Modelling Peroxisomal Disorders in Zebrafish
Abstract
1. Introduction

2. Visualisation of Peroxisomes in Zebrafish
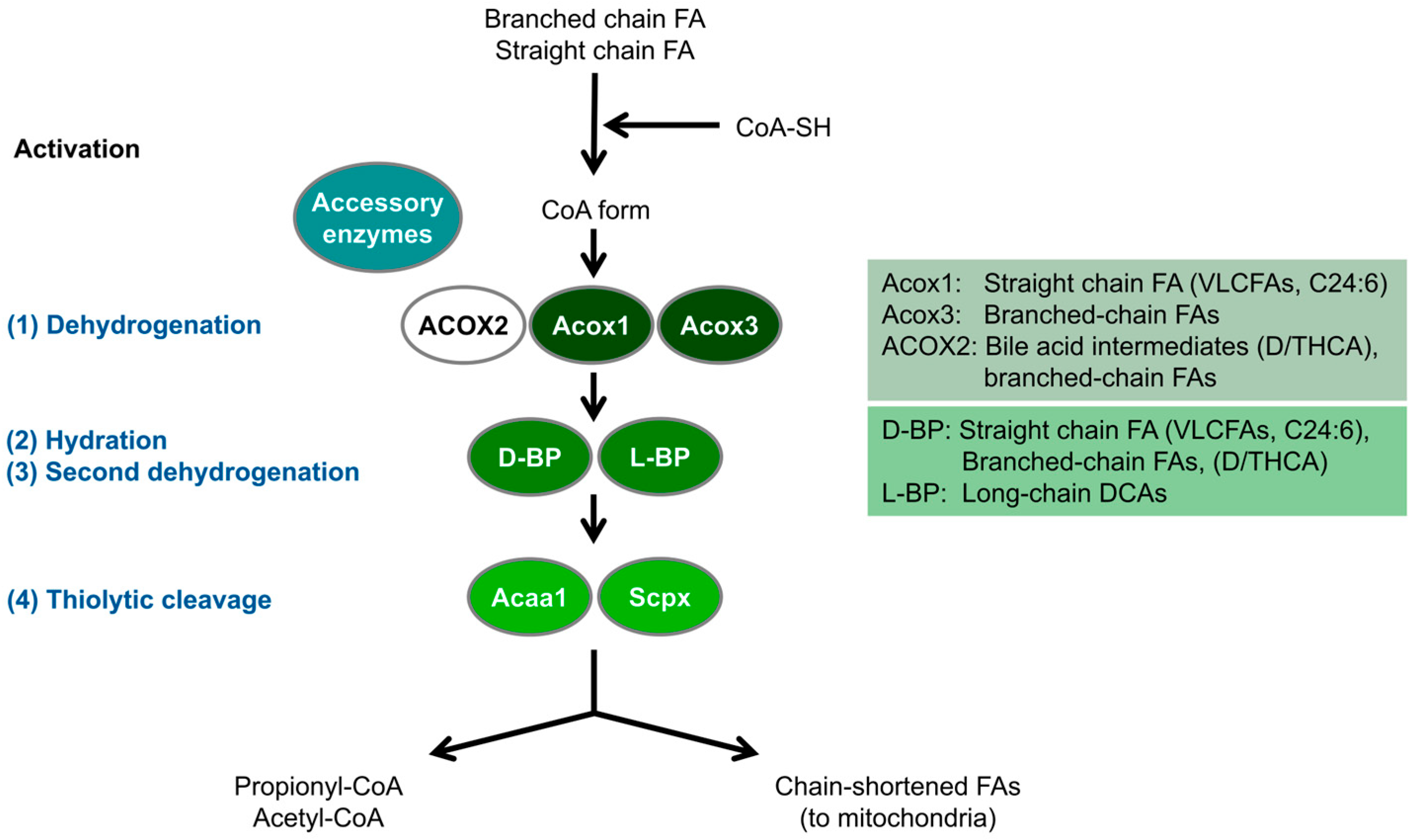
3. Peroxisomal Protein Inventory and Metabolic Pathways in Zebrafish
4. Zebrafish Models of Peroxisomal Disorders
4.1. Peroxisome Biogenesis Disorders/Zellweger Spectrum Disorders
4.1.1. pex2 Mutant Zebrafish
| Human Disorder | Targeted Gene | Method | Studied Tissues | Peroxisomal Phenotypes | Phenotypes Related to Clinical Features | Ref. |
|---|---|---|---|---|---|---|
| ZSD | pex2 | TALENs- mediated knockout | Liver Brain Eyes Muscles | ↓ Peroxisomal matrix protein import ↓ Catalase and glutathione peroxidase activities ↑ VLCFAs ↓ Ether phospholipids | △ Motor activity Hypotonia ↑ Hepatic lipid △ Neuronal and muscle function △ Gametogenesis ↓ Crystallin genes ↓ Survival | [63] |
| ZSD | pex5 | CRISPR/Cas9-mediated knockout | Liver Nervous system Whole larvae | ↓ Peroxisomal matrix protein import ↓ Peroxisome abundance | △ Motor activity Demyelination ↑ Hepatic lipid Edema Deflated swim bladder Shrunken liver ↓ Survival Expedited death under fasting conditions | [69] |
| ZSD | pex13 | CRISPR/Cas9-mediated knockout | Liver Whole larvae | ↓ Peroxisomal matrix protein import ↓ Peroxisome abundance ↑ Ubiquitinated PEX5 ↑ Peroxisome-dependent ROS ↑ Pexophagy | △ Motor activity ↑ Hepatic lipid accumulation △ Neuronal function Liver steatosis Laval mortality | [38] |
| pex13 | Morpholino- mediated knockout (mosaic) | Liver Pronephric duct Yolk sac | Partial elimination of peroxisomes | Not assessed | [30] | |
| ALD | abcd1 | TALENs- mediated knockout | CNS Adrenal glands Whole embryo | ↑ VLCFAs ↑ Cholesterol | △ Motor activity Hypomyelination △ Oligodendrocyte patterning △ CNS development ↑ Apoptosis ↓ Survival | [70] |
| Mitchell Syndrome | acox1 | Transient overexpression (N237S) | Brain Spinal cord Whole embryo | ↓ Peroxisome density ↑ Oxidative stress | △ Motor activity Activation of ISR ↓ Survival | [71] |
| D-BPD | dbp | Morpholino- mediated knockdown | Whole embryo Liver Pancreas | ↓ beta-oxidation ↓ Ether phospholipids synthesis ↓ pex5 expression | △ Yolk lipid consumption Growth retardation Morphological malformation △ Neuronal, liver, pancreas, cartilage, blood, blood vessels, digestive organ development Abnormal vascular patterning Embryonic lethality | [55] |
| Osteoarthritis | fis1 | Morpholino- mediated knockdown | Whole embryo | ↓ Peroxisome abundance ↓ Catalase and glutathione peroxidase activities ↓ beta-oxidation gene expression | ↑ Apoptosis ↑ Lipid ↓ Survival | [37] |
| Unspecified | vwa8 | Morpholino- mediated knockdown | Whole embryo | Not assessed | △ Motor activity Developmental delays and defects Light sensitivity Facial dysmorphism ↓ Survival | [72] |
| Autosomal-dominant retinitis pigmentosa | vwa8 | Morpholino- mediated knockdown | Retina Photoreceptor layer | Not assessed | △ Visual function Retinal pigment deposition Thinning of the retinal photoreceptor layer | [73] |
4.1.2. pex3 Mutant Zebrafish
4.1.3. pex5 Mutant Zebrafish
4.1.4. pex13 Mutant Zebrafish
4.2. Peroxisomal Single Enzyme Deficiency
Adrenoleukodystrophy (ALD) Model Zebrafish
4.3. Peroxisomal Beta-Oxidation Deficiency
4.3.1. ACOX1 Mutant Disease Model Zebrafish
4.3.2. D-Bifunctional Protein (Dbp) Deficiency Model Zebrafish
4.4. Dually Targeted Peroxisomal/Mitochondrial Protein Deficiency
4.4.1. Fission 1 (FIS1) Deficiency Model Zebrafish
4.4.2. VWA8 Deficiency Model Zebrafish
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rhodin, J. Correlation of Ultrastructural Organization and Function in Normal Experimentally Changed Convoluted Tubule Cells of the Mouse Kidney; Aktiebolaget Godvil: Stockholm, Sweden, 1954. [Google Scholar]
- De Duve, C.; Baudhuin, P. Peroxisomes (Microbodies and Related Particles). Physiol. Rev. 1966, 46, 323–357. [Google Scholar] [CrossRef] [PubMed]
- Fransen, M.; Nordgren, M.; Wang, B.; Apanasets, O.; Van Veldhoven, P.P. Aging, Age-Related Diseases and Peroxisomes. Subcell. Biochem. 2013, 69, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Islinger, M.; Voelkl, A.; Fahimi, H.D.; Schrader, M. The Peroxisome: An Update on Mysteries 2.0. Histochem. Cell Biol. 2018, 150, 443–471. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.M.; Sharma, M.; Naidu, S.; Bömmel, H.; Prabhuswamimath, S.C.; Madhusudhan, T.; Wihadmadyatami, H.; Bachhuka, A.; Karnati, S. Peroxisomes and PPARs: Emerging Role as Master Regulators of Cancer Metabolism. Mol. Metab. 2024, 90, 102044. [Google Scholar] [CrossRef] [PubMed]
