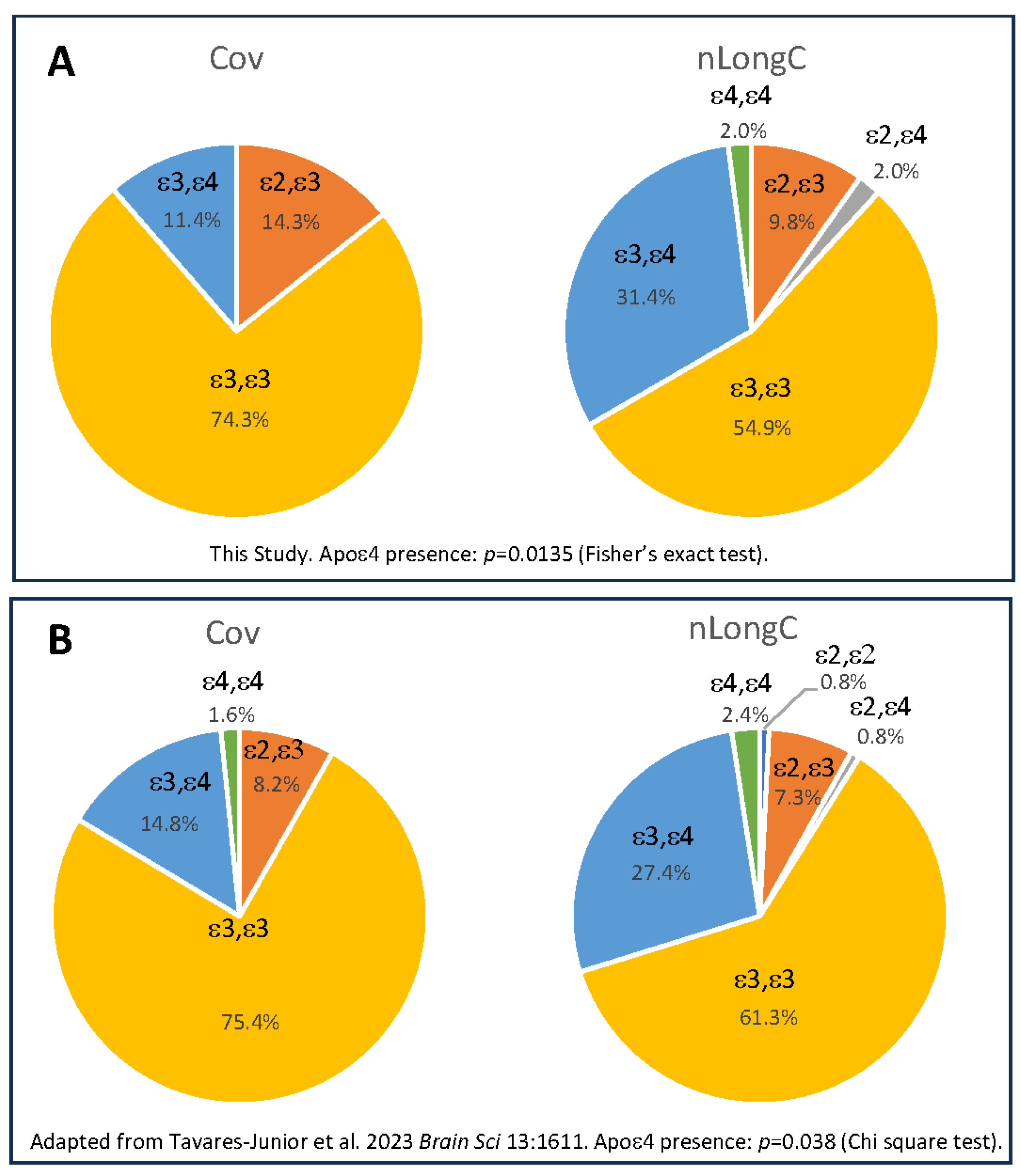
2.4. APOE Genotyping
2.5. Plasma Cytokine Analysis Using Meso Scale Discovery (MSD) Assays
2.6. Fibrinogen Quantification
2.7. Plasma Neural Protein Analysis Using High Sensitivity MSD Assays
2.8. Validation Using Ella Automated ELISA System
2.9. Statistical Analysis and Bioinformatics
3. Results
3.1. Particicpant Demographics, Clinical Measures and APOE Genotype

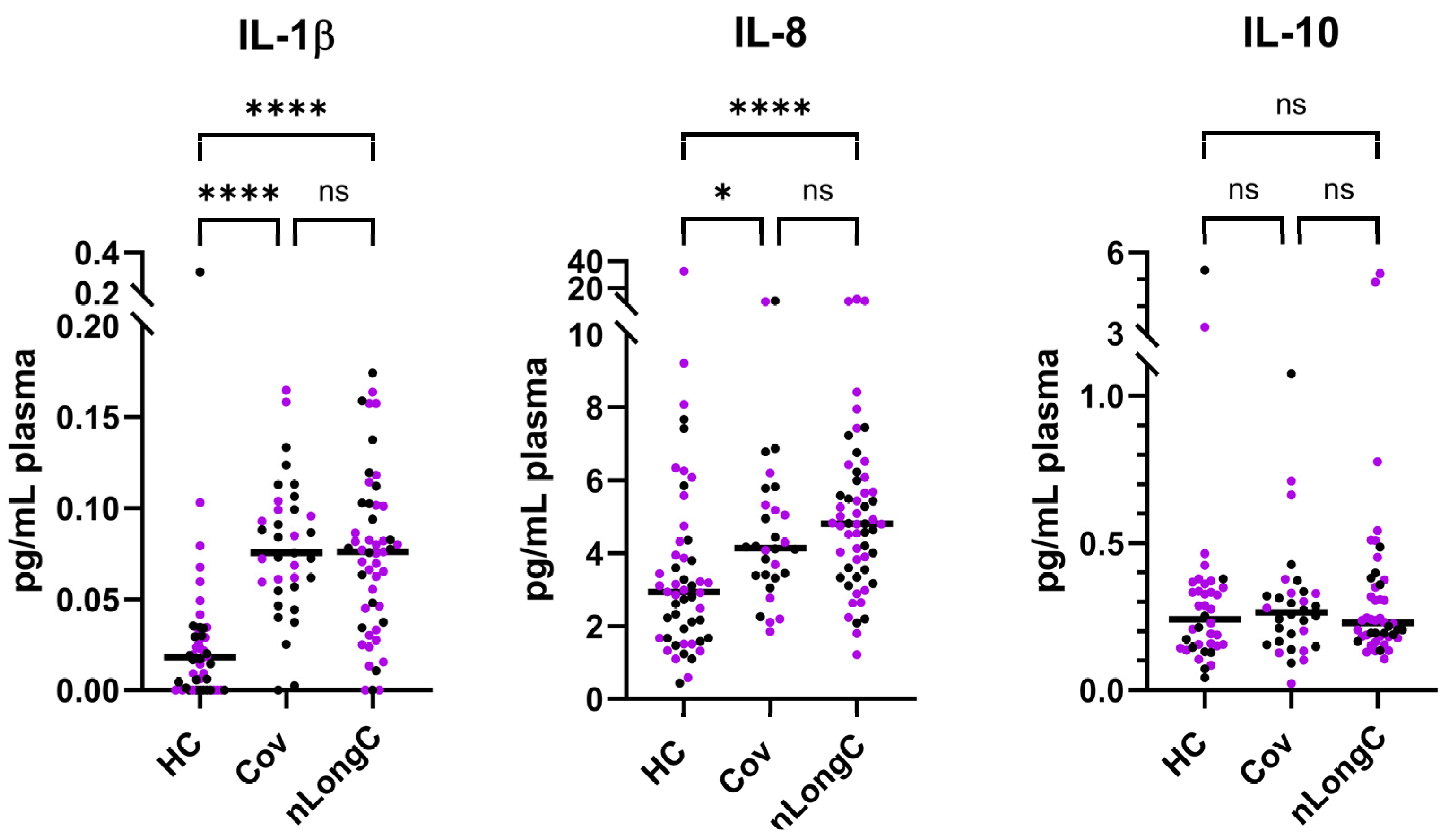
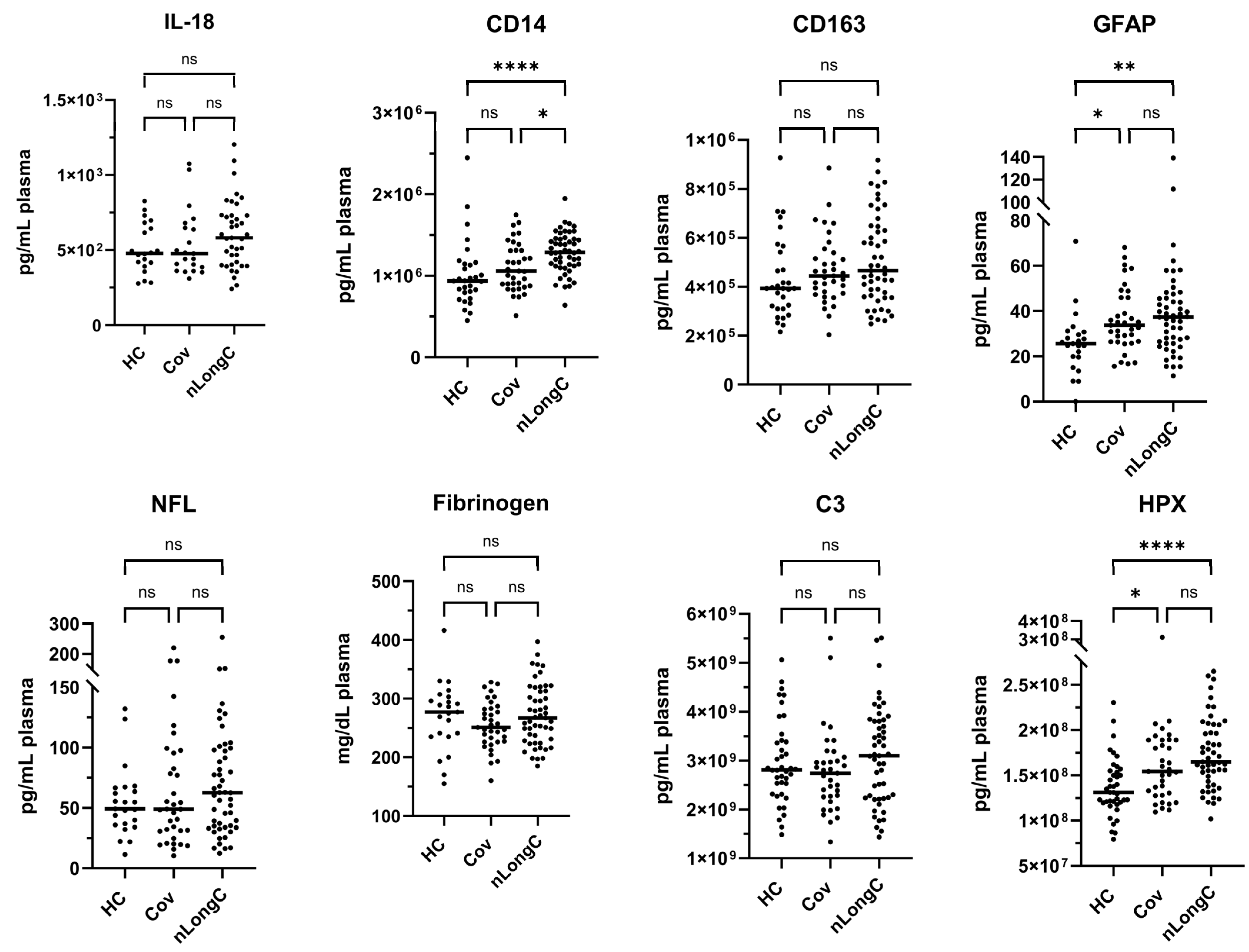
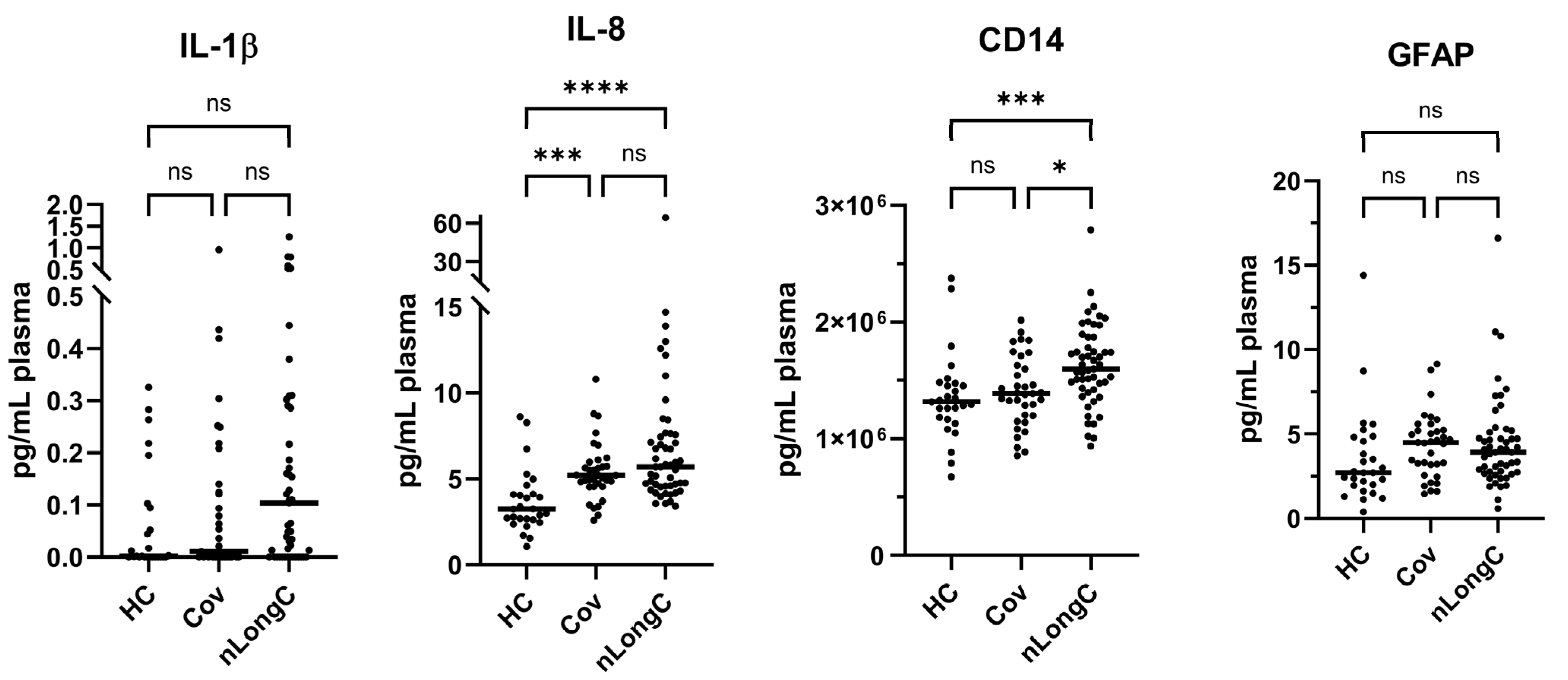
3.2. Cytokine and Plasma Biomarkers for Peripheral Inflammation, Neuroinflammation and Vascular Health
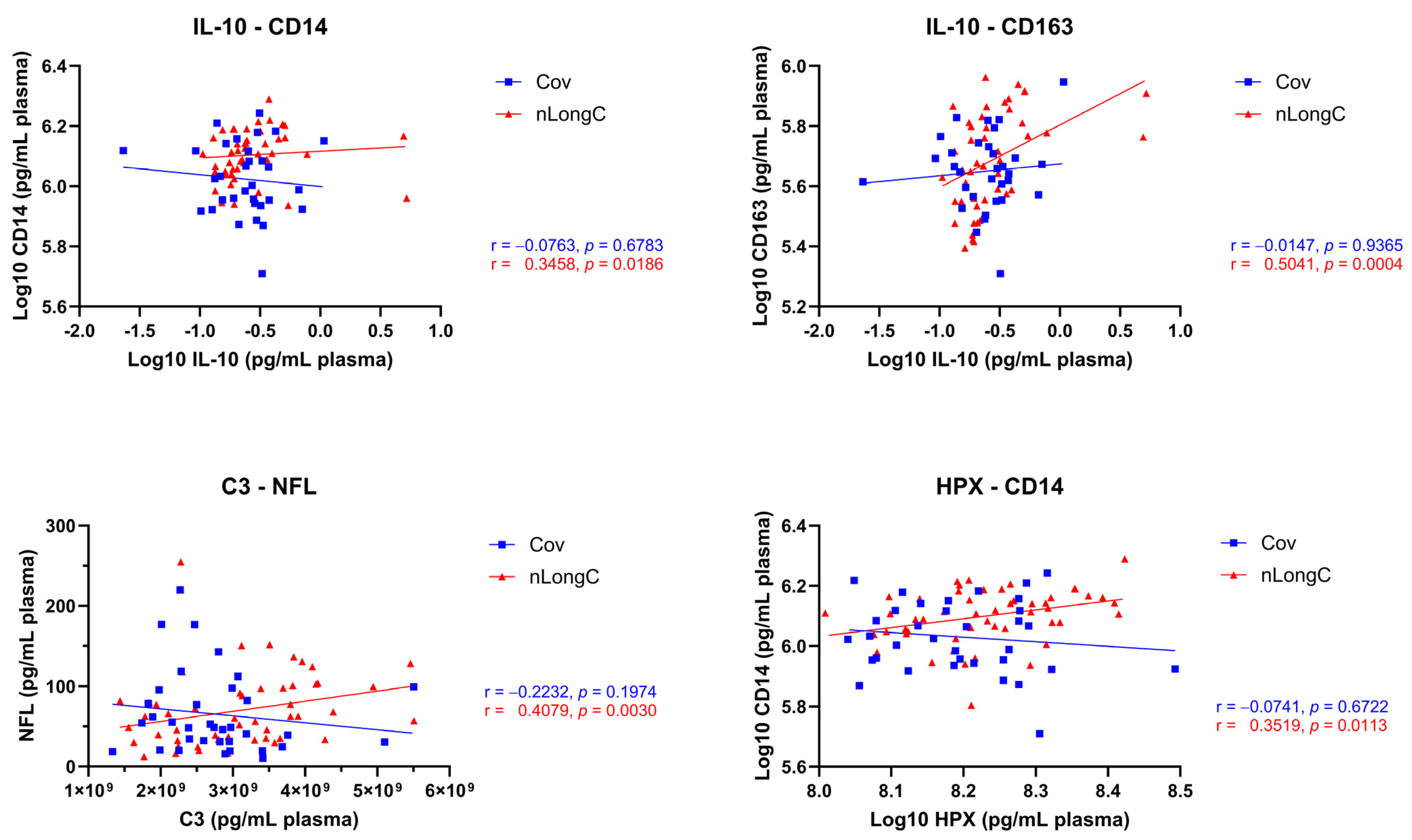
3.3. Correlation Matrices and Patterns
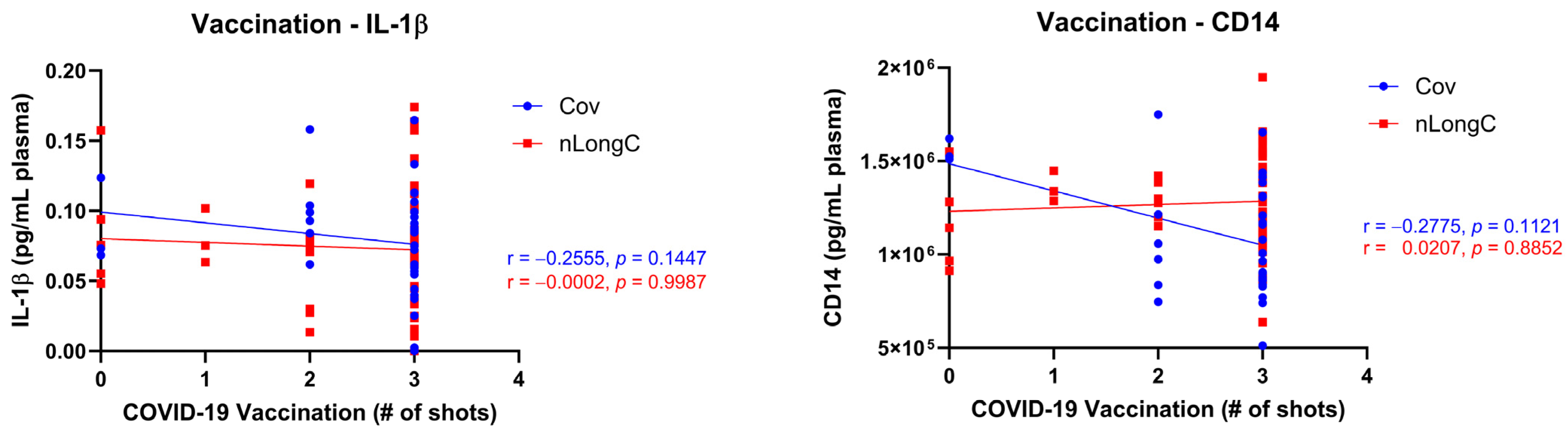
3.4. Chronic Inflammation and Altered Immune Responses in nLongC
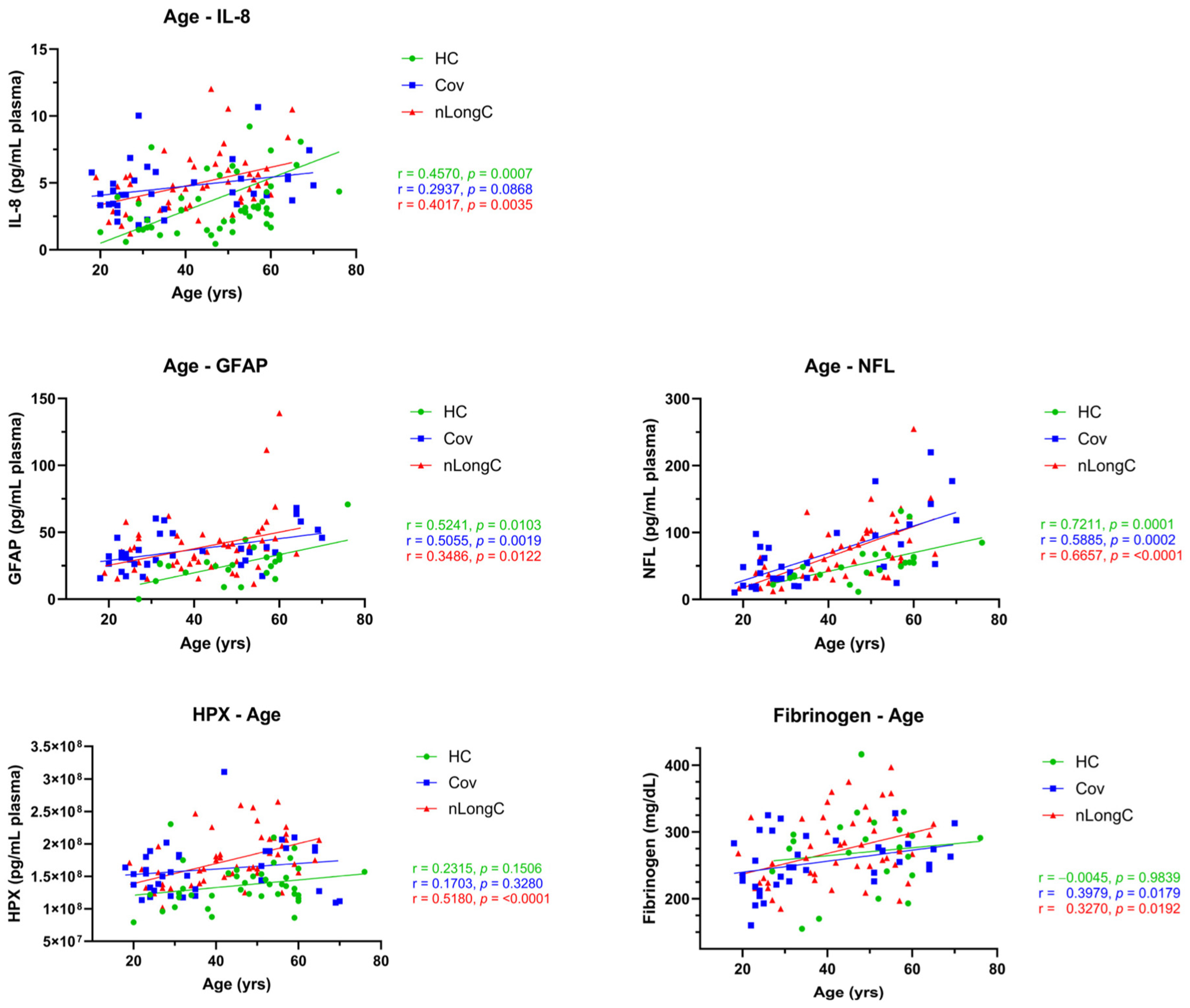
3.5. Association of Protein Markers to Aging
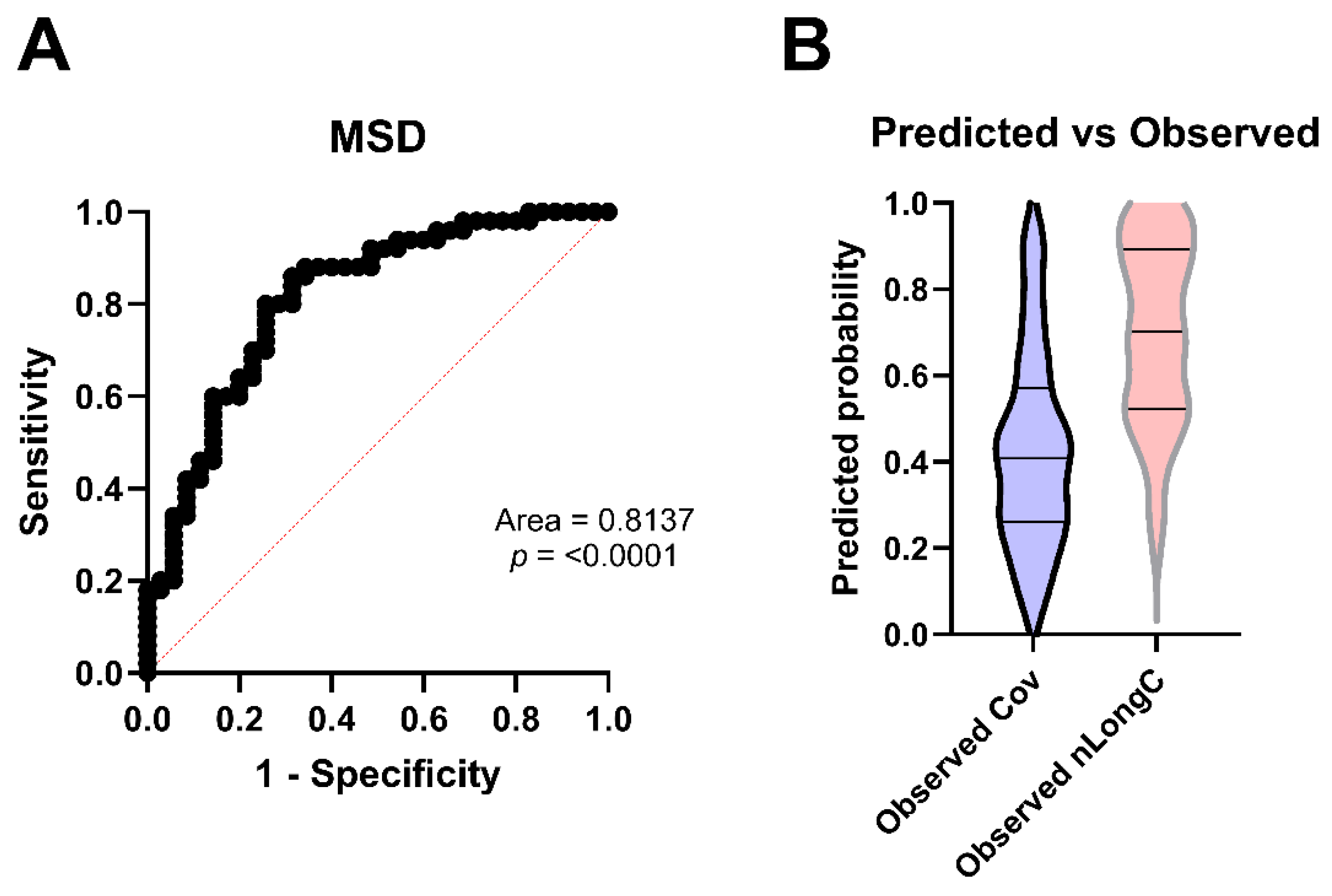
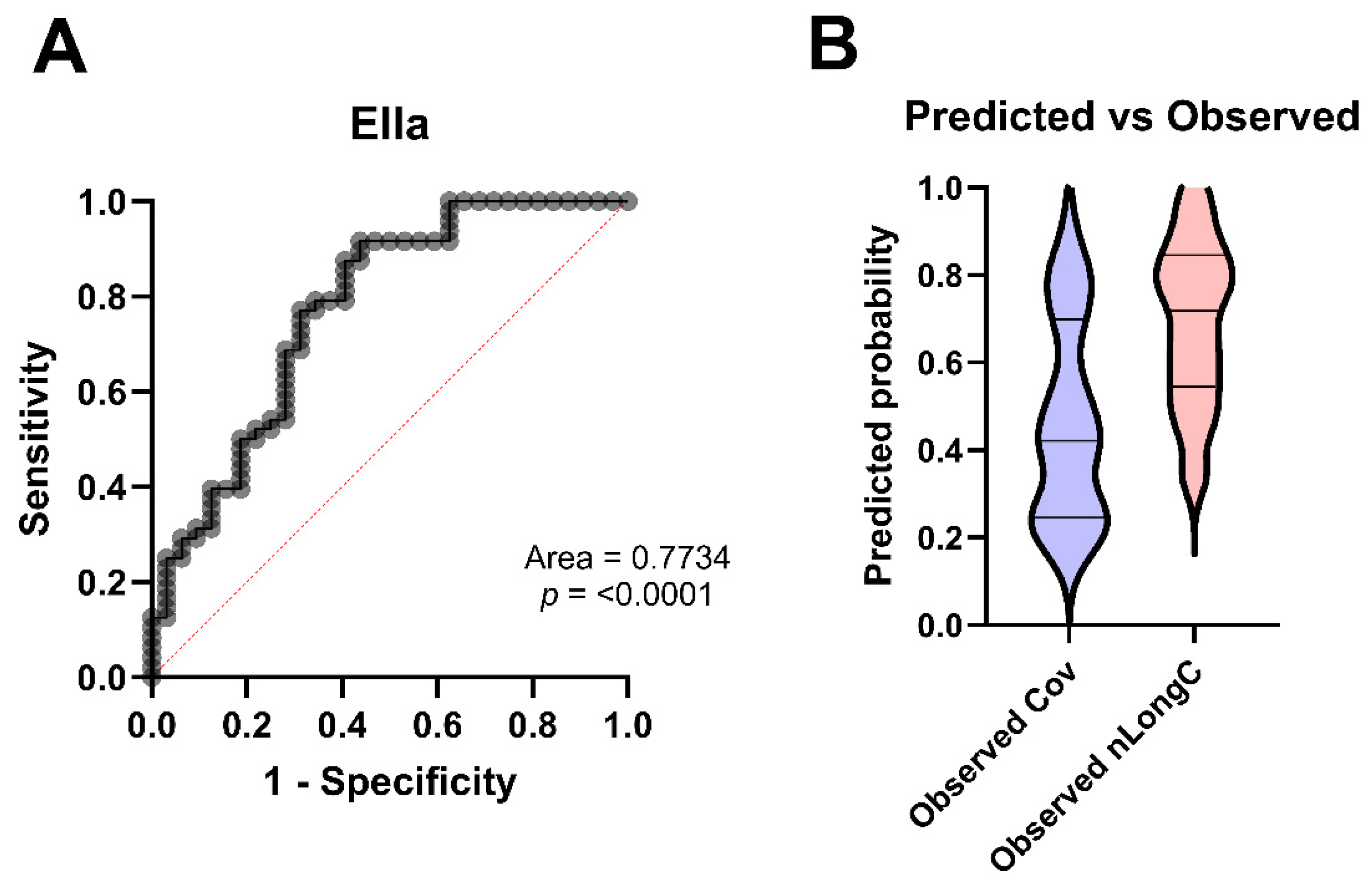
3.6. Predicting nLongC
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APOE | Apolipoprotein E |
| APOE4 | Apolipoprotein E ε4 allele |
| BMI | Body mass index |
| C3 | Complement C3 |
| CD14 | (Soluble) Cluster of differentiation 14 |
| CD163 | (Soluble) Cluster of differentiation 163 |
| Cov | People fully recovered from SARS-Cov-2 infection with no lingering symptoms >3 months post infection |
| Days P.I. | Days post infection |
| GFAP | Glial fibrillary acidic protein |
| HCs | Healthy controls, pre-pandemic |
| HPX | Hemopexin |
| HIV | Human immunodeficiency virus |
| IL-1β | Interleukin 1 beta |
| IL-8 | Interleukin 8 |
| IL-10 | Interleukin 10 |
| LongC | LongCOVID-19 |
| NFL | Neurofilament light chain |
| nLongC | People with continued neurological sequelae >3 months post SARS-Cov-2 infection |
| PASC | Post-acute sequelae of COVID-19 |
| RBC | Red blood cell |
Appendix A
| Age | Days P.I. | BMI | Fibrinogen | C3 | HPX | IL-1β | IL-8 | IL-10 | IL-18 | GFAP | NFL | CD14 | CD163 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HC | ||||||||||||||
| Age | 0.984 | 0.310 | 0.151 | 0.914 | 0.001 | 0.338 | 0.142 | 0.010 | 0.000 | 0.784 | 0.120 | |||
| Days P.I. | ||||||||||||||
| BMI | ||||||||||||||
| Fibrinogen | 0.983899 | 0.480 | 0.188 | 0.540 | 0.270 | 0.838 | 0.333 | 0.178 | 0.700 | 0.309 | 0.071 | |||
| C3 | 0.3099399 | 0.480 | 5.894 × 10−7 | 0.688 | 0.054 | 0.229 | 0.435 | 0.048 | 0.461 | 0.026 | 0.748 | |||
| HPX | 0.1506363 | 0.188 | 5.894 × 10−7 | 0.312 | 0.132 | 0.410 | 0.557 | 0.414 | 0.267 | 0.298 | 0.749 | |||
| IL-1β | 0.914376 | 0.540 | 0.688 | 0.312 | 0.975 | 0.305 | 0.929 | 0.939 | 0.491 | 0.166 | 0.529 | |||
| IL-8 | 0.0007487 | 0.270 | 0.054 | 0.132 | 0.975 | 0.060 | 0.346 | 0.047 | 0.155 | 0.807 | 0.302 | |||
| IL-10 | 0.3380212 | 0.838 | 0.229 | 0.410 | 0.305 | 0.060 | 0.843 | 0.247 | 0.313 | 0.109 | 0.438 | |||
| IL-18 | 0.1419747 | 0.333 | 0.435 | 0.557 | 0.929 | 0.346 | 0.843 | 1.000 | 1.000 | 1.000 | 1.000 | |||
| GFAP | 0.0102523 | 0.178 | 0.048 | 0.414 | 0.939 | 0.047 | 0.247 | 1.000 | 0.077 | 0.394 | 0.396 | |||
| NFL | 0.0001034 | 0.700 | 0.461 | 0.267 | 0.491 | 0.155 | 0.313 | 1.000 | 0.077 | 0.210 | 0.142 | |||
| CD14 | 0.7842992 | 0.309 | 0.026 | 0.298 | 0.166 | 0.807 | 0.109 | 1.000 | 0.394 | 0.210 | 0.000 | |||
| CD163 | 0.1203823 | 0.071 | 0.748 | 0.749 | 0.529 | 0.302 | 0.438 | 1.000 | 0.396 | 0.142 | 0.000 | |||
| Cov | ||||||||||||||
| Age | 0.387 | 0.648 | 0.018 | 0.789 | 0.328 | 0.646 | 0.087 | 0.068 | 0.278 | 0.002 | 0.000 | 0.001 | 0.096 | |
| Days P.I. | 0.387 | 0.096 | 0.485 | 0.606 | 0.474 | 0.005 | 0.665 | 0.459 | 0.555 | 0.187 | 0.666 | 0.411 | 0.302 | |
| BMI | 0.648 | 0.096 | 0.004 | 2.902 × 10−6 | 0.005 | 0.065 | 0.264 | 0.648 | 0.002 | 0.008 | 0.469 | 0.749 | 0.143 | |
| Fibrinogen | 0.018 | 0.485 | 0.004 | 0.006 | 0.195 | 0.128 | 0.697 | 0.470 | 0.104 | 0.715 | 0.914 | 0.013 | 0.008 | |
| C3 | 0.789 | 0.606 | 2.902 × 10−6 | 0.006 | 6.254 × 10−7 | 0.055 | 0.018 | 0.199 | 0.046 | 0.124 | 0.197 | 0.929 | 0.122 | |
| HPX | 0.328 | 0.474 | 0.005 | 0.195 | 6.254 × 10−7 | 0.511 | 0.197 | 0.888 | 0.007 | 0.363 | 0.517 | 0.672 | 0.538 | |
| IL-1β | 0.646 | 0.005 | 0.065 | 0.128 | 0.055 | 0.511 | 0.050 | 0.260 | 0.124 | 0.280 | 0.194 | 0.381 | 0.556 | |
| IL-8 | 0.087 | 0.665 | 0.264 | 0.697 | 0.018 | 0.197 | 0.050 | 0.177 | 0.808 | 0.360 | 0.373 | 0.034 | 0.432 | |
| IL-10 | 0.068 | 0.459 | 0.648 | 0.470 | 0.199 | 0.888 | 0.260 | 0.177 | 1.000 | 0.589 | 0.124 | 0.678 | 0.937 | |
| IL-18 | 0.278 | 0.555 | 0.002 | 0.104 | 0.046 | 0.007 | 0.124 | 0.808 | 1.000 | 0.773 | 0.750 | 0.492 | 0.141 | |
| GFAP | 0.002 | 0.187 | 0.008 | 0.715 | 0.124 | 0.363 | 0.280 | 0.360 | 0.589 | 0.773 | 0.045 | 0.065 | 0.743 | |
| NFL | 0.000 | 0.666 | 0.469 | 0.914 | 0.197 | 0.517 | 0.194 | 0.373 | 0.124 | 0.750 | 0.045 | 0.544 | 0.699 | |
| CD14 | 0.001 | 0.411 | 0.749 | 0.013 | 0.929 | 0.672 | 0.381 | 0.034 | 0.678 | 0.492 | 0.065 | 0.544 | 0.203 | |
| CD163 | 0.096 | 0.302 | 0.143 | 0.008 | 0.122 | 0.538 | 0.556 | 0.432 | 0.937 | 0.141 | 0.743 | 0.699 | 0.203 | |
| nLongC | ||||||||||||||
| Age | 0.024 | 0.004 | 0.019 | 2.308 × 10−5 | 9.902 × 10−5 | 0.061 | 0.003 | 0.986 | 0.009 | 0.012 | 9.82 × 10−8 | 0.226 | 0.686 | |
