Gene–Environment Interaction: Small Deletions (DELs) and Transcriptomic Profiles in Non-Melanoma Skin Cancer (NMSC) and Potential Implications for Therapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Biological Samples
2.2. Arsenic Exposure Measurement
2.3. Somatic Mutation Assay
2.4. Gene Expression Assay
2.5. Mutation Detection
2.6. Statistical Methods
3. Results
- NMSC-associated DELs: A total of 965 DEL events representing 617 unique genomic coordinates (light pink region of the Venn diagram in Figure 1A).
- DELs common in NMSC and healthy skin: A total of 1729 DEL events representing 460 unique genomic coordinates (light green and light pink overlapping region in Figure 1A).
- DELs associated with healthy skin: A total of 170 DEL events representing 145 unique genomic coordinates (light green region in Venn diagram in Figure 1A).
3.1. BCC-Associated DELs
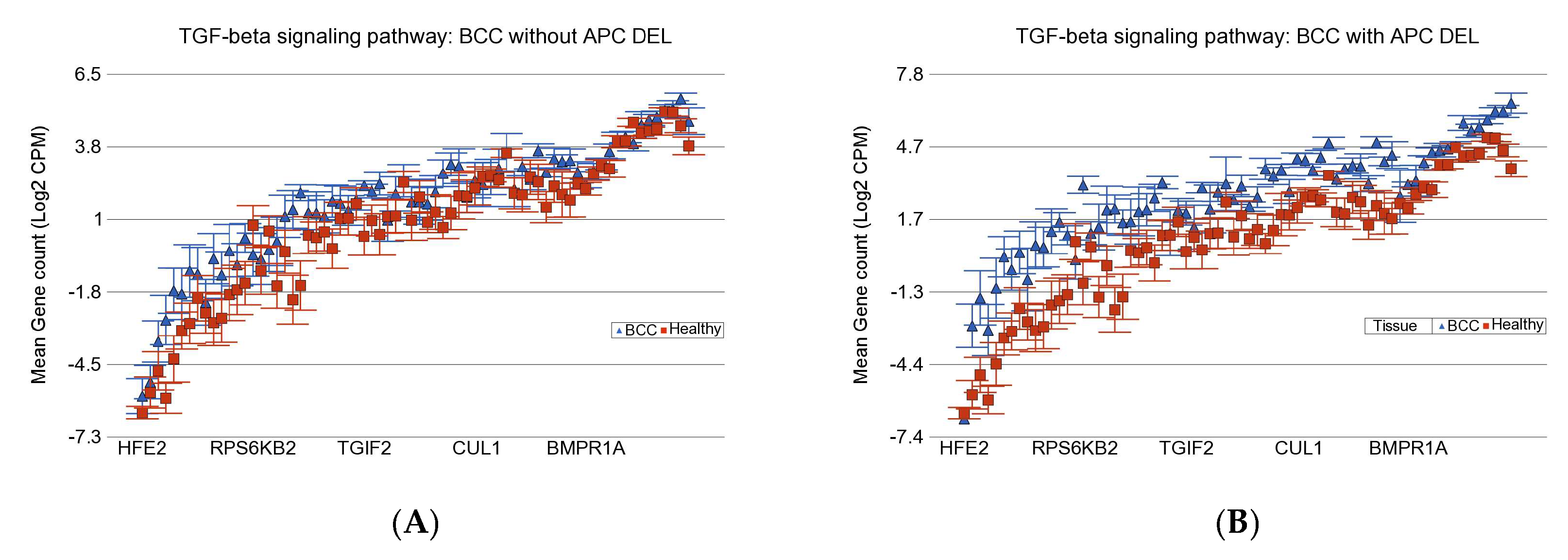
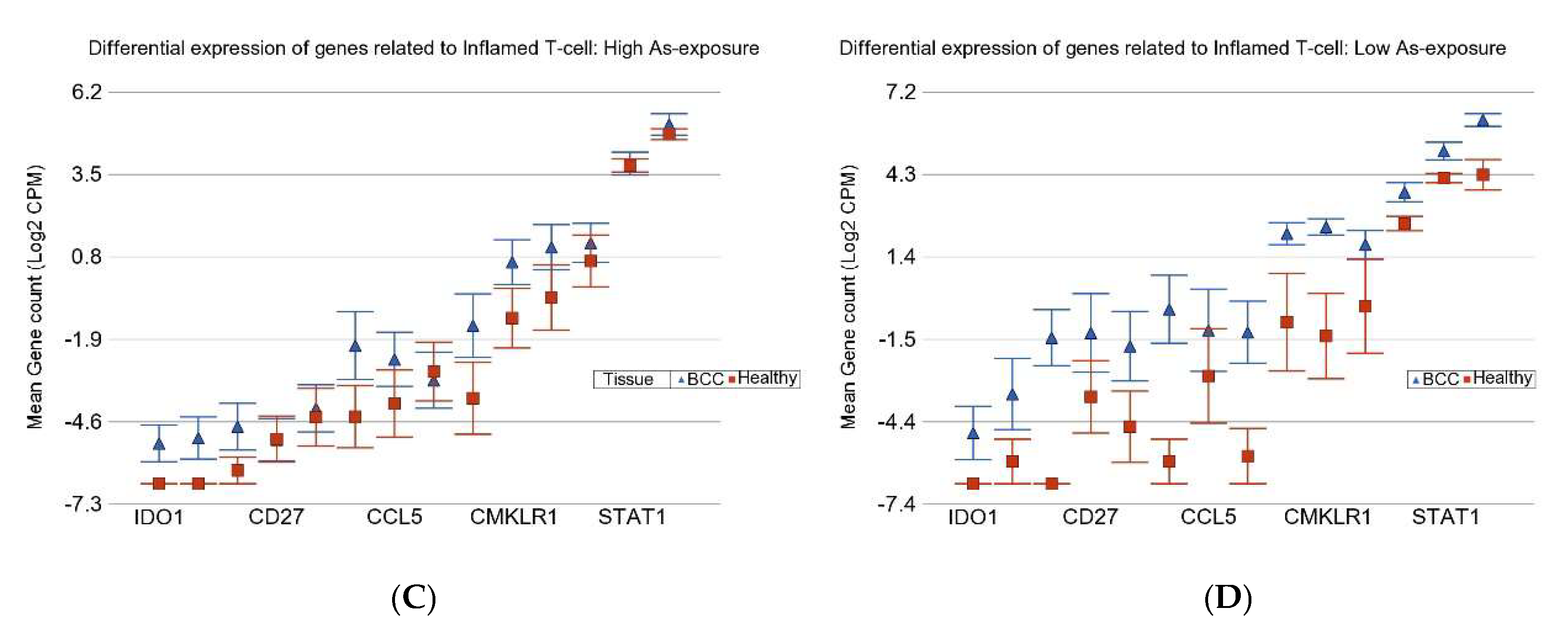
3.2. Gene–Environment Interaction
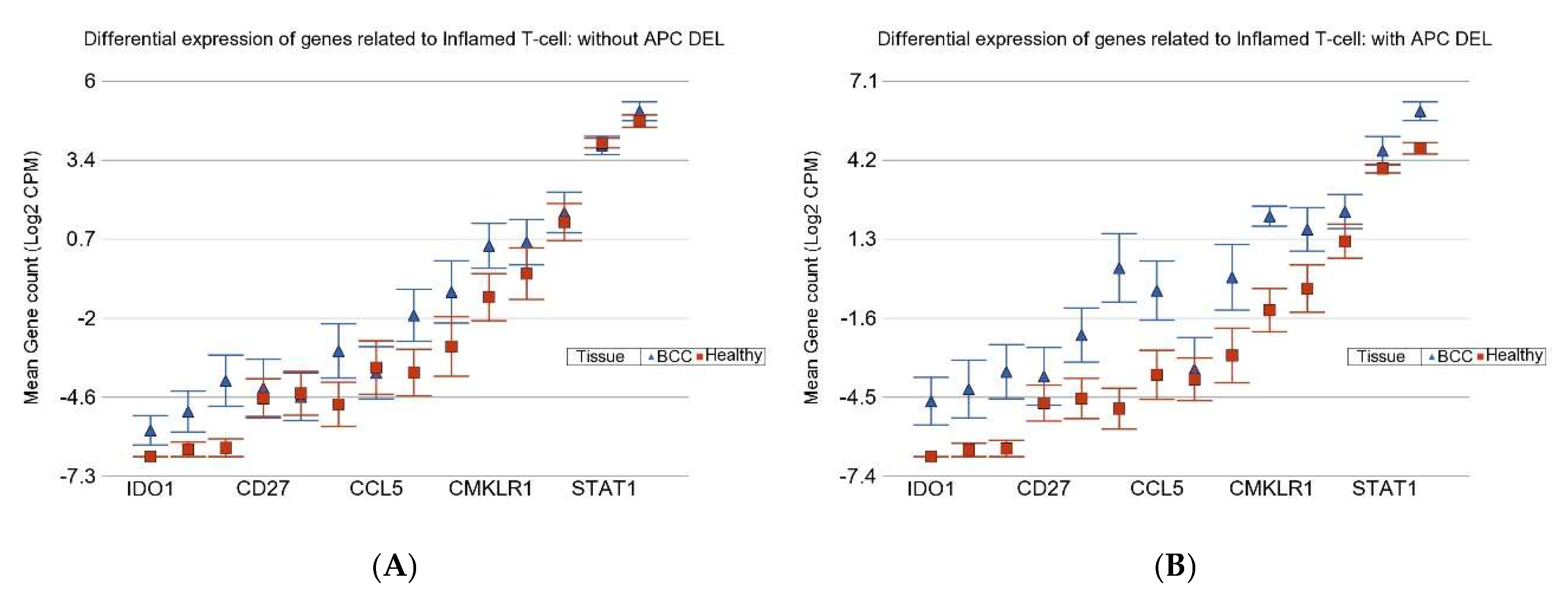
3.3. Inflamed T Cell Markers
3.4. APC DELs and PTCH1 Mutation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NMSC | Non-melanoma skin cancer |
| IPPH | Institute for Population and Precision Health |
| URB | UChicago Research Bangladesh |
| As | Arsenic |
| DEL | Small deletion |
| FC | Fold change |
| CI | Confidence interval |
| BD | Bowen’s disease |
| BCC | Basal-cell carcinoma |
| SCC | Squamous-cell carcinoma |
| SNV | Single-nucleotide variation |
| ASIR | Age-standardized incidence rate |
| CAM | Cell adhesion molecules |
| HEALS | Health Effects of Arsenic Longitudinal Study |
| SHS | Strong Heart Study |
| NHSCS | New Hampshire Skin Cancer Study |
| DMA% | Percentage of dimethylarsinic acid |
| Hh | Hedgehog signaling pathway |
| BEST | Bangladesh Vitamin E and Selenium Trial |
| UACR | Urinary As–creatinine ratio |
| ICP-MS | Inductively coupled plasma mass spectrometry |
| DNA | Deoxyribonucleic acid |
| RNA | Ribonucleic acid |
| TAS | Targeted amplicon sequencing |
| SNP | Single-nucleotide polymorphism |
| CPM | Counts per million |
| INS | Insertion |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ICI | Immune checkpoint inhibitors |
| OR | Odds ratio |
| RA | Rheumatoid arthritis |
| RR | Relative risk |
| Con A | Concanavalin A |
| TGF | Transforming growth factor |
| FN | Fibronectin |
References