- Fransen, M.; Lismont, C. Peroxisomal Hydrogen Peroxide Signaling: A New Chapter in Intracellular Communication Research. Curr. Opin. Chem. Biol. 2024, 78, 102426. [Google Scholar] [CrossRef]
- Lismont, C.; Revenco, I.; Fransen, M. Peroxisomal Hydrogen Peroxide Metabolism and Signaling in Health and Disease. Int. J. Mol. Sci. 2019, 20, 3673. [Google Scholar] [CrossRef]
- Dixit, E.; Boulant, S.; Zhang, Y.; Lee, A.S.Y.; Odendall, C.; Shum, B.; Hacohen, N.; Chen, Z.J.; Whelan, S.P.; Fransen, M.; et al. Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell 2010, 141, 668–681. [Google Scholar] [CrossRef]
- Weinhofer, I.; Buda, A.; Kunze, M.; Palfi, Z.; Traunfellner, M.; Hesse, S.; Villoria-Gonzalez, A.; Hofmann, J.; Hametner, S.; Regelsberger, G.; et al. Peroxisomal Very Long-Chain Fatty Acid Transport Is Targeted by Herpesviruses and the Antiviral Host Response. Commun. Biol. 2022, 5, 944. [Google Scholar] [CrossRef]
- Pellegrino, E.; Aylan, B.; Bussi, C.; Fearns, A.; Bernard, E.M.; Athanasiadi, N.; Santucci, P.; Botella, L.; Gutierrez, M.G. Peroxisomal ROS Control Cytosolic Mycobacterium Tuberculosis Replication in Human Macrophages. J. Cell Biol. 2023, 222, e202303066. [Google Scholar] [CrossRef]
- Di Cara, F.; Savary, S.; Kovacs, W.J.; Kim, P.; Rachubinski, R.A. The Peroxisome: An up-and-Coming Organelle in Immunometabolism. Trends Cell Biol. 2023, 33, 70–86. [Google Scholar] [CrossRef]
- Wanders, R.J.A.; Waterham, H.R. Biochemistry of Mammalian Peroxisomes Revisited. Annu. Rev. Biochem. 2006, 75, 295–332. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.A.; Baes, M.; Ribeiro, D.; Ferdinandusse, S.; Waterham, H.R. The Physiological Functions of Human Peroxisomes. Physiol. Rev. 2023, 103, 957–1024. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M.; Costello, J.; Godinho, L.F.; Islinger, M. Peroxisome-Mitochondria Interplay and Disease. J. Inherit. Metab. Dis. 2015, 38, 681–702. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.A.; Vaz, F.M.; Waterham, H.R.; Ferdinandusse, S. Fatty Acid Oxidation in Peroxisomes: Enzymology, Metabolic Crosstalk with Other Organelles and Peroxisomal Disorders. Adv. Exp. Med. Biol. 2020, 1299, 55–70. [Google Scholar] [CrossRef]
- Dorninger, F.; Werner, E.R.; Berger, J.; Watschinger, K. Regulation of Plasmalogen Metabolism and Traffic in Mammals: The Fog Begins to Lift. Front. Cell Dev. Biol. 2022, 10, 946393. [Google Scholar] [CrossRef]
- Silva, B.S.C.; DiGiovanni, L.; Kumar, R.; Carmichael, R.E.; Kim, P.K.; Schrader, M. Maintaining Social Contacts: The Physiological Relevance of Organelle Interactions. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118800. [Google Scholar] [CrossRef]
- Costello, J.L.; Castro, I.G.; Hacker, C.; Schrader, T.A.; Metz, J.; Zeuschner, D.; Azadi, A.S.; Godinho, L.F.; Costina, V.; Findeisen, P.; et al. ACBD5 and VAPB Mediate Membrane Associations between Peroxisomes and the ER. J. Cell Biol. 2017, 216, 331–342. [Google Scholar] [CrossRef]
- Kors, S.; Hacker, C.; Bolton, C.; Maier, R.; Reimann, L.; Kitchener, E.J.A.; Warscheid, B.; Costello, J.L.; Schrader, M. Regulating Peroxisome–ER Contacts via the ACBD5-VAPB Tether by FFAT Motif Phosphorylation and GSK3β. J. Cell Biol. 2022, 221, e202003143. [Google Scholar] [CrossRef]
- Kumar, R.; Islinger, M.; Worthy, H.; Carmichael, R.; Schrader, M. The Peroxisome: An Update on Mysteries 3.0. Histochem. Cell Biol. 2024, 161, 99–132. [Google Scholar] [CrossRef]
- Kamoshita, M.; Kumar, R.; Anteghini, M.; Kunze, M.; Islinger, M.; Martins Dos Santos, V.; Schrader, M. Insights into the Peroxisomal Protein Inventory of Zebrafish. Front. Physiol. 2022, 13, 822509. [Google Scholar] [CrossRef]
- Steinberg, S.J.; Raymond, G.V.; Braverman, N.E.; Moser, A.B. Zellweger Spectrum Disorder. In GeneReviews®; University of Washington: Seattle, WA, USA, 2020. [Google Scholar]
- Engelen, M. Peroxisomal Leukodystrophy. Handb. Clin. Neurol. 2024, 204, 139–145. [Google Scholar] [CrossRef]
- Kocherlakota, S.; Swinkels, D.; Van Veldhoven, P.P.; Baes, M. Mouse Models to Study Peroxisomal Functions and Disorders: Overview, Caveats, and Recommendations. Methods Mol. Biol. 2023, 2643, 469–500. [Google Scholar] [CrossRef] [PubMed]