| Days P.I. | 0.024 | 0.123 | 0.937 | 0.256 | 0.182 | 0.395 | 0.359 | 0.962 | 0.680 | 0.939 | 0.117 | 0.899 | 0.201 | |
| BMI | 0.004 | 0.123 | 0.002 | 7.365 × 10−6 | 0.023 | 0.942 | 0.023 | 0.837 | 0.031 | 0.440 | 0.093 | 0.945 | 0.080 | |
| Fibrinogen | 0.019 | 0.937 | 0.002 | 1.888 × 10−5 | 0.001 | 0.313 | 0.214 | 0.396 | 0.000 | 0.582 | 0.076 | 0.104 | 0.055 | |
| C3 | 0.000 | 0.256 | 7.365 × 10−6 | 1.888 × 10−5 | 8.374 × 10−12 | 0.249 | 0.001 | 0.039 | 4.71 × 10−5 | 0.912 | 0.003 | 0.205 | 0.008 | |
| HPX | 0.000 | 0.182 | 0.023 | 0.001 | 8.374 × 10−12 | 0.340 | 0.061 | 0.222 | 0.001 | 0.700 | 0.005 | 0.011 | 0.047 | |
| IL-1β | 0.061 | 0.395 | 0.942 | 0.313 | 0.249 | 0.340 | 5.47 × 10−5 | 0.001 | 0.938 | 0.987 | 0.144 | 0.181 | 0.731 | |
| IL-8 | 0.003 | 0.359 | 0.023 | 0.214 | 0.001 | 0.061 | 5.467 × 10−5 | 0.003 | 0.591 | 0.973 | 0.060 | 0.336 | 0.097 | |
| IL-10 | 0.986 | 0.962 | 0.837 | 0.396 | 0.039 | 0.222 | 0.001 | 0.003 | 0.799 | 0.638 | 0.525 | 0.019 | 0.000 | |
| IL-18 | 0.009 | 0.680 | 0.031 | 0.000 | 0.000 | 0.001 | 0.938 | 0.591 | 0.799 | 0.239 | 0.020 | 0.234 | 0.867 | |
| GFAP | 0.012 | 0.939 | 0.440 | 0.582 | 0.912 | 0.700 | 0.987 | 0.973 | 0.638 | 0.239 | 0.034 | 0.553 | 0.718 | |
| NFL | 9.819 × 10−8 | 0.117 | 0.093 | 0.076 | 0.003 | 0.005 | 0.144 | 0.060 | 0.525 | 0.020 | 0.034 | 0.554 | 0.784 | |
| CD14 | 0.226 | 0.899 | 0.945 | 0.104 | 0.205 | 0.011 | 0.181 | 0.336 | 0.019 | 0.234 | 0.553 | 0.554 | 0.009 | |
| CD163 | 0.686 | 0.201 | 0.080 | 0.055 | 0.008 | 0.047 | 0.731 | 0.097 | 0.000 | 0.867 | 0.718 | 0.784 | 0.009 |
Appendix B

Appendix C
| Category | Term | Genes | Count | p-Value | Benjamini |
|---|---|---|---|---|---|
| GAD Disease Class | Aging | CXCL8, IL1B, APOE, GFAP | 4 | 0.0035 | 0.0325 |
| GAD Disease Class | Reproduction | CXCL8, IL1B, CD14, APOE | 4 | 0.0036 | 0.0325 |
| GAD Disease Class | Pharmacogenomic | HPX, CXCL8, IL1B, CD14, APOE | 5 | 0.0081 | 0.0358 |
| GAD Disease Class | Neurological | CXCL8, IL1B, CD14, APOE, GFAP | 5 | 0.0101 | 0.0358 |
| GAD Disease Class | Renal | CXCL8, IL1B, CD14, APOE | 4 | 0.0110 | 0.0358 |
| GAD Disease Class | Normal Variation | CXCL8, IL1B, APOE | 3 | 0.0146 | 0.0358 |
| GAD Disease Class | Other | CXCL8, IL1B, CD14, APOE | 4 | 0.0153 | 0.0358 |
| GAD Disease Class | Unknown | CXCL8, IL1B, CD14, APOE | 4 | 0.0159 | 0.0358 |
| GAD Disease Class | Vision | CXCL8, IL1B, APOE | 3 | 0.0254 | 0.0509 |
| GAD Disease Class | Infection | CXCL8, IL1B, CD14, APOE | 4 | 0.0353 | 0.0636 |
| Category | Term | Genes | Count | p-Value | Benjamini |
| GOTERM BP All | Regulation of response to external stimulus | HPX, CXCL8, IL1B, CD14, APOE | 5 | 0.0001 | 0.0369532 |
| GOTERM BP All | Response to stress | HPX, CXCL8, IL1B, CD14, APOE, GFAP | 6 | 0.0002 | 0.0369532 |
| GOTERM BP All | Regulation of signaling | HPX, CXCL8, IL1B, CD14, APOE, GFA | 6 | 0.0003 | 0.0369532 |
| GOTERM BP All | Regulation of cell communication | HPX, CXCL8, IL1B, CD14, APOE, GFAP | 6 | 0.0003 | 0.0369532 |
| GOTERM BP All | Regulation of response to biotic stimulus | HPX, IL1B, CD14, APOE | 4 | 0.0003 | 0.0369532 |
| GOTERM BP All | Biological process involved in interspecies interaction between organisms | HPX, CXCL8, IL1B, CD14, APOE | 5 | 0.0003 | 0.0369532 |
| GOTERM BP All | Positive regulation of response to external stimulus | HPX, CXCL8, IL1B, CD14 | 4 | 0.0003 | 0.0369532 |
| GOTERM BP All | Regulation of immune process | HPX, CXCL8, IL1B, CD14, APOE | 5 | 0.0003 | 0.0369532 |
| GOTERM BP All | Positive regulation of multicellular organismal process | CXCL8, IL1B, CD14, APOE, GFAP | 5 | 0.0003 | 0.0369532 |
| GOTERM BP All | Positive regulation of cell communication | HPX, IL1B, CD14, APOE, GFAP | 5 | 0.0004 | 0.0386044 |
| GOTERM BP All | Positive regulation of cell signaling | HPX, IL1B, CD14, APOE, GFAP | 5 | 0.0004 | 0.0386044 |
| GOTERM BP All | Import into cell | CXCL8, CD14, APOE, GFAP | 4 | 0.0005 | 0.0410767 |
| Annotation Cluster 1 | Enrichment Score: 2.8 | Genes | Count | p-Value | Benjamini |
| GOTERM BP All | Regulation of response to biotic stimulus | HPX, IL1B, CD14, APOE | 4 | 0.0003 | 0.0370 |
| GOTERM BP All | Regulation of defense response | HPX, IL1B, CD14, APOE | 4 | 0.0010 | 0.0663 |
| GOTERM BP All | Regulation of immune response | HPX, IL1B, CD14, APOE | 4 | 0.0016 | 0.0748 |
| GOTERM BP All | Regulation of response to stress | HPX, IL1B, CD14, APOE | 4 | 0.0043 | 0.1161 |
| GOTERM BP All | Positive regulation of signal transduction | HPX, IL1B, CD14, APOE | 4 | 0.0057 | 0.1200 |
| Annotation Cluster 2 | Enrichment Score: 2.66 | Genes | Count | p-Value | Benjamini |
| GOTERM BP All | Positive regulation of multicellular organismal process | CXCL8, IL1B, CD14, APOE, GFAP | 5 | 0.0003 | 0.0370 |
| GOTERM BP All | Regulation of multicellular organismal process | CXCL8, IL1B, CD14, APOE, GFAP | 5 | 0.0032 | 0.1135 |
| GAD Disease Class | Neurological | CXCL8, IL1B, CD14, APOE, GFAP | 5 | 0.0101 | 0.0358 |
| Source | Term Name | Genes | Count | Padj | |
| WP | LTF danger signal response pathway | CD14, CXCL8, IL1B | 3 | 8.357 × 10−6 | |
| WP | Post COVID neuroinflammation | CXCL8, GFAP, IL1B | 3 | 1.526 × 10−5 | |
| WP | Toll like receptor signaling | CD14, CXCL8, IL1B | 3 | 1.178 × 10−3 | |