- Jasmine, F.; Argos, M.; Khamkevych, Y.; Islam, T.; Rakibuz-Zaman, M.; Shahriar, M.; Shea, C.R.; Ahsan, H.; Kibriya, M.G. Molecular Profiling and the Interaction of Somatic Mutations with Transcriptomic Profiles in Non-Melanoma Skin Cancer (NMSC) in a Population Exposed to Arsenic. Cells 2024, 13, 1056. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdelalim, A.; Abdoli, A.; Abdollahpour, I.; Abdulle, A.S.M.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oh, C.C.; Jin, A.; Koh, W.P. Trends of cutaneous basal cell carcinoma, squamous cell carcinoma, and melanoma among the Chinese, Malays, and Indians in Singapore from 1968–2016. JAAD Int. 2021, 4, 39–45. [Google Scholar] [CrossRef] [PubMed Central]
- Ogata, D.; Namikawa, K.; Nakano, E.; Fujimori, M.; Uchitomi, Y.; Higashi, T.; Yamazaki, N.; Kawai, A. Epidemiology of skin cancer based on Japan’s National Cancer Registry 2016–2017. Cancer Sci. 2023, 114, 2986–2992. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kibriya, M.G.; Jasmine, F.; Munoz, A.; Islam, T.; Ahmed, A.; Tong, L.; Rakibuz-Zaman, M.; Shahriar, M.; Kamal, M.; Shea, C.R.; et al. Interaction of Arsenic Exposure and Transcriptomic Profile in Basal Cell Carcinoma. Cancers 2022, 14, 5598. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, Y.; Parvez, F.; Gamble, M.; Islam, T.; Ahmed, A.; Argos, M.; Graziano, J.H.; Ahsan, H. Arsenic exposure at low-to-moderate levels and skin lesions, arsenic metabolism, neurological functions, and biomarkers for respiratory and cardiovascular diseases: Review of recent findings from the Health Effects of Arsenic Longitudinal Study (HEALS) in Bangladesh. Toxicol. Appl. Pharmacol. 2009, 239, 184–192. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Naujokas, M.F.; Anderson, B.; Ahsan, H.; Aposhian, H.V.; Graziano, J.H.; Thompson, C.; Suk, W.A. The broad scope of health effects from chronic arsenic exposure: Update on a worldwide public health problem. Environ. Health Perspect. 2013, 121, 295–302. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- IARC Working Group. Arsenic, metals, fibres, and dusts. IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100 Pt C, 11–465. [Google Scholar] [PubMed]
- García-Esquinas, E.; Pollán, M.; Umans, J.G.; Francesconi, K.A.; Goessler, W.; Guallar, E.; Howard, B.; Farley, J.; Best, L.G.; Navas-Acien, A. Arsenic exposure and cancer mortality in a US-based prospective cohort: The strong heart study. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1944–1953. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yu, R.C.; Hsu, K.H.; Chen, C.J.; Froines, J.R. Arsenic methylation capacity and skin cancer. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1259–1262. [Google Scholar] [PubMed]