- Hölttä-Vuori, M.; Salo, V.T.V.; Nyberg, L.; Brackmann, C.; Enejder, A.; Panula, P.; Ikonen, E. Zebrafish: Gaining Popularity in Lipid Research. Biochem. J. 2010, 429, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Turchini, G.M.; Francis, D.S.; Du, Z.-Y.; Olsen, R.E.; Ringø, E.; Tocher, D.R. The Lipids. In Fish Nutrition; Elsevier: Amsterdam, The Netherlands, 2022; pp. 303–467. [Google Scholar]
- Dawes, M.L.; Soeller, C.; Scholpp, S. Studying Molecular Interactions in the Intact Organism: Fluorescence Correlation Spectroscopy in the Living Zebrafish Embryo. Histochem. Cell Biol. 2020, 154, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Raas, Q.; van de Beek, M.-C.; Forss-Petter, S.; Dijkstra, I.M.; Deschiffart, A.; Freshner, B.C.; Stevenson, T.J.; Jaspers, Y.R.; Nagtzaam, L.; Wanders, R.J.; et al. Metabolic Rerouting via SCD1 Induction Impacts X-Linked Adrenoleukodystrophy. J. Clin. Investig. 2021, 131, e142500. [Google Scholar] [CrossRef] [PubMed]
- Braunbeck, T.; Storch, V.; Bresch, H. Species-Specific Reaction of Liver Ultrastructure in Zebrafish (Brachydanio Rerio) and Trout (Salmo Gairdneri) after Prolonged Exposure to 4-Chloroaniline. Arch. Environ. Contam. Toxicol. 1990, 19, 405–418. [Google Scholar] [CrossRef]
- Krysko, O.; Stevens, M.; Langenberg, T.; Fransen, M.; Espeel, M.; Baes, M. Peroxisomes in Zebrafish: Distribution Pattern and Knockdown Studies. Histochem. Cell Biol. 2010, 134, 39–51. [Google Scholar] [CrossRef]
- Dariush Fahimi, H. Peroxisomes: 40 Years of Histochemical Staining, Personal Reminiscences. Histochem. Cell Biol. 2009, 131, 437–440. [Google Scholar] [CrossRef]
- Venkatachalam, A.B.; Lall, S.P.; Denovan-Wright, E.M.; Wright, J.M. Tissue-Specific Differential Induction of Duplicated Fatty Acid-Binding Protein Genes by the Peroxisome Proliferator, Clofibrate, in Zebrafish (Danio Rerio). BMC Evol. Biol. 2012, 12, 112. [Google Scholar] [CrossRef]
- Ortiz-Zarragoitia, M.; Trant, J.M.; Cajaravillet, M.P. Effects of Dibutylphthalate and Ethynylestradiol on Liver Peroxisomes, Reproduction, and Development of Zebrafish (Danio Rerio). Environ. Toxicol. Chem. 2006, 25, 2394–2404. [Google Scholar] [CrossRef]
- Olivares-Rubio, H.F.; Vega-López, A. Fatty Acid Metabolism in Fish Species as a Biomarker for Environmental Monitoring. Environ. Pollut. 2016, 218, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Cancio, I.; Cajaraville, M. Cell Biology of Peroxisomes and Their Characteristics in Aquatic Organisms. Int. Rev. Cytol. 2000, 199, 201–293. [Google Scholar] [CrossRef] [PubMed]
- Den Broeder, M.J.; Kopylova, V.A.; Kamminga, L.M.; Legler, J. Zebrafish as a Model to Study the Role of Peroxisome Proliferating-Activated Receptors in Adipogenesis and Obesity. PPAR Res. 2015, 2015, 358029. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, J.; Kang, Y.; Park, S.; Kim, Y.-I.; Kwak, S.; Lim, D.; Park, R.; Chun, C.-H.; Choe, S.-K.; et al. Fis1 Depletion in Osteoarthritis Impairs Chondrocyte Survival and Peroxisomal and Lysosomal Function. J. Mol. Med. 2016, 94, 1373–1384. [Google Scholar] [CrossRef]
- Demers, N.D.; Riccio, V.; Jo, D.S.; Bhandari, S.; Law, K.B.; Liao, W.; Kim, C.; McQuibban, G.A.; Choe, S.-K.; Cho, D.-H.; et al. PEX13 Prevents Pexophagy by Regulating Ubiquitinated PEX5 and Peroxisomal ROS. Autophagy 2023, 19, 1781–1802. [Google Scholar] [CrossRef]
- Pap, E.H.; Dansen, T.B.; Wirtz, K.W. Peptide-Based Targeting of Fluorophores to Peroxisomes in Living Cells. Trends Cell Biol. 2001, 11, 10–12. [Google Scholar] [CrossRef]
- Korotkova, D.; Borisyuk, A.; Guihur, A.; Bardyn, M.; Kuttler, F.; Reymond, L.; Schuhmacher, M.; Amen, T. Fluorescent Fatty Acid Conjugates for Live Cell Imaging of Peroxisomes. Nat. Commun. 2024, 15, 4314. [Google Scholar] [CrossRef]
- Amaral, I.; Antunes, S.C.; Rebelo, D.; Carvalho, A.P.; Rodrigues, S. Biopesticide Spinosad: Unraveling Ecotoxicological Effects on Zebrafish, Danio Rerio. Environ. Toxicol. Pharmacol. 2024, 108, 104458. [Google Scholar] [CrossRef]
- Hagey, L.R.; Møller, P.R.; Hofmann, A.F.; Krasowski, M.D. Diversity of Bile Salts in Fish and Amphibians: Evolution of a Complex Biochemical Pathway. Physiol. Biochem. Zool. 2010, 83, 308–321. [Google Scholar] [CrossRef]