| WP | Spinal cord injury | CXCL8, GFAP, IL1B | 3 | 2.094 × 10−3 | |
| WP | COVID 19 adverse outcome pathway | CXCL8, IL1B | 2 | 2.632 × 10−3 | |
| WP | LDL influence on CD14 and TLR4 | CD14, IL1B | 2 | 6.329 × 10−3 | |
| WP | Antiviral and anti inflammatory effects on Nrf2 on SARS CoV 2 pathway | CXCL8, IL1B | 2 | 1.238 × 10−2 | |
| WP | Prostaglandin signaling | CXCL8, IL1B | 2 | 1.318 × 10−2 | |
| WP | Photodynamic therapy induced NF kB survival signaling | CXCL8, IL1B | 2 | 1.484 × 10−2 | |
| WP | Immune infiltration in pancreatic cancer | CXCL8, IL1B | 2 | 1.847 × 10−2 | |
| WP | Network map of SARS CoV 2 signaling | CD14, CXCL8, IL1B | 3 | 2.208 × 10−2 | |
| WP | IL26 signaling | CXCL8, IL1B | 2 | 2.576 × 10−2 | |
| WP | Vitamin B12 metabolism | APOE, IL1B | 2 | 3.554 × 10−2 | |
| WP | Lung fibrosis | CXCL8, IL1B | 2 | 4.841 × 10−2 |
Appendix D
| Parameter Estimates | Variable | Estimate | Standard Error | 95% CI (Profile Likelihood) |
|---|---|---|---|---|
| β0 | Intercept | −6.152 | 2.648 | −12.03 to −1.506 |
| β1 | Age | −0.001986 | 0.023 | −0.04825 to 0.04290 |
| β2 | ApoE4[Yes] | 1.241 | 0.7098 | −0.07853 to 2.761 |
| β3 | BMI | 0.1129 | 0.06046 | 0.008530 to 0.2432 |
| β4 | IL-1β | −8.766 | 8.521 | −26.62 to 7.141 |
| β5 | IL-8 | 0.1661 | 0.2802 | −0.3453 to 0.7678 |
| β6 | GFAP | 0.01614 | 0.03287 | −0.03937 to 0.09608 |
| β7 | CD14 | 1.367 × 10−6 | 1.832 × 10−6 | −2.289 × 10−6 to 5.006 × 10−6 |
| β8 | CD14/IL-1β | 7.482 × 10−9 | 3.143 × 10−8 | −5.655 × 10−8 to 6.792 × 10−8 |
| β9 | IL-8/IL-1β | −0.0102 | 0.0174 | −0.04457 to ??? |
| β10 | GFAP/IL-1β | 0.0007072 | 0.0007783 | 3.462 × 10−5 to 0.002890 |
| β11 | CD14/IL-8 | 1.176 × 10−6 | 4.248 × 10−6 | −6.629 × 10−6 to 1.039 × 10−5 |
| β12 | CD14/GFAP | 2.520 × 10−5 | 3.204 × 10−5 | −2.951 × 10−5 to 0.0001009 |
| Odds ratios | Variable | Estimate | 95% CI (profile likelihood) | |
| β0 | Intercept | 0.002129 | 5.969 × 10−6 to 0.2219 | |
| β1 | Age | 0.998 | 0.9529 to 1.044 | |
| β2 | ApoE4[Yes] | 3.461 | 0.9245 to 15.82 | |
| β3 | BMI | 1.119 | 1.009 to 1.275 | |
| β4 | IL-1β | 0.000156 | 2.745 × 10−12 to 1262 | |
| β5 | IL-8 | 1.181 | 0.7080 to 2.155 | |
| β6 | GFAP | 1.016 | 0.9614 to 1.101 | |
| β7 | CD14 | 1.000 | 1.000 to 1.000 | |
| β8 | CD14/IL-1β | 1.000 | 1.000 to 1.000 | |
| β9 | IL-8/IL-1β | 0.9899 | 0.9564 to ??? | |
| β10 | GFAP/IL-1β | 1.001 | 1.000 to 1.003 | |
| β11 | CD14/IL-8 | 1.000 | 1.000 to 1.000 | |
| β12 | CD14/GFAP | 1.000 | 1.000 to 1.000 | |
| Area under the ROC curve | ||||
| Area | 0.8137 | |||
| Std. Error | 0.04798 | |||
| 95% confidence interval | 0.7197 to 0.9078 | |||
| p value | <0.0001 | |||
| Classification table | Predicted Cov | Predicted nLongC | Total | % Correctly classified |
| Observed Cov | 24 | 11 | 35 | 68.57 |
| Observed nLongC | 9 | 41 | 50 | 82 |
| Total | 33 | 52 | 85 | 76.47 |
| Negative predictive power (%) | 72.73 | |||
| Positive predictive power (%) | 78.85 | |||
| Classification cutoff | 0.5 | |||
| Data summary | ||||
| Number of nLongC | 50 | |||
| Number of Cov | 35 |
| Parameter Estimates | Variable | Estimate | Standard Error | 95% CI (Profile Likelihood) |
|---|---|---|---|---|
| β0 | Intercept | −5.537 | 2.727 | −11.41 to −0.7910 |
| β1 | Age | 0.01469 | 0.02542 | −0.03554 to 0.06524 |
| β2 | ApoE4[Yes] | 0.91 | 0.7308 | −0.4703 to 2.457 |
| β3 | BMI | 0.07305 | 0.05881 | −0.01032 to 0.1962 |
| β4 | IL-1b | −0.2131 | 1.327 | −2.795 to 2.720 |
| β5 | IL-8 | 0.2633 | 0.316 | −0.07788 to 0.9661 |
| β6 | GFAP | 0.009413 | 0.2191 | −0.3920 to 0.5003 |
| β7 | sCD14 | 2.258 × 10−7 | 1.706 × 10−6 | −3.229 × 10−6 to 3.428 × 10−6 |
| β8 | CD14/IL-1b | 6.73 × 10−11 | 8.565 × 10−10 | −1.610 × 10−9 to 1.855 × 10−9 |
| β9 | IL-8/IL-1b | −0.001359 | 0.0008232 | −0.003074 to 0.0002059 |
| β10 | sGFAP/IL-1b | 0.0009888 | 0.000924 | −0.001090 to 0.002764 |
| β11 | CD14/IL-8 | 3.416 × 10−6 | 7.709 × 10−6 | −9.011 × 10−6 to 1.953 × 10−5 |
| β12 | CD14/GFAP | 1.607 × 10−6 | 1.839 × 10−6 | −1.038 × 10−6 to 6.298 × 10−6 |
| Odds ratios | Variable | Estimate | 95% CI (profile likelihood) | |
| β0 | Intercept | 0.003939 | 1.105 × 10−5 to 0.4534 | |
| β1 | Age | 1.015 | 0.9651 to 1.067 | |
| β2 | ApoE4[Yes] | 2.484 | 0.6248 to 11.67 | |
| β3 | BMI | 1.076 | 0.9897 to 1.217 | |
| β4 | IL-1b | 0.808 | 0.06110 to 15.18 | |
| β5 | IL-8 | 1.301 | 0.9251 to 2.628 | |
| β6 | GFAP | 1.009 | 0.6757 to 1.649 | |
| β7 | sCD14 | 1.000 | 1.000 to 1.000 | |
| β8 | CD14/IL-1b | 1.000 | 1.000 to 1.000 | |
| β9 | IL-8/IL-1b | 0.9986 | 0.9969 to 1.000 | |
| β10 | sGFAP/IL-1b | 1.001 | 0.9989 to 1.003 | |
| β11 | CD14/IL-8 | 1.000 | 1.000 to 1.000 | |
| β12 | CD14/GFAP | 1.000 | 1.000 to 1.000 | |
| Area under the ROC curve | ||||
| Area | 0.7734 | |||
| Std. Error | 0.05519 | |||
| 95% confidence interval | 0.6653 to 0.8816 | |||
| p value | <0.0001 | |||
| Classification table | Predicted Cov | Predicted nLongC | Total | % Correctly classified |
| Observed Cov | 19 | 13 | 32 | 59.38 |
| Observed nLongC | 8 | 40 | 48 | 83.33 |
| Total | 27 | 53 | 80 | 73.75 |
| Negative predictive power (%) | 70.37 | |||
| Positive predictive power (%) | 75.47 | |||
| Classification cutoff | 0.5 | |||
| Data summary | ||||