- Delgado, D.A.; Chernoff, M.; Huang, L.; Tong, L.; Chen, L.; Jasmine, F.; Shinkle, J.; Cole, S.A.; Haack, K.; Kent, J.; et al. Rare, Protein-Altering Variants in AS3MT and Arsenic Metabolism Efficiency: A Multi-Population Association Study. Environ. Health Perspect. 2021, 129, 47007. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pierce, B.L.; Tong, L.; Dean, S.; Argos, M.; Jasmine, F.; Rakibuz-Zaman, M.; Sarwar, G.; Islam, M.T.; Shahriar, H.; Islam, T.; et al. A missense variant in FTCD is associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet. 2019, 15, e1007984. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reichard, J.F.; Puga, A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics 2010, 2, 87–104. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jee, B.A.; Lim, H.; Kwon, S.M.; Jo, Y.; Park, M.C.; Lee, I.J.; Woo, H.G. Molecular classification of basal cell carcinoma of skin by gene expression profiling. Mol. Carcinog. 2015, 54, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, X.; Parmentier, L.; King, B.; Bezrukov, F.; Kaya, G.; Zoete, V.; Seplyarskiy, V.B.; Sharpe, H.J.; McKee, T.; Letourneau, A.; et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat. Genet. 2016, 48, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Bian, Q. Identification of Potential Biomarkers Associated with Basal Cell Carcinoma. Biomed Res. Int. 2020, 2020, 2073690. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pellegrini, C.; Maturo, M.G.; Di Nardo, L.; Ciciarelli, V.; Gutierrez Garcia-Rodrigo, C.; Fargnoli, M.C. Understanding the Molecular Genetics of Basal Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 2485. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Argos, M.; Rahman, M.; Parvez, F.; Dignam, J.; Islam, T.; Quasem, I.; Samar, K.H.; Haider, A.T.; Hossain, Z.; Patwary, T.I.; et al. Baseline comorbidities in a skin cancer prevention trial in Bangladesh. Eur. J. Clin. Investig. 2013, 43, 579–588. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nixon, D.E.; Mussmann, G.V.; Eckdahl, S.J.; Moyer, T.P. Total arsenic in urine: Palladium-persulfate vs nickel as a matrix modifier for graphite furnace atomic absorption spectrophotometry. Clin. Chem. 1991, 37, 1575–1579. [Google Scholar] [CrossRef] [PubMed]