- Islinger, M.; Cardoso, M.J.R.; Schrader, M. Be Different—The Diversity of Peroxisomes in the Animal Kingdom. Biochim. Biophys. Acta 2010, 1803, 881–897. [Google Scholar] [CrossRef]
- Hayashi, S.; Fujiwara, S.; Noguchi, T. Degradation of Uric Acid in Fish Liver Peroxisomes. Intraperoxisomal Localization of Hepatic Allantoicase and Purification of Its Peroxisomal Membrane-Bound Form. J. Biol. Chem. 1989, 264, 3211–3215. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Fujiwara, S.; Noguchi, T. Evolution of Urate-Degrading Enzymes in Animal Peroxisomes. Cell Biochem. Biophys. 2000, 32, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R. Metabolism and Functions of Lipids and Fatty Acids in Teleost Fish. Rev. Fish. Sci. 2003, 11, 107–184. [Google Scholar] [CrossRef]
- Yang, G.; Sun, S.; He, J.; Wang, Y.; Ren, T.; He, H.; Gao, J. Enoyl-CoA Hydratase/3-Hydroxyacyl CoA Dehydrogenase Is Essential for the Production of DHA in Zebrafish. J. Lipid Res. 2023, 64, 100326. [Google Scholar] [CrossRef]
- Ferdinandusse, S.; Denis, S.; Van Roermund, C.W.T.; Wanders, R.J.A.; Dacremont, G. Identification of the Peroxisomal Beta-Oxidation Enzymes Involved in the Degradation of Long-Chain Dicarboxylic Acids. J. Lipid Res. 2004, 45, 1104–1111. [Google Scholar] [CrossRef]
- Houten, S.M.; Denis, S.; Argmann, C.A.; Jia, Y.; Ferdinandusse, S.; Reddy, J.K.; Wanders, R.J.A. Peroxisomal L-Bifunctional Enzyme (Ehhadh) Is Essential for the Production of Medium-Chain Dicarboxylic Acids. J. Lipid Res. 2012, 53, 1296–1303. [Google Scholar] [CrossRef]
- Ranea-Robles, P.; Violante, S.; Argmann, C.; Dodatko, T.; Bhattacharya, D.; Chen, H.; Yu, C.; Friedman, S.L.; Puchowicz, M.; Houten, S.M. Murine Deficiency of Peroxisomal L-Bifunctional Protein (EHHADH) Causes Medium-Chain 3-Hydroxydicarboxylic Aciduria and Perturbs Hepatic Cholesterol Homeostasis. Cell. Mol. Life Sci. 2021, 78, 5631–5646. [Google Scholar] [CrossRef]
- Klootwijk, E.D.; Reichold, M.; Helip-Wooley, A.; Tolaymat, A.; Broeker, C.; Robinette, S.L.; Reinders, J.; Peindl, D.; Renner, K.; Eberhart, K.; et al. Mistargeting of Peroxisomal EHHADH and Inherited Renal Fanconi’s Syndrome. N. Engl. J. Med. 2014, 370, 129–138. [Google Scholar] [CrossRef]
- Ranea-Robles, P.; Portman, K.; Bender, A.S.; Lee, K.; He, J.; Mulholland, D.; Argmann, C.; Houten, S. Peroxisomal L-Bifunctional Protein Deficiency Causes Male-Specific Kidney Hypertrophy and Proximal Tubular Injury in Mice. Kidney360 2021, 2, 1441–1454. [Google Scholar] [CrossRef]
- Baes, M.; Huyghe, S.; Carmeliet, P.; Declercq, P.E.; Collen, D.; Mannaerts, G.P.; Van Veldhoven, P.P. Inactivation of the Peroxisomal Multifunctional Protein-2 in Mice Impedes the Degradation of Not Only 2-Methyl-Branched Fatty Acids and Bile Acid Intermediates but Also of Very Long Chain Fatty Acids. J. Biol. Chem. 2000, 275, 16329–16336. [Google Scholar] [CrossRef]
- Ferdinandusse, S.; Denis, S.; Overmars, H.; Van Eeckhoudt, L.; Van Veldhoven, P.P.; Duran, M.; Wanders, R.J.A.; Baes, M. Developmental Changes of Bile Acid Composition and Conjugation in L- and D-Bifunctional Protein Single and Double Knockout Mice. J. Biol. Chem. 2005, 280, 18658–18666. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-I.; Bhandari, S.; Lee, J.N.; Yoo, K.-W.; Kim, S.-J.; Oh, G.-S.; Kim, H.-J.; Cho, M.; Kwak, J.-Y.; So, H.-S.; et al. Developmental Roles of D-Bifunctional Protein-A Zebrafish Model of Peroxisome Dysfunction. Mol. Cells 2014, 37, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Klouwer, F.C.C.; Berendse, K.; Ferdinandusse, S.; Wanders, R.J.A.; Engelen, M.; Poll-The, B.T. Zellweger Spectrum Disorders: Clinical Overview and Management Approach. Orphanet J. Rare Dis. 2015, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; El Magraoui, F.; Bäumer, B.E.; Schlee, D.; Girzalsky, W.; Erdmann, R. Pex2 and Pex12 Function as Protein-Ubiquitin Ligases in Peroxisomal Protein Import. Mol. Cell. Biol. 2009, 29, 5505–5516. [Google Scholar] [CrossRef]
- Fujiki, Y.; Abe, Y.; Imoto, Y.; Tanaka, A.J.; Okumoto, K.; Honsho, M.; Tamura, S.; Miyata, N.; Yamashita, T.; Chung, W.K.; et al. Recent Insights into Peroxisome Biogenesis and Associated Diseases. J. Cell Sci. 2020, 133, jcs236943. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Imanaka, T. Peroxisome Biogenesis. In Peroxisomes: Biogenesis, Function, and Role in Human Disease; Springer: Singapore, 2019; pp. 15–42. ISBN 9789811511684. [Google Scholar]