| Number of nLongC | 48 | |||
| Number of Cov | 32 |
Appendix E
References
- Soriano, J.B.; Murthy, S.; Marshall, J.C.; Relan, P.; Diaz, J.V. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect. Dis. 2022, 22, e102–e107. [Google Scholar] [CrossRef]
- Hastie, C.E.; Lowe, D.J.; McAuley, A.; Winter, A.J.; Mills, N.L.; Black, C.; Scott, J.T.; O’Donnell, C.A.; Blane, D.N.; Browne, S.; et al. Outcomes among confirmed cases and a matched comparison group in the Long-COVID in Scotland study. Nat. Commun. 2022, 13, 5663. [Google Scholar] [CrossRef]
- Woodrow, M.; Carey, C.; Ziauddeen, N.; Thomas, R.; Akrami, A.; Lutje, V.; Greenwood, D.C.; Alwan, N.A. Systematic Review of the Prevalence of Long COVID. Open Forum Infect. Dis. 2023, 10, ofad233. [Google Scholar] [CrossRef] [PubMed]
- Moen, J.K.; Baker, C.A.; Iwasaki, A. Neuroimmune pathophysiology of long COVID. Psychiatry Clin. Neurosci. 2025, 79, 514–530. [Google Scholar] [CrossRef]
- Chen, C.; Haupert, S.R.; Zimmermann, L.; Shi, X.; Fritsche, L.G.; Mukherjee, B. Global Prevalence of Post-Coronavirus Disease 2019 (COVID-19) Condition or Long COVID: A Meta-Analysis and Systematic Review. J. Infect. Dis. 2022, 226, 1593–1607. [Google Scholar] [CrossRef]
- Ceban, F.; Ling, S.; Lui, L.M.W.; Lee, Y.; Gill, H.; Teopiz, K.M.; Rodrigues, N.B.; Subramaniapillai, M.; Di Vincenzo, J.D.; Cao, B.; et al. Fatigue and cognitive impairment in Post-COVID-19 Syndrome: A systematic review and meta-analysis. Brain Behav. Immun. 2022, 101, 93–135. [Google Scholar] [CrossRef]
- Ziauddeen, N.; Gurdasani, D.; O’Hara, M.E.; Hastie, C.; Roderick, P.; Yao, G.; Alwan, N.A. Characteristics and impact of Long Covid: Findings from an online survey. PLoS ONE 2022, 17, e0264331. [Google Scholar] [CrossRef]
- Sartori, A.C.; Vance, D.E.; Slater, L.Z.; Crowe, M. The impact of inflammation on cognitive function in older adults: Implications for healthcare practice and research. J. Neurosci. Nurs. 2012, 44, 206–217. [Google Scholar] [CrossRef]
- Shi, F.-D.; Yong, V.W. Neuroinflammation across neurological diseases. Science 2025, 388, eadx0043. [Google Scholar] [CrossRef] [PubMed]
- Henn, R.E.; Noureldein, M.H.; Elzinga, S.E.; Kim, B.; Savelieff, M.G.; Feldman, E.L. Glial-neuron crosstalk in health and disease: A focus on metabolism, obesity, and cognitive impairment. Neurobiol. Dis. 2022, 170, 105766. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, N.A.; Mukherjee, S.; Singer, T.; Venkatesh, A.; Perez Giraldo, G.S.; Jimenez, M.; Miller, J.; Lopez, M.; Hanson, B.A.; Bawa, A.P.; et al. Neurologic Manifestations of Long COVID Disproportionately Affect Young and Middle-Age Adults. Ann. Neurol. 2025, 97, 369–383. [Google Scholar] [CrossRef]
- Tang, N.; Kido, T.; Shi, J.; McCafferty, E.; Ford, J.M.; Dal Bon, K.; Pulliam, L. Blood Markers Show Neural Consequences of LongCOVID-19. Cells 2024, 13, 478. [Google Scholar] [CrossRef]
- Pase, M.P.; Himali, J.J.; Beiser, A.S.; DeCarli, C.; McGrath, E.R.; Satizabal, C.L.; Aparicio, H.J.; Adams, H.H.H.; Reiner, A.P.; Longstreth, W.T., Jr.; et al. Association of CD14 with incident dementia and markers of brain aging and injury. Neurology 2020, 94, e254–e266. [Google Scholar] [CrossRef]
- Abondio, P.; Sazzini, M.; Garagnani, P.; Boattini, A.; Monti, D.; Franceschi, C.; Luiselli, D.; Giuliani, C. The Genetic Variability of APOE in Different Human Populations and Its Implications for Longevity. Genes 2019, 10, 222. [Google Scholar] [CrossRef]
- Database for Annotation, Visualization, and Integrated Discovery (DAVID). Available online: https://davidbioinformatics.nih.gov/ (accessed on 17 June 2025).
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- g:Profiler. Available online: https://biit.cs.ut.ee/gprofiler/gost (accessed on 17 June 2025).
- Tavares-Júnior, J.W.L.; Oliveira, D.N.; da Silva, J.B.S.; Queiroz Feitosa, W.L.; Sousa, A.V.M.; Marinho, S.C.; Cunha, L.C.V.; Gaspar, S.B.; Gomes, C.M.P.; de Oliveira, L.L.B.; et al. Post-COVID-19 Cognitive Decline and Apoe Polymorphism: Towards a Possible Link? Brain Sci. 2023, 13, 1611. [Google Scholar] [CrossRef]
- Sudre, C.H.; Murray, B.; Varsavsky, T.; Graham, M.S.; Penfold, R.S.; Bowyer, R.C.; Pujol, J.C.; Klaser, K.; Antonelli, M.; Canas, L.S.; et al. Attributes and predictors of long COVID. Nat. Med. 2021, 27, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Chambliss, A.B.; Aljehani, M.; Tran, B.; Chen, X.; Elton, E.; Garri, C.; Ung, N.; Matasci, N.; Gross, M.E. Immune biomarkers associated with COVID-19 disease severity in an urban, hospitalized population. Pract. Lab. Med. 2023, 36, e00323. [Google Scholar] [CrossRef]
- Hill, E.L.; Mehta, H.B.; Sharma, S.; Mane, K.; Singh, S.K.; Xie, C.; Cathey, E.; Loomba, J.; Russell, S.; Spratt, H.; et al. Risk factors associated with post-acute sequelae of SARS-CoV-2: An N3C and NIH RECOVER study. BMC Public Health 2023, 23, 2103. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.-Y.; Yuen, C.-K.; Ip, J.D.; Wong, W.-M.; To, K.K.-W.; Yuen, K.-Y.; Kok, K.-H. Loss of orf3b in the circulating SARS-CoV-2 strains. Emerg. Microbes Infect. 2020, 9, 2685–2696. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kainulainen, M.H.; Jiang, N.; Di, H.; Bonenfant, G.; Mills, L.; Currier, M.; Shrivastava-Ranjan, P.; Calderon, B.M.; Sheth, M.; et al. Differential neutralization and inhibition of SARS-CoV-2 variants by antibodies elicited by COVID-19 mRNA vaccines. Nat. Commun. 2022, 13, 4350. [Google Scholar] [CrossRef]