- Heinegård, D.; Tiderström, G. Determination of serum creatinine by a direct colorimetric method. Clin. Chim. Acta 1973, 43, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Kibriya, M.G.; Jasmine, F.; Parvez, F.; Argos, M.; Roy, S.; Paul-Brutus, R.; Islam, T.; Ahmed, A.; Rakibuz-Zaman, M.; Shinkle, J.; et al. Association between genome-wide copy number variation and arsenic-induced skin lesions: A prospective study. Environ. Health 2017, 16, 75. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, Y.; Bizzaro, J.W.; Marx, K.A. Homopolymer tract length dependent enrichments in functional regions of 27 eukaryotes and their novel dependence on the organism DNA (G+C)% composition. BMC Genom. 2004, 5, 95. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Halman, A.; Lonsdale, A.; Oshlack, A. Analysis of Tandem Repeats in Short-Read Sequencing Data: From Genotyping Known Pathogenic Repeats to Discovering Novel Expansions. Curr. Protoc. 2024, 4, e70010. [Google Scholar] [CrossRef] [PubMed]
- Tanudisastro, H.A.; Deveson, I.W.; Dashnow, H.; MacArthur, D.G. Sequencing and characterizing short tandem repeats in the human genome. Nat. Rev. Genet. 2024, 25, 460–475. [Google Scholar] [CrossRef] [PubMed]
- Carless, M.A.; Griffiths, L.R. Cytogenetics of melanoma and nonmelanoma skin cancer. Adv. Exp. Med. Biol. 2014, 810, 160–181. [Google Scholar] [CrossRef] [PubMed]
- Ashton, K.J.; Carless, M.A.; Griffiths, L.R. Cytogenetic alterations in nonmelanoma skin cancer: A review. Genes. Chromosomes Cancer 2005, 43, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Krzysztofik, M.; Brzewski, P.; Cuber, P.; Kacprzyk, A.; Kulbat, A.; Richter, K.; Wojewoda, T.; Wysocki, W.M. Risk of Melanoma and Non-Melanoma Skin Cancer in Patients with Psoriasis and Psoriatic Arthritis Treated with Targeted Therapies: A Systematic Review and Meta-Analysis. Pharmaceuticals 2023, 17, 14. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, X.; Cai, G.; Liu, C.; Zhao, J.; Gu, C.; Wu, L.; Hamilton, T.A.; Zhang, C.J.; Ko, J.; Zhu, L.; et al. IL-17R-EGFR axis links wound healing to tumorigenesis in Lrig1(+) stem cells. J. Exp. Med. 2019, 216, 195–214. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kibriya, M.G.; Jasmine, F.; Khamkevych, Y.; Raza, M.; Kamal, M.; Bissonnette, M.; Ahsan, H. Association of Microsatellite Instability and Gene Expression Profile in Colorectal Carcinoma and Potential Implications for Therapy. Medicina 2024, 60, 348. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, S.; Na, R.; Li, X.; Zhang, Y.; Zheng, T. Targeting interleukin-17 enhances tumor response to immune checkpoint inhibitors in colorectal cancer. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188758. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Kang, H.; Fung, A.; Zhao, H.; Wang, T.; Ma, D. The role of interleukin 17 in tumour proliferation, angiogenesis, and metastasis. Mediat. Inflamm. 2014, 2014, 623759. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, J.; Chen, X.; Herjan, T.; Li, X. The role of interleukin-17 in tumor development and progression. J. Exp. Med. 2020, 217, e20190297. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chung, L.; Thiele Orberg, E.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e5. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grivennikov, S.I.; Wang, K.; Mucida, D.; Stewart, C.A.; Schnabl, B.; Jauch, D.; Taniguchi, K.; Yu, G.Y.; Osterreicher, C.H.; Hung, K.E.; et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012, 491, 254–258. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, K.; Kim, M.K.; Di Caro, G.; Wong, J.; Shalapour, S.; Wan, J.; Zhang, W.; Zhong, Z.; Sanchez-Lopez, E.; Wu, L.W.; et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity 2014, 41, 1052–1063. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, L.; Chen, X.; Zhao, J.; Martin, B.; Zepp, J.A.; Ko, J.S.; Gu, C.; Cai, G.; Ouyang, W.; Sen, G.; et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J. Exp. Med. 2015, 212, 1571–1587. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, R.; Wan, Q.; Zhao, R.; Xiao, H.; Cen, Y.; Xu, X. Risk of non-melanoma skin cancer with biological therapy in common inflammatory diseases: A systemic review and meta-analysis. Cancer Cell Int. 2021, 21, 614. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, J.; Song, Y.; Liu, L.; Yang, S.; Fang, B. Effect of arsenic trioxide on the Tregs ratio and the levels of IFN-γ, IL-4, IL-17 and TGF-β1 in the peripheral blood of severe aplastic anemia patients. Medicine 2020, 99, e20630. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dai, J.; Xu, M.; Zhang, X.; Niu, Q.; Hu, Y.; Li, Y.; Li, S. Bi-directional regulation of TGF-β/Smad pathway by arsenic: A systemic review and meta-analysis of in vivo and in vitro studies. Life Sci. 2019, 220, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Fijałkowska, M.; Bonczar, M.; Jastrzębski, I.; Ostrowski, P.; Antoszewski, B.; Koziej, M. The role of TGF-β and antimicrobial peptides in basal cell carcinoma: A systematic review. Postepy Dermatol. Alergol. 