- Faust, P.L.; Hatten, M.E. Targeted Deletion of the PEX2 Peroxisome Assembly Gene in Mice Provides a Model for Zellweger Syndrome, a Human Neuronal Migration Disorder. J. Cell Biol. 1997, 139, 1293–1305. [Google Scholar] [CrossRef]
- Santos, M.J.; Imanaka, T.; Shio, H.; Small, G.M.; Lazarow, P.B. Peroxisomal Membrane Ghosts in Zellweger Syndrome—Aberrant Organelle Assembly. Science 1988, 239, 1536–1538. [Google Scholar] [CrossRef]
- Trompier, D.; Vejux, A.; Zarrouk, A.; Gondcaille, C.; Geillon, F.; Nury, T.; Savary, S.; Lizard, G. Brain Peroxisomes. Biochimie 2014, 98, 102–110. [Google Scholar] [CrossRef]
- Takashima, S.; Takemoto, S.; Toyoshi, K.; Ohba, A.; Shimozawa, N. Zebrafish Model of Human Zellweger Syndrome Reveals Organ-Specific Accumulation of Distinct Fatty Acid Species and Widespread Gene Expression Changes. Mol. Genet. Metab. 2021, 133, 307–323. [Google Scholar] [CrossRef]
- Lebeaupin, C.; Vallée, D.; Hazari, Y.; Hetz, C.; Chevet, E.; Bailly-Maitre, B. Endoplasmic Reticulum Stress Signalling and the Pathogenesis of Non-Alcoholic Fatty Liver Disease. J. Hepatol. 2018, 69, 927–947. [Google Scholar] [CrossRef]
- Han, J.; Kaufman, R.J. The Role of ER Stress in Lipid Metabolism and Lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Malhi, H.; Kaufman, R.J. Endoplasmic Reticulum Stress in Liver Disease. J. Hepatol. 2011, 54, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, W.J.; Tape, K.N.; Shackelford, J.E.; Wikander, T.M.; Richards, M.J.; Fliesler, S.J.; Krisans, S.K.; Faust, P.L. Peroxisome Deficiency Causes a Complex Phenotype Because of Hepatic SREBP/Insig Dysregulation Associated with Endoplasmic Reticulum Stress. J. Biol. Chem. 2009, 284, 7232–7245. [Google Scholar] [CrossRef] [PubMed]
- Folz, S.J.; Trobe, J.D. The Peroxisome and the Eye. Surv. Ophthalmol. 1991, 35, 353–368. [Google Scholar] [CrossRef]
- Bhandari, S.; Kim, Y.-I.; Nam, I.-K.; Hong, K.; Jo, Y.; Yoo, K.-W.; Liao, W.; Lim, J.-Y.; Kim, S.-J.; Um, J.-Y.; et al. Loss of Pex5 Sensitizes Zebrafish to Fasting due to Deregulated Mitochondria, MTOR, and Autophagy. Cell. Mol. Life Sci. 2023, 80, 69. [Google Scholar] [CrossRef]
- Strachan, L.R.; Stevenson, T.J.; Freshner, B.; Keefe, M.D.; Miranda Bowles, D.; Bonkowsky, J.L. A Zebrafish Model of X-Linked Adrenoleukodystrophy Recapitulates Key Disease Features and Demonstrates a Developmental Requirement for Abcd1 in Oligodendrocyte Patterning and Myelination. Hum. Mol. Genet. 2017, 26, 3600–3614. [Google Scholar] [CrossRef]
- Raas, Q.; Wood, A.; Stevenson, T.J.; Swartwood, S.; Liu, S.; Kannan, R.M.; Kannan, S.; Bonkowsky, J.L. Generation and Characterization of a Zebrafish Gain-of-Function ACOX1 Mitchell Disease Model. Front. Pediatr. 2024, 12, 1326886. [Google Scholar] [CrossRef]
- Umair, M.; Farooq Khan, M.; Aldrees, M.; Nashabat, M.; Alhamoudi, K.M.; Bilal, M.; Alyafee, Y.; Al Tuwaijri, A.; Aldarwish, M.; Al-Rumayyan, A.; et al. Mutated VWA8 Is Associated with Developmental Delay, Microcephaly, and Scoliosis and Plays a Novel Role in Early Development and Skeletal Morphogenesis in Zebrafish. Front. Cell Dev. Biol. 2021, 9, 736960. [Google Scholar] [CrossRef]
- Kong, L.; Chu, G.; Ma, W.; Liang, J.; Liu, D.; Liu, Q.; Wei, X.; Jia, S.; Gu, H.; He, Y.; et al. Mutations in VWA8 Cause Autosomal-Dominant Retinitis Pigmentosa via Aberrant Mitophagy Activation. J. Med. Genet. 2023, 60, 939–950. [Google Scholar] [CrossRef]
- Fujiki, Y.; Okumoto, K.; Mukai, S.; Honsho, M.; Tamura, S. Peroxisome Biogenesis in Mammalian Cells. Front. Physiol. 2014, 5, 307. [Google Scholar] [CrossRef]
- Muntau, A.C.; Mayerhofer, P.U.; Paton, B.C.; Kammerer, S.; Roscher, A.A. Defective Peroxisome Membrane Synthesis due to Mutations in Human PEX3 Causes Zellweger Syndrome, Complementation Group G. Am. J. Hum. Genet. 2000, 67, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Soukupova, M.; Sprenger, C.; Gorgas, K.; Kunau, W.H.; Dodt, G. Identification and Characterization of the Human Peroxin PEX3. Eur. J. Cell Biol. 1999, 78, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Shimozawa, N.; Zhang, Z.; Suzuki, Y.; Imamura, A.; Tsukamoto, T.; Osumi, T.; Fujiki, Y.; Orii, T.; Barth, P.G.; Wanders, R.J.; et al. Functional Heterogeneity of C-Terminal Peroxisome Targeting Signal 1 in PEX5-Defective Patients. Biochem. Biophys. Res. Commun. 1999, 262, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Baes, M.; Gressens, P.; Baumgart, E.; Carmeliet, P.; Casteels, M.; Fransen, M.; Evrard, P.; Fahimi, D.; Declercq, P.E.; Collen, D.; et al. A Mouse Model for Zellweger Syndrome. Nat. Genet. 1997, 17, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, E.; Vanhorebeek, I.; Grabenbauer, M.; Borgers, M.; Declercq, P.E.; Fahimi, H.D.; Baes, M. Mitochondrial Alterations Caused by Defective Peroxisomal Biogenesis in a Mouse Model for Zellweger Syndrome (PEX5 Knockout Mouse). Am. J. Pathol. 2001, 159, 1477–1494. [Google Scholar] [CrossRef]