- Berber, E.; Ross, T.M. Factors Predicting COVID-19 Vaccine Effectiveness and Longevity of Humoral Immune Responses. Vaccines 2024, 12, 1284. [Google Scholar] [CrossRef]
- Nasser, S.M.T.; Rana, A.A.; Doffinger, R.; Kafizas, A.; Khan, T.A.; Nasser, S. Elevated free interleukin-18 associated with severity and mortality in prospective cohort study of 206 hospitalised COVID-19 patients. Intensive Care Med. Exp. 2023, 11, 9. [Google Scholar] [CrossRef]
- Satış, H.; Özger, H.S.; Aysert Yıldız, P.; Hızel, K.; Gulbahar, Ö.; Erbaş, G.; Aygencel, G.; Guzel Tunccan, O.; Öztürk, M.A.; Dizbay, M.; et al. Prognostic value of interleukin-18 and its association with other inflammatory markers and disease severity in COVID-19. Cytokine 2021, 137, 155302. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Shahbaz, S.; Luo, X.; Osman, M.; Redmond, D.; Cohen Tervaert, J.W.; Li, L.; Elahi, S. Metabolomic and immune alterations in long COVID patients with chronic fatigue syndrome. Front. Immunol. 2024, 15, 1341843. [Google Scholar] [CrossRef]
- Attia, H.; El Nagdy, M.; Abdel Halim, R.M. Preliminary Study of sCD14 and sCD163 as Predictors of Disease Severity and ICU Admission in COVID-19: Relation to Hematological Parameters, Blood Morphological Changes and Inflammatory Biomarkers. Mediterr. J. Hematol. Infect. Dis. 2023, 15, e2023046. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Rial, J.; Currás-Tuala, M.J.; Rivero-Calle, I.; Gómez-Carballa, A.; Cebey-López, M.; Rodríguez-Tenreiro, C.; Dacosta-Urbieta, A.; Rivero-Velasco, C.; Rodríguez-Núñez, N.; Trastoy-Pena, R.; et al. Increased Serum Levels of sCD14 and sCD163 Indicate a Preponderant Role for Monocytes in COVID-19 Immunopathology. Front. Immunol. 2020, 11, 560381. [Google Scholar] [CrossRef]
- Wang, X.; Shi, Z.; Qiu, Y.; Sun, D.; Zhou, H. Peripheral GFAP and NfL as early biomarkers for dementia: Longitudinal insights from the UK Biobank. BMC Med. 2024, 22, 192. [Google Scholar] [CrossRef]
- Plantone, D.; Stufano, A.; Righi, D.; Locci, S.; Iavicoli, I.; Lovreglio, P.; De Stefano, N. Neurofilament light chain and glial fibrillary acid protein levels are elevated in post-mild COVID-19 or asymptomatic SARS-CoV-2 cases. Sci. Rep. 2024, 14, 6429. [Google Scholar] [CrossRef] [PubMed]
- Wruck, W.; Adjaye, J. Meta-analysis of human prefrontal cortex reveals activation of GFAP and decline of synaptic transmission in the aging brain. Acta Neuropathol. Commun. 2020, 8, 26. [Google Scholar] [CrossRef]
- Chatterjee, P.; Pedrini, S.; Stoops, E.; Goozee, K.; Villemagne, V.L.; Asih, P.R.; Verberk, I.M.W.; Dave, P.; Taddei, K.; Sohrabi, H.R.; et al. Plasma glial fibrillary acidic protein is elevated in cognitively normal older adults at risk of Alzheimer’s disease. Transl. Psychiatry 2021, 11, 27. [Google Scholar] [CrossRef] [PubMed]
- Capo, X.; Galmes-Panades, A.M.; Navas-Enamorado, C.; Ortega-Moral, A.; Marín, S.; Cascante, M.; Sánchez-Polo, A.; Masmiquel, L.; Torrens-Mas, M.; Gonzalez-Freire, M. Circulating Neurofilament Light Chain Levels Increase with Age and Are Associated with Worse Physical Function and Body Composition in Men but Not in Women. Int. J. Mol. Sci. 2023, 24, 12751. [Google Scholar] [CrossRef]
- Jung, Y.; Damoiseaux, J.S. The potential of blood neurofilament light as a marker of neurodegeneration for Alzheimer’s disease. Brain 2024, 147, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Gutman, E.G.; Salvio, A.L.; Fernandes, R.A.; Duarte, L.A.; Raposo-Vedovi, J.V.; Alcaraz, H.F.; Teixeira, M.A.; Passos, G.F.; de Medeiros, K.Q.M.; Hammerle, M.B.; et al. Long COVID: Plasma levels of neurofilament light chain in mild COVID-19 patients with neurocognitive symptoms. Mol. Psychiatry 2024, 29, 3106–3116. [Google Scholar] [CrossRef]
- Yong, S.J.; Halim, A.; Halim, M.; Liu, S.; Aljeldah, M.; Al Shammari, B.R.; Alwarthan, S.; Alhajri, M.; Alawfi, A.; Alshengeti, A.; et al. Inflammatory and vascular biomarkers in post-COVID-19 syndrome: A systematic review and meta-analysis of over 20 biomarkers. Rev. Med. Virol. 2023, 33, e2424. [Google Scholar] [CrossRef]
- Ryu, J.K.; Yan, Z.; Montano, M.; Sozmen, E.G.; Dixit, K.; Suryawanshi, R.K.; Matsui, Y.; Helmy, E.; Kaushal, P.; Makanani, S.K.; et al. Fibrin drives thromboinflammation and neuropathology in COVID-19. Nature 2024, 633, 905–913. [Google Scholar] [CrossRef]
- Obeagu, E.I.; Obeagu, G.U. Thromboinflammation in COVID-19: Unraveling the interplay of coagulation and inflammation. Medicine 2024, 103, e38922. [Google Scholar] [CrossRef]
- Sui, J.; Noubouossie, D.F.; Gandotra, S.; Cao, L. Elevated Plasma Fibrinogen Is Associated With Excessive Inflammation and Disease Severity in COVID-19 Patients. Front. Cell Infect. Microbiol. 2021, 11, 734005. [Google Scholar] [CrossRef]
- Conway, E.M.; Mackman, N.; Warren, R.Q.; Wolberg, A.S.; Mosnier, L.O.; Campbell, R.A.; Gralinski, L.E.; Rondina, M.T.; van de Veerdonk, F.L.; Hoffmeister, K.M.; et al. Understanding COVID-19-associated coagulopathy. Nat. Rev. Immunol. 2022, 22, 639–649. [Google Scholar] [CrossRef]