2023, 40, 384–389. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Heller, E.R.; Gor, A.; Wang, D.; Hu, Q.; Lucchese, A.; Kanduc, D.; Katdare, M.; Liu, S.; Sinha, A.A. Molecular signatures of basal cell carcinoma susceptibility and pathogenesis: A genomic approach. Int. J. Oncol. 2013, 42, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Siljamäki, E.; Rappu, P.; Riihilä, P.; Nissinen, L.; Kähäri, V.M.; Heino, J. H-Ras activation and fibroblast-induced TGF-β signaling promote laminin-332 accumulation and invasion in cutaneous squamous cell carcinoma. Matrix Biol. 2020, 87, 26–47. [Google Scholar] [CrossRef] [PubMed]
- Siljamäki, E.; Riihilä, P.; Suwal, U.; Nissinen, L.; Rappu, P.; Kallajoki, M.; Kähäri, V.M.; Heino, J. Inhibition of TGF-β signaling, invasion, and growth of cutaneous squamous cell carcinoma by PLX8394. Oncogene 2023, 42, 3633–3647. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Papageorgis, P. TGFβ Signaling in Tumor Initiation, Epithelial-to-Mesenchymal Transition, and Metastasis. J. Oncol. 2015, 2015, 587193. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Huang, C.Y.; Chung, C.L.; Hu, T.H.; Chen, J.J.; Liu, P.F.; Chen, C.L. Recent progress in TGF-β inhibitors for cancer therapy. Biomed. Pharmacother. 2021, 134, 111046. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Kang, K.; Chen, P.; Zeng, Z.; Li, G.; Xiong, W.; Yi, M.; Xiang, B. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol. Cancer 2024, 23, 108. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chang, J.; Zhu, G.A.; Cheung, C.; Li, S.; Kim, J.; Chang, A.L. Association Between Programmed Death Ligand 1 Expression in Patients With Basal Cell Carcinomas and the Number of Treatment Modalities. JAMA Dermatol. 2017, 153, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Gompertz-Mattar, M.; Perales, J.; Sahu, A.; Mondaca, S.; Gonzalez, S.; Uribe, P.; Navarrete-Dechent, C. Differential expression of programmed cell death ligand 1 (PD-L1) and inflammatory cells in basal cell carcinoma subtypes. Arch. Dermatol. Res. 2022, 314, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Ren, J.; Lu, X.; Ma, S.; Wu, C. Gene-Environment Interaction: A Variable Selection Perspective. Methods Mol. Biol. 2021, 2212, 191–223. [Google Scholar] [CrossRef] [PubMed]