- Peeters, A.; Fraisl, P.; van den Berg, S.; Ver Loren van Themaat, E.; Van Kampen, A.; Rider, M.H.; Takemori, H.; van Dijk, K.W.; Van Veldhoven, P.P.; Carmeliet, P.; et al. Carbohydrate Metabolism Is Perturbed in Peroxisome-Deficient Hepatocytes due to Mitochondrial Dysfunction, AMP-Activated Protein Kinase (AMPK) Activation, and Peroxisome Proliferator-Activated Receptor γ Coactivator 1α (PGC-1α) Suppression. J. Biol. Chem. 2011, 286, 42162–42179. [Google Scholar] [CrossRef]
- Tanaka, H.; Okazaki, T.; Aoyama, S.; Yokota, M.; Koike, M.; Okada, Y.; Fujiki, Y.; Gotoh, Y. Peroxisomes Control Mitochondrial Dynamics and the Mitochondrion-Dependent Apoptosis Pathway. J. Cell Sci. 2019, 132, jcs224766. [Google Scholar] [CrossRef]
- Jiang, C.; Okazaki, T. Control of Mitochondrial Dynamics and Apoptotic Pathways by Peroxisomes. Front. Cell Dev. Biol. 2022, 10, 938177. [Google Scholar] [CrossRef]
- Elgersma, Y.; Kwast, L.; Klein, A.; Voorn-Brouwer, T.; van den Berg, M.; Metzig, B.; America, T.; Tabak, H.F.; Distel, B. The SH3 Domain of the Saccharomyces Cerevisiae Peroxisomal Membrane Protein Pex13p Functions as a Docking Site for Pex5p, a Mobile Receptor for the Import PTS1-Containing Proteins. J. Cell Biol. 1996, 135, 97–109. [Google Scholar] [CrossRef]
- Gould, S.J.; Kalish, J.E.; Morrell, J.C.; Bjorkman, J.; Urquhart, A.J.; Crane, D.I. Pex13p Is an SH3 Protein of the Peroxisome Membrane and a Docking Factor for the Predominantly Cytoplasmic PTs1 Receptor. J. Cell Biol. 1996, 135, 85–95. [Google Scholar] [CrossRef]
- Krause, C.; Rosewich, H.; Woehler, A.; Gärtner, J. Functional Analysis of PEX13 Mutation in a Zellweger Syndrome Spectrum Patient Reveals Novel Homooligomerization of PEX13 and Its Role in Human Peroxisome Biogenesis. Hum. Mol. Genet. 2013, 22, 3844–3857. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gaussmann, S.; Peschel, R.; Ott, J.; Zak, K.M.; Sastre, J.; Delhommel, F.; Popowicz, G.M.; Boekhoven, J.; Schliebs, W.; Erdmann, R.; et al. Modulation of Peroxisomal Import by the PEX13 SH3 Domain and a Proximal FxxxF Binding Motif. Nat. Commun. 2024, 15, 3317. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Skowyra, M.L.; Feng, P.; Rapoport, T.A. Protein Import into Peroxisomes Occurs through a Nuclear Pore-like Phase. Science 2022, 378, eadf3971. [Google Scholar] [CrossRef] [PubMed]
- Barros-Barbosa, A.; Ferreira, M.J.; Rodrigues, T.A.; Pedrosa, A.G.; Grou, C.P.; Pinto, M.P.; Fransen, M.; Francisco, T.; Azevedo, J.E. Membrane Topologies of PEX13 and PEX14 Provide New Insights on the Mechanism of Protein Import into Peroxisomes. FEBS J. 2019, 286, 205–222. [Google Scholar] [CrossRef]
- Meinecke, M.; Cizmowski, C.; Schliebs, W.; Krüger, V.; Beck, S.; Wagner, R.; Erdmann, R. The Peroxisomal Importomer Constitutes a Large and Highly Dynamic Pore. Nat. Cell Biol. 2010, 12, 273–277. [Google Scholar] [CrossRef]
- Maxwell, M.; Bjorkman, J.; Nguyen, T.; Sharp, P.; Finnie, J.; Paterson, C.; Tonks, I.; Paton, B.C.; Kay, G.F.; Crane, D.I. Pex13 Inactivation in the Mouse Disrupts Peroxisome Biogenesis and Leads to a Zellweger Syndrome Phenotype. Mol. Cell. Biol. 2003, 23, 5947–5957. [Google Scholar] [CrossRef]
- Liu, Y.; Björkman, J.; Urquhart, A.; Wanders, R.J.; Crane, D.I.; Gould, S.J. PEX13 Is Mutated in Complementation Group 13 of the Peroxisome-Biogenesis Disorders. Am. J. Hum. Genet. 1999, 65, 621–634. [Google Scholar] [CrossRef]
- Shimozawa, N.; Suzuki, Y.; Zhang, Z.; Imamura, A.; Toyama, R.; Mukai, S.; Fujiki, Y.; Tsukamoto, T.; Osumi, T.; Orii, T.; et al. Nonsense and Temperature-Sensitive Mutations in PEX13 Are the Cause of Complementation Group H of Peroxisome Biogenesis Disorders. Hum. Mol. Genet. 1999, 8, 1077–1083. [Google Scholar] [CrossRef][Green Version]