- Zinellu, A.; Mangoni, A.A. Serum Complement C3 and C4 and COVID-19 Severity and Mortality: A Systematic Review and Meta-Analysis With Meta-Regression. Front. Immunol. 2021, 12, 696085. [Google Scholar] [CrossRef] [PubMed]
- Tomo, S.; Kiran Kumar, P.; Yadav, D.; Sankanagoudar, S.; Charan, J.; Purohit, A.; Nag, V.L.; Bhatia, P.K.; Singh, K.; Dutt, N.; et al. Association of Serum Complement C3 Levels with Severity and Mortality in COVID 19. Indian. J. Clin. Biochem. 2023, 38, 447–456. [Google Scholar] [CrossRef]
- Chauhan, A.J.; Wiffen, L.J.; Brown, T.P. COVID-19: A collision of complement, coagulation and inflammatory pathways. J. Thromb. Haemost. 2020, 18, 2110–2117. [Google Scholar] [CrossRef]
- Baumann, H.; Gauldie, J. The acute phase response. Immunol. Today 1994, 15, 74–80. [Google Scholar] [CrossRef]
- de Lima, F.; Moraes, C.R.P.; Barbosa, M.S.; Bombassaro, B.; Palma, A.C.; Dertkigil, S.S.J.; Moretti, M.L.; Orsi, F.A.; Annichino-Bizzacchi, J.M.; Mansour, E.; et al. Association of heme-oxygenase 1, hemopexin, and heme levels with markers of disease severity in COVID-19. Exp. Biol. Med. 2023, 248, 309–316. [Google Scholar] [CrossRef]
- Castaño, E.M.; Roher, A.E.; Esh, C.L.; Kokjohn, T.A.; Beach, T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer’s disease and non-demented elderly subjects. Neurol. Res. 2006, 28, 155–163. [Google Scholar] [CrossRef]
- Ashraf, A.; Ashton, N.J.; Chatterjee, P.; Goozee, K.; Shen, K.; Fripp, J.; Ames, D.; Rowe, C.; Masters, C.L.; Villemagne, V.; et al. Plasma transferrin and hemopexin are associated with altered Aβ uptake and cognitive decline in Alzheimer’s disease pathology. Alzheimers Res. Ther. 2020, 12, 72. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, L.M.; Escandón, K.; Ulrich, A.K.; Rasmussen, A.L.; Roy, C.J.; Bix, G.J.; Popescu, S.V.; Moore, K.A.; Osterholm, M.T. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Dose, Infection, and Disease Outcomes for Coronavirus Disease 2019 (COVID-19): A Review. Clin Infect Dis 2022, 75, e1195–e1201. [Google Scholar] [CrossRef] [PubMed]
- Zsichla, L.; Müller, V. Risk Factors of Severe COVID-19: A Review of Host, Viral and Environmental Factors. Viruses 2023, 15, 175. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ma, F.; Yokota, T.; Garcia, G., Jr.; Palermo, A.; Wang, Y.; Farrell, C.; Wang, Y.-C.; Wu, R.; Zhou, Z.; et al. Metabolic reprogramming and epigenetic changes of vital organs in SARS-CoV-2–induced systemic toxicity. JCI Insight 2023, 6, e145027. [Google Scholar] [CrossRef]
- Cao, X.; Li, W.; Wang, T.; Ran, D.; Davalos, V.; Planas-Serra, L.; Pujol, A.; Esteller, M.; Wang, X.; Yu, H. Accelerated biological aging in COVID-19 patients. Nat. Commun. 2022, 13, 2135. [Google Scholar] [CrossRef]
- Franceschi, C.; Bonafè, M.; Valensin, S.; Olivieri, F.; De Luca, M.; Ottaviani, E.; De Benedictis, G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci. 2000, 908, 244–254. [Google Scholar] [CrossRef]
- Quach, H.Q.; Goergen, K.M.; Grill, D.E.; Ovsyannikova, I.G.; Poland, G.A.; Kennedy, R.B. Interleukin 8 as a Causal Link Between Aging and Impaired Influenza Antibody Responses in Older Adults. J. Infect. Dis. 2025, 232, 718–726. [Google Scholar] [CrossRef]
- Nichols, N.R.; Day, J.R.; Laping, N.J.; Johnson, S.A.; Finch, C.E. GFAP mRNA increases with age in rat and human brain. Neurobiol. Aging 1993, 14, 421–429. [Google Scholar] [CrossRef]
- David, J.P.; Ghozali, F.; Fallet-Bianco, C.; Wattez, A.; Delaine, S.; Boniface, B.; Di Menza, C.; Delacourte, A. Glial reaction in the hippocampal formation is highly correlated with aging in human brain. Neurosci. Lett. 1997, 235, 53–56. [Google Scholar] [CrossRef]
- Hu, H.; Chen, K.L.; Ou, Y.N.; Cao, X.P.; Chen, S.D.; Cui, M.; Dong, Q.; Tan, L.; Yu, J.T. Neurofilament light chain plasma concentration predicts neurodegeneration and clinical progression in nondemented elderly adults. Aging 2019, 11, 6904–6914. [Google Scholar] [CrossRef] [PubMed]
- Khalil, M.; Pirpamer, L.; Hofer, E.; Voortman, M.M.; Barro, C.; Leppert, D.; Benkert, P.; Ropele, S.; Enzinger, C.; Fazekas, F.; et al. Serum neurofilament light levels in normal aging and their association with morphologic brain changes. Nat. Commun. 2020, 11, 812. [Google Scholar] [CrossRef]
- Harp, C.; Thanei, G.A.; Jia, X.; Kuhle, J.; Leppert, D.; Schaedelin, S.; Benkert, P.; von Büdingen, H.C.; Hendricks, R.; Herman, A. Development of an age-adjusted model for blood neurofilament light chain. Ann. Clin. Transl. Neurol. 2022, 9, 444–453. [Google Scholar] [CrossRef]
- Needham, E.J.; Ren, A.L.; Digby, R.J.; Norton, E.J.; Ebrahimi, S.; Outtrim, J.G.; Chatfield, D.A.; Manktelow, A.E.; Leibowitz, M.M.; Newcombe, V.F.J.; et al. Brain injury in COVID-19 is associated with dysregulated innate and adaptive immune responses. Brain 2022, 145, 4097–4107. [Google Scholar] [CrossRef]
- Michael, B.D.; Dunai, C.; Needham, E.J.; Tharmaratnam, K.; Williams, R.; Huang, Y.; Boardman, S.A.; Clark, J.J.; Sharma, P.; Subramaniam, K.; et al. Para-infectious brain injury in COVID-19 persists at follow-up despite attenuated cytokine and autoantibody responses. Nat. Commun. 2023, 14, 8487. [Google Scholar] [CrossRef] [PubMed]