| Coordinate | Type | Reference | Allele | Average Quality | Exact Match | Coding Region Change | Amino Acid Change |
|---|---|---|---|---|---|---|---|
| chr5:112111310 | Deletion | A | - | 37.00 | clinvar_20210828_hg19 | NM_000038.5:c.423-4delA; NM_001127510.2:c.423-4delA; NM_001127511.2:c.453-4delA | |
| chr5:112164717 | Deletion | T | - | 35.29 | NM_000038.5:c.1743+55delT; NM_001127510.2:c.1743+55delT; NM_001127511.2:c.1689+55delT | ||
| chr5:112178312 | Deletion | A | - | 35.29 | NM_000038.5:c.7023delA; NM_001127510.2:c.7023delA; NM_001127511.2:c.6969delA | NP_000029.2:p.Lys2341fs; NP_001120982.1:p.Lys2341fs; NP_001120983.2:p.Lys2323fs | |
| chr5:112164717 | Deletion | T | - | 36.48 | NM_000038.5:c.1743+55delT; NM_001127510.2:c.1743+55delT; NM_001127511.2:c.1689+55delT | ||
| chr5:112111310 | Deletion | A | - | 36.54 | clinvar_20210828_hg19 | NM_000038.5:c.423-4delA; NM_001127510.2:c.423-4delA; NM_001127511.2:c.453-4delA | |
| chr5:112111310..112111312 | Deletion | AAA | - | 33.00 | NM_000038.5:c.423-6_423-4delAAA; NM_001127510.2:c.423-6_423-4delAAA; NM_001127511.2:c.453-6_453-4delAAA | ||
| chr5:112163028 | Deletion | T | - | 37.00 | NM_000038.5:c.1548+91delT; NM_001127510.2:c.1548+91delT; NM_001127511.2:c.1494+91delT | ||
| chr5:112163028 | Deletion | T | - | 37.00 | NM_000038.5:c.1548+91delT; NM_001127510.2:c.1548+91delT; NM_001127511.2:c.1494+91delT | ||
| chr5:112164717 | Deletion | T | - | 35.50 | NM_000038.5:c.1743+55delT; NM_001127510.2:c.1743+55delT; NM_001127511.2:c.1689+55delT | ||
| chr5:112164717 | Deletion | T | - | 35.67 | NM_000038.5:c.1743+55delT; NM_001127510.2:c.1743+55delT; NM_001127511.2:c.1689+55delT | ||
| chr5:112164717 | Deletion | T | - | 37.00 | NM_000038.5:c.1743+55delT; NM_001127510.2:c.1743+55delT; NM_001127511.2:c.1689+55delT | ||
| chr5:112164717 | Deletion | T | - | 36.11 | NM_000038.5:c.1743+55delT; NM_001127510.2:c.1743+55delT; NM_001127511.2:c.1689+55delT |
| TGF-β Signaling Pathway | IL-17 Signaling Pathway | PD-L1 Expression and PD-1 Checkpoint Pathway in Cancer | ||||||
|---|---|---|---|---|---|---|---|---|
| UACR | APC DEL − ve | APC DEL + ve | APC DEL − ve | APC DEL + ve | APC DEL − ve | APC DEL + ve | ||
| >192 µg/g | FC | 1.3 | 2.33 | 1.13 | 1.86 | −1.03 | 2.02 | |
| 95% CI | (1.12 to 1.52) | (1.93 to 2.81) | (−1.05 to 1.34) | (1.52 to 2.29) | (−1.20 to 1.12) | (1.68 to 2.41) | ||
| BCC Cases | n = 11 | n = 6 | n = 11 | n = 6 | n = 11 | n = 6 | ||
| ≤192 µg/g | FC | 3.91 | 5.62 | 5.06 | 6.22 | 3.3 | 4.86 | |