- Krause, C.; Rosewich, H.; Thanos, M.; Gärtner, J. Identification of Novel Mutations in PEX2, PEX6, PEX10, PEX12, and PEX13 in Zellweger Spectrum Patients. Hum. Mutat. 2006, 27, 1157. [Google Scholar] [CrossRef]
- Al-Dirbashi, O.Y.; Shaheen, R.; Al-Sayed, M.; Al-Dosari, M.; Makhseed, N.; Abu Safieh, L.; Santa, T.; Meyer, B.F.; Shimozawa, N.; Alkuraya, F.S. Zellweger Syndrome Caused by PEX13 Deficiency: Report of Two Novel Mutations. Am. J. Med. Genet. A 2009, 149, 1219–1223. [Google Scholar] [CrossRef]
- Shih, H.-Y.; Raas, Q.; Bonkowsky, J.L. Progress in Leukodystrophies with Zebrafish. Dev. Growth Differ. 2024, 66, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Engelen, M.; Barbier, M.; Dijkstra, I.M.E.; Schür, R.; de Bie, R.M.A.; Verhamme, C.; Dijkgraaf, M.G.W.; Aubourg, P.A.; Wanders, R.J.A.; van Geel, B.M.; et al. X-Linked Adrenoleukodystrophy in Women: A Cross-Sectional Cohort Study. Brain 2014, 137, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Montoro, R.; Heine, V.M.; Kemp, S.; Engelen, M. Evolution of Adrenoleukodystrophy Model Systems. J. Inherit. Metab. Dis. 2021, 44, 544–553. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, H.I.; Vluggens, A.; Andreoletti, P.; Ragot, K.; Mandard, S.; Kersten, S.; Waterham, H.R.; Lizard, G.; Wanders, R.J.A.; Reddy, J.K.; et al. The Inflammatory Response in Acyl-CoA Oxidase 1 Deficiency (Pseudoneonatal Adrenoleukodystrophy). Endocrinology 2012, 153, 2568–2575. [Google Scholar] [CrossRef]
- Chung, H.-L.; Wangler, M.F.; Marcogliese, P.C.; Jo, J.; Ravenscroft, T.A.; Zuo, Z.; Duraine, L.; Sadeghzadeh, S.; Li-Kroeger, D.; Schmidt, R.E.; et al. Loss- or Gain-of-Function Mutations in ACOX1 Cause Axonal Loss via Different Mechanisms. Neuron 2020, 106, 589–606.e6. [Google Scholar] [CrossRef]
- Sarkar, C.; Lipinski, M.M. Role and Function of Peroxisomes in Neuroinflammation. Cells 2024, 13, 1655. [Google Scholar] [CrossRef]
- Huang, J.; Viswakarma, N.; Yu, S.; Jia, Y.; Bai, L.; Vluggens, A.; Cherkaoui-Malki, M.; Khan, M.; Singh, I.; Yang, G.; et al. Progressive Endoplasmic Reticulum Stress Contributes to Hepatocarcinogenesis in Fatty Acyl-CoA Oxidase 1-Deficient Mice. Am. J. Pathol. 2011, 179, 703–713. [Google Scholar] [CrossRef]
- Shen, M.; Chen, Q.; Gao, Y.; Yan, H.; Feng, S.; Ji, X.; Zhang, X. A de Novo Heterozygous Variant in ACOX1 Gene Cause Mitchell Syndrome: The First Case in China and Literature Review. BMC Med. Genom. 2023, 16, 156. [Google Scholar] [CrossRef]
- Mehtälä, M.L.; Lensink, M.F.; Pietikäinen, L.P.; Hiltunen, J.K.; Glumoff, T. On the Molecular Basis of D-Bifunctional Protein Deficiency Type III. PLoS ONE 2013, 8, e53688. [Google Scholar] [CrossRef]
- Möller, G.; van Grunsven, E.G.; Wanders, R.J.; Adamski, J. Molecular Basis of D-Bifunctional Protein Deficiency. Mol. Cell. Endocrinol. 2001, 171, 61–70. [Google Scholar] [CrossRef]
- Suzuki, Y.; Jiang, L.L.; Souri, M.; Miyazawa, S.; Fukuda, S.; Zhang, Z.; Une, M.; Shimozawa, N.; Kondo, N.; Orii, T.; et al. D-3-Hydroxyacyl-CoA Dehydratase/D-3-Hydroxyacyl-CoA Dehydrogenase Bifunctional Protein Deficiency: A Newly Identified Peroxisomal Disorder. Am. J. Hum. Genet. 1997, 61, 1153–1162. [Google Scholar] [CrossRef] [PubMed]
- Van Grunsven, E.G.; van Berkel, E.; Lemonde, H.; Clayton, P.T.; Wanders, R.J. Bifunctional Protein Deficiency: Complementation within the Same Group Suggesting Differential Enzyme Defects and Clues to the Underlying Basis. J. Inherit. Metab. Dis. 1998, 21, 298–301. [Google Scholar] [CrossRef] [PubMed]
- van Grunsven, E.G.; Mooijer, P.A.; Aubourg, P.; Wanders, R.J. Enoyl-CoA Hydratase Deficiency: Identification of a New Type of D-Bifunctional Protein Deficiency. Hum. Mol. Genet. 1999, 8, 1509–1516. [Google Scholar] [CrossRef] [PubMed]
- Wanders, R.J.A. Metabolic Functions of Peroxisomes in Health and Disease. Biochimie 2014, 98, 36–44. [Google Scholar] [CrossRef]
- Huyghe, S.; Schmalbruch, H.; De Gendt, K.; Verhoeven, G.; Guillou, F.; Van Veldhoven, P.P.; Baes, M. Peroxisomal Multifunctional Protein 2 Is Essential for Lipid Homeostasis in Sertoli Cells and Male Fertility in Mice. Endocrinology 2006, 147, 2228–2236. [Google Scholar] [CrossRef]
- Costello, J.L.; Passmore, J.B.; Islinger, M.; Schrader, M. Multi-Localized Proteins: The Peroxisome-Mitochondria Connection. Subcell. Biochem. 2018, 89, 383–415. [Google Scholar] [CrossRef]