- Abdelhak, A.; Huss, A.; Kassubek, J.; Tumani, H.; Otto, M. Serum GFAP as a biomarker for disease severity in multiple sclerosis. Sci. Rep. 2018, 8, 14798. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.; Stukas, S.; Hoiland, R.L.; Fergusson, N.A.; Thiara, S.; Foster, D.; Mitra, A.; Stoessl, J.A.; Panenka, W.J.; Sekhon, M.S.; et al. Quantification of Neurological Blood-Based Biomarkers in Critically Ill Patients With Coronavirus Disease 2019. Crit. Care Explor. 2020, 2, e0238. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Nakamura, Y.; Michalak, Z.; Isobe, N.; Barro, C.; Leppert, D.; Matsushita, T.; Hayashi, F.; Yamasaki, R.; Kuhle, J.; et al. Serum GFAP and neurofilament light as biomarkers of disease activity and disability in NMOSD. Neurology 2019, 93, e1299–e1311. [Google Scholar] [CrossRef] [PubMed]
- Ameres, M.; Brandstetter, S.; Toncheva, A.A.; Kabesch, M.; Leppert, D.; Kuhle, J.; Wellmann, S. Association of neuronal injury blood marker neurofilament light chain with mild-to-moderate COVID-19. J. Neurol. 2020, 267, 3476–3478. [Google Scholar] [CrossRef]
- Poillerat, V.; Gentinetta, T.; Leon, J.; Wassmer, A.; Edler, M.; Torset, C.; Luo, D.; Tuffin, G.; Roumenina, L.T. Hemopexin as an Inhibitor of Hemolysis-Induced Complement Activation. Front. Immunol. 2020, 11, 1684. [Google Scholar] [CrossRef]
- Muller-Eberhard, U.; Liem, H.H.; Cox, K.H.; Conway, T.P. Hemopexin synthesis in vitro by human fetal tissues. Pediatr. Res. 1975, 9, 519–521. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, X.; Luo, Y.; He, S.; Lu, Y.; Gong, Y.; Gao, L.; Mao, S.; Liu, X.; Jiang, N.; Pu, Q.; et al. Age-, sex- and proximal–distal-resolved multi-omics identifies regulators of intestinal aging in non-human primates. Nat. Aging 2024, 4, 414–433. [Google Scholar] [CrossRef]
- Donkin, R.; Fung, Y.L.; Singh, I. Fibrinogen, Coagulation, and Ageing. In Biochemistry and Cell Biology of Ageing: Part III Biomedical Science; Harris, J.R., Korolchuk, V.I., Eds.; Springer International Publishing: Cham, Switzerland, 2023; pp. 313–342. [Google Scholar]
- Jackson, R.J.; Hyman, B.T.; Serrano-Pozo, A. Multifaceted roles of APOE in Alzheimer disease. Nat. Rev. Neurol. 2024, 20, 457–474. [Google Scholar] [CrossRef]
- Byambasuren, O.; Stehlik, P.; Clark, J.; Alcorn, K.; Glasziou, P. Effect of COVID-19 vaccination on long covid: Systematic review. BMJ Med. 2023, 2, e000385. [Google Scholar] [CrossRef]
- Mukherjee, S.; Singer, T.; Venkatesh, A.; Choudhury, N.A.; Perez Giraldo, G.S.; Jimenez, M.; Miller, J.; Lopez, M.; Hanson, B.A.; Bawa, A.P.; et al. Vaccination prior to SARS-CoV-2 infection does not affect the neurologic manifestations of long COVID. Brain Commun. 2025, 7, fcae448. [Google Scholar] [CrossRef]
- Soung, A.L.; Vanderheiden, A.; Nordvig, A.S.; Sissoko, C.A.; Canoll, P.; Mariani, M.B.; Jiang, X.; Bricker, T.; Rosoklija, G.B.; Arango, V.; et al. COVID-19 induces CNS cytokine expression and loss of hippocampal neurogenesis. Brain 2022, 145, 4193–4201. [Google Scholar] [CrossRef]
- Wu, M.D.; Hein, A.M.; Moravan, M.J.; Shaftel, S.S.; Olschowka, J.A.; O’Banion, M.K. Adult murine hippocampal neurogenesis is inhibited by sustained IL-1β and not rescued by voluntary running. Brain Behav. Immun. 2012, 26, 292–300. [Google Scholar] [CrossRef]
- Vanderheiden, A.; Hill, J.D.; Jiang, X.; Deppen, B.; Bamunuarachchi, G.; Soudani, N.; Joshi, A.; Cain, M.D.; Boon, A.C.M.; Klein, R.S. Vaccination reduces central nervous system IL-1β and memory deficits after COVID-19 in mice. Nat. Immunol. 2024, 25, 1158–1171. [Google Scholar] [CrossRef]
- Heneka, M.T.; van der Flier, W.M.; Jessen, F.; Hoozemanns, J.; Thal, D.R.; Boche, D.; Brosseron, F.; Teunissen, C.; Zetterberg, H.; Jacobs, A.H.; et al. Neuroinflammation in Alzheimer disease. Nat. Rev. Immunol. 2025, 25, 321–352. [Google Scholar] [CrossRef] [PubMed]
- Rea, I.M.; Gibson, D.S.; McGilligan, V.; McNerlan, S.E.; Alexander, H.D.; Ross, O.A. Age and Age-Related Diseases: Role of Inflammation Triggers and Cytokines. Front. Immunol. 2018, 9, 586. [Google Scholar] [CrossRef]
- Xu, E.; Xie, Y.; Al-Aly, Z. Long-term neurologic outcomes of COVID-19. Nat. Med. 2022, 28, 2406–2415. [Google Scholar] [CrossRef]
- Müller, L.; Di Benedetto, S. Inflammaging, immunosenescence, and cardiovascular aging: Insights into long COVID implications. Front. Cardiovasc. Med. 2024, 11, 1384996. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Pawelec, G.; Khalil, A.; Cohen, A.A.; Hirokawa, K.; Witkowski, J.M.; Franceschi, C. Immunology of Aging: The Birth of Inflammaging. Clin. Rev. Allergy Immunol. 2023, 64, 109–122. [Google Scholar] [CrossRef]
- Greene, C.; Connolly, R.; Brennan, D.; Laffan, A.; O’Keeffe, E.; Zaporojan, L.; O’Callaghan, J.; Thomson, B.; Connolly, E.; Argue, R.; et al. Blood–brain barrier disruption and sustained systemic inflammation in individuals with long COVID-associated cognitive impairment. Nat. Neurosci. 2024, 27, 421–432. [Google Scholar] [CrossRef]
- Bowman, G.L.; Dayon, L.; Kirkland, R.; Wojcik, J.; Peyratout, G.; Severin, I.C.; Henry, H.; Oikonomidi, A.; Migliavacca, E.; Bacher, M.; et al. Blood-brain barrier breakdown, neuroinflammation, and cognitive decline in older adults. Alzheimer’s Dement. 2018, 14, 1640–1650. [Google Scholar] [CrossRef]