| 95% CI | (3.21 to 4.77) | (4.53 to 6.98) | (4.06 to 6.30) | (4.90 to 7.89) | (2.72 to 4.00) | (3.94 to 5.99) | ||
| BCC Cases | n = 5 | n = 4 | n = 5 | n = 4 | n = 5 | n = 4 | ||
| Interaction p | 3.44 × 10−44 | 8.11 × 10−57 | 5.29 × 10−58 | |||||
| Genes Related to Inflamed T-Cells | ||||
|---|---|---|---|---|
| UACR | APC DEL − ve | APC DEL + ve | ||
| ≥192 µg/g | FC | 1.2 | 5.84 | |
| 95% CI | (−1.29 to 1.85) | (3.48 to 9.79) | ||
| BCC Cases | n = 11 | n = 6 | ||
| ≤192 µg/g | FC | 5.66 | 8.88 | |
| 95% CI | (2.97 to 10.79) | (4.48 to 17.60) | ||
| BCC Cases | n = 5 | n = 4 | ||
| Interaction p | 4.49 × 10−15 | |||
| Genes Related to Inflamed T-cell | |||
|---|---|---|---|
| APC DEL − ve | APC DEL + ve | ||
| PTCH1 SNV − ve | FC | 1.59 | 6.27 |
| 95% CI | (1.05 to 2.40) | (2.77 to 14.20) | |
| BCC Cases | n = 12 | n = 2 | |
| PTCH1 SNV + ve | FC | 6.04 | 7.51 |
| 95% CI | (3.28 to 11.10) | (4.68 to 12.04) | |
| BCC Cases | n = 4 | n = 8 | |
| Interaction p | 1.79 × 10−9 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jasmine, F.; Almazan, A.; Khamkevych, Y.; Argos, M.; Shahriar, M.; Islam, T.; Shea, C.R.; Ahsan, H.; Kibriya, M.G. Gene–Environment Interaction: Small Deletions (DELs) and Transcriptomic Profiles in Non-Melanoma Skin Cancer (NMSC) and Potential Implications for Therapy. Cells 2025, 14, 95. https://doi.org/10.3390/cells14020095
Jasmine F, Almazan A, Khamkevych Y, Argos M, Shahriar M, Islam T, Shea CR, Ahsan H, Kibriya MG. Gene–Environment Interaction: Small Deletions (DELs) and Transcriptomic Profiles in Non-Melanoma Skin Cancer (NMSC) and Potential Implications for Therapy. Cells. 2025; 14(2):95. https://doi.org/10.3390/cells14020095
Chicago/Turabian StyleJasmine, Farzana, Armando Almazan, Yuliia Khamkevych, Maria Argos, Mohammad Shahriar, Tariqul Islam, Christopher R. Shea, Habibul Ahsan, and Muhammad G. Kibriya. 2025. "Gene–Environment Interaction: Small Deletions (DELs) and Transcriptomic Profiles in Non-Melanoma Skin Cancer (NMSC) and Potential Implications for Therapy" Cells 14, no. 2: 95. https://doi.org/10.3390/cells14020095
APA StyleJasmine, F., Almazan, A., Khamkevych, Y., Argos, M., Shahriar, M., Islam, T., Shea, C. R., Ahsan, H., & Kibriya, M. G. (2025). Gene–Environment Interaction: Small Deletions (DELs) and Transcriptomic Profiles in Non-Melanoma Skin Cancer (NMSC) and Potential Implications for Therapy. Cells, 14(2), 95. https://doi.org/10.3390/cells14020095