- Montagne, A.; Barnes, S.R.; Sweeney, M.D.; Halliday, M.R.; Sagare, A.P.; Zhao, Z.; Toga, A.W.; Jacobs, R.E.; Liu, C.Y.; Amezcua, L.; et al. Blood-brain barrier breakdown in the aging human hippocampus. Neuron 2015, 85, 296–302. [Google Scholar] [CrossRef]
- Akhter, N.; Ahmad, S.; Alzahrani, F.A.; Dar, S.A.; Wahid, M.; Haque, S.; Bhatia, K.; Almalki, S., Sr.; Alharbi, R.A.; Sindi, A.A.A. Impact of COVID-19 on the cerebrovascular system and the prevention of RBC lysis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10267–10278. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, F.; Cheng, D.; Wang, Z.; Xing, N.; Yuan, J.; Zhang, W.; Xing, F. Free heme induces neuroinflammation and cognitive impairment by microglial activation via the TLR4/MyD88/NF-κB signaling pathway. Cell Commun. Signal 2024, 22, 16. [Google Scholar] [CrossRef]
- Gao, Q.; Zhou, Y.; Chen, Y.; Hu, W.; Jin, W.; Zhou, C.; Yuan, H.; Li, J.; Lin, Z.; Lin, W. Role of iron in brain development, aging, and neurodegenerative diseases. Ann. Med. 2025, 57, 2472871. [Google Scholar] [CrossRef]
- Remesal, L.; Sucharov-Costa, J.; Wu, Y.; Pratt, K.J.B.; Bieri, G.; Philp, A.; Phan, M.; Aghayev, T.; White, C.W.; Wheatley, E.G.; et al. Targeting iron-associated protein Ftl1 in the brain of old mice improves age-related cognitive impairment. Nat. Aging 2025, 5, 1957–1969. [Google Scholar] [CrossRef]
- Borish, L. IL-10: Evolving concepts. J. Allergy Clin. Immunol. 1998, 101, 293–297. [Google Scholar] [CrossRef]
- Etzerodt, A.; Moestrup, S.K. CD163 and inflammation: Biological, diagnostic, and therapeutic aspects. Antioxid. Redox Signal 2013, 18, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Lévêque, M.; Jeune, K.S.-L.; Jouneau, S.; Moulis, S.; Desrues, B.; Belleguic, C.; Brinchault, G.; Le Trionnaire, S.; Gangneux, J.-P.; Dimanche-Boitrel, M.-T.; et al. Soluble CD14 acts as a DAMP in human macrophages: Origin and involvement in inflammatory cytokine/chemokine production. FASEB J. 2017, 31, 1891–1902. [Google Scholar] [CrossRef]
- Takahashi, N.; Eltalkhawy, Y.M.; Nasu, K.; Abdelnaser, R.A.; Monde, K.; Habash, S.A.; Nasser, H.; Hiyoshi, M.; Ishimoto, T.; Suzu, S. IL-10 induces activated phenotypes of monocytes observed in virally-suppressed HIV-1-infected individuals. Biochem. Biophys. Res. Commun. 2024, 729, 150342. [Google Scholar] [CrossRef]
- Proal, A.D.; Aleman, S.; Bomsel, M.; Brodin, P.; Buggert, M.; Cherry, S.; Chertow, D.S.; Davies, H.E.; Dupont, C.L.; Deeks, S.G.; et al. Targeting the SARS-CoV-2 reservoir in long COVID. Lancet Infect. Dis. 2025, 25, e294–e306. [Google Scholar] [CrossRef] [PubMed]




| HC (% or SD) n = 51 | Cov (% or SD) n = 35 | nLongC (% or SD) n = 51 | p Value | ||
|---|---|---|---|---|---|
| Sex (%) | Female | 17 (33.3) | 19 (54.3) | 25 (49.0) | 0.1150 a |
| Male | 34 (66.7) | 16 (45.7) | 26 (51.0) | ||
| Race (%) | White | 27 (52.9) | 14 (40.0) | 29 (56.9) | 0.0003 a,# |
| African American | 10 (19.6) | 3 (8.6) | 9 (17.6) | ||
| Asian | 2 (3.9) | 15 (42.9) | 6 (11.8) | ||
| Other | 0 (0) | 3 (8.6) | 4 (7.8) | ||
| Unknown | 12 (23.5) | 0 (0) | 3 (5.9) | ||
| Ethnicity (%) | Hispanic | 5 (9.8) | 4 (11.4) | 9 (17.6) | 0.6882 a,# |
| Non-Hispanic | 38 (74.5) | 31 (88.6) | 42 (82.4) | ||
| Unknown | 8 (15.7) | 0 (0) | 0 (0) | ||
| Apolipoprotein E genotype (%) | ε2, ε3 | NA | 5 (14.3) | 5 (9.8) | 0.1046 a |
| ε2, ε4 | NA | 0 (0) | 1 (2.0) | ||
| ε3, ε3 | NA | 26 (74.3) | 28 (54.9) | ||
| ε3, ε4 | NA | 4 (11.4) | 16 (31.4) | ||
| ε4, ε4 | NA | 0 (0) | 1 (2.0) | ||
| ε4 positive | NA | 4 (11.4) | 18 (35.3) | 0.0135 a | |
| Comorbidity § (%) | NA | 3 (5.7) | 19 (37.3) | 0.0027 a | |
| Vaccination status * (%) | Vn | NA | 3 (8.6) | 5 (9.8) | 0.4387 a |
| Vb | NA | 4 (11.4) | 12 (23.5) | ||
| Va | NA | 28 (80.0) | 33 (64.7) | ||
| Vu | NA | 0 (0) | 1 (2.0) | ||
| Multiple infections till visit (%) | NA | 2 (5.7) | 2 (3.9) | >0.9999 a | |
| Days till visit, Mean (SD) | NA | 462 (340) | 469 (351) | 0.9182 b | |
| Body Mass Index, Mean (SD) | NA | 24.2 (4.1) | 29.2 (20.6) | 0.1159 b | |
| Age in years, Range Mean (SD) | 20–76 47.7 (13.1) | 18–69 38.1 (16.6) | 19–64 42.9 (12.6) | 0.0086 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, N.; Ford, J.M.; Dal Bon, K.; Pulliam, L. Chronic Inflammation and Altered Immune Responses in LongCOVID Associate with Neurological Manifestations and Accelerated Aging. Cells 2025, 14, 1875. https://doi.org/10.3390/cells14231875
Tang N, Ford JM, Dal Bon K, Pulliam L. Chronic Inflammation and Altered Immune Responses in LongCOVID Associate with Neurological Manifestations and Accelerated Aging. Cells. 2025; 14(23):1875. https://doi.org/10.3390/cells14231875
Chicago/Turabian StyleTang, Norina, Judith M. Ford, Kaitlyn Dal Bon, and Lynn Pulliam. 2025. "Chronic Inflammation and Altered Immune Responses in LongCOVID Associate with Neurological Manifestations and Accelerated Aging" Cells 14, no. 23: 1875. https://doi.org/10.3390/cells14231875
APA StyleTang, N., Ford, J. M., Dal Bon, K., & Pulliam, L. (2025). Chronic Inflammation and Altered Immune Responses in LongCOVID Associate with Neurological Manifestations and Accelerated Aging. Cells, 14(23), 1875. https://doi.org/10.3390/cells14231875