- Schrader, T.A.; Carmichael, R.E.; Islinger, M.; Costello, J.L.; Hacker, C.; Bonekamp, N.A.; Weishaupt, J.H.; Andersen, P.M.; Schrader, M. PEX11β and FIS1 Cooperate in Peroxisome Division Independently of Mitochondrial Fission Factor. J. Cell Sci. 2022, 135, jcs259924. [Google Scholar] [CrossRef]
- Koch, A.; Yoon, Y.; Bonekamp, N.A.; McNiven, M.A.; Schrader, M. A Role for Fis1 in Both Mitochondrial and Peroxisomal Fission in Mammalian Cells. Mol. Biol. Cell 2005, 16, 5077–5086. [Google Scholar] [CrossRef]
- Gandre-Babbe, S.; van der Bliek, A.M. The Novel Tail-Anchored Membrane Protein Mff Controls Mitochondrial and Peroxisomal Fission in Mammalian Cells. Mol. Biol. Cell 2008, 19, 2402–2412. [Google Scholar] [CrossRef]
- Niwa, H.; Miyauchi-Nanri, Y.; Okumoto, K.; Mukai, S.; Noi, K.; Ogura, T.; Fujiki, Y. A Newly Isolated Pex7-Binding, Atypical PTS2 Protein P7BP2 Is a Novel Dynein-Type AAA+ Protein. J. Biochem. 2018, 164, 437–447. [Google Scholar] [CrossRef]
- Wiese, S.; Gronemeyer, T.; Brites, P.; Ofman, R.; Bunse, C.; Renz, C.; Meyer, H.E.; Wanders, R.J.A.; Warscheid, B. Comparative Profiling of the Peroxisomal Proteome of Wildtype and Pex7 Knockout Mice by Quantitative Mass Spectrometry. Int. J. Mass Spectrom. 2012, 312, 30–40. [Google Scholar] [CrossRef]
- Luo, M.; Mengos, A.E.; Ma, W.; Finlayson, J.; Bustos, R.Z.; Xiao Zhu, Y.; Shi, C.-X.; Stubblefield, T.M.; Willis, W.T.; Mandarino, L.J. Characterization of the Novel Protein KIAA0564 (Von Willebrand Domain-Containing Protein 8). Biochem. Biophys. Res. Commun. 2017, 487, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Ma, W.; Sand, Z.; Finlayson, J.; Wang, T.; Brinton, R.D.; Willis, W.T.; Mandarino, L.J. Von Willebrand Factor A Domain-Containing Protein 8 (VWA8) Localizes to the Matrix Side of the Inner Mitochondrial Membrane. Biochem. Biophys. Res. Commun. 2020, 521, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Willis, W.T.; Coletta, D.K.; Langlais, P.R.; Mengos, A.; Ma, W.; Finlayson, J.; Wagner, G.R.; Shi, C.-X.; Mandarino, L.J. Deletion of the Mitochondrial Protein VWA8 Induces Oxidative Stress and an HNF4α Compensatory Response in Hepatocytes. Biochemistry 2019, 58, 4983–4996. [Google Scholar] [CrossRef]
- Luo, M.; Ma, W.; Zapata-Bustos, R.; Willis, W.T.; Mandarino, L.J. Deletion of Von Willebrand A Domain Containing Protein (VWA8) Raises Activity of Mitochondrial Electron Transport Chain Complexes in Hepatocytes. Biochem. Biophys. Rep. 2021, 26, 100928. [Google Scholar] [CrossRef]
- Imanaka, T.; Kawaguchi, K. A Novel Dynein-Type AAA+ Protein with Peroxisomal Targeting Signal Type 2. J. Biochem. 2020, 167, 429–432. [Google Scholar] [CrossRef]
- Key, J.; Gispert, S.; Koepf, G.; Steinhoff-Wagner, J.; Reichlmeir, M.; Auburger, G. Translation Fidelity and Respiration Deficits in CLPP-Deficient Tissues: Mechanistic Insights from Mitochondrial Complexome Profiling. Int. J. Mol. Sci. 2023, 24, 17503. [Google Scholar] [CrossRef]
- Quiñonez-Silvero, C.; Hübner, K.; Herzog, W. Development of the Brain Vasculature and the Blood-Brain Barrier in Zebrafish. Dev. Biol. 2020, 457, 181–190. [Google Scholar] [CrossRef]
| Function/Pathway | Comments | Dr | Hs |
|---|---|---|---|
| Fatty acid beta-oxidation | Acox2 is absent in zebrafish ACOXL/ACOX4 is present in humans and zebrafish, but not well defined | ||
| Bile acid synthesis | Bile acid-CoA:amino acid N-acyltransferase (Baat) is absent in zebrafish | - | |
| Fatty acid alpha-oxidation | |||
| Saturation of PUFAs | |||
| Ether lipid synthesis | |||
| Glycolate/ Glyoxylate metabolism | Hydroxyacid oxidase 3 (Hao3) is absent in zebrafish | ||
| Amino acid catabolism | D-amino acid oxidases 2 and 3 (Dao2, Dao3) are absent in humans | ||
| Amine metabolism | |||
| Purine and pyrimidine metabolism | Urate oxidase (Uricase) (Uox), Allantoicase (Allc), Urate (5-hydroxyiso-) hydrolase a (Uraha), Ureidoimidazoline decarboxylase (Urad) are absent or inactivated in humans | - | |
| Oxygen metabolism/ Oxidation redox equivalents | |||
| Proteases | |||
| Carbohydrate metabolism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, C.S.; Schrader, M. Modelling Peroxisomal Disorders in Zebrafish. Cells 2025, 14, 147. https://doi.org/10.3390/cells14020147
Jiang CS, Schrader M. Modelling Peroxisomal Disorders in Zebrafish. Cells. 2025; 14(2):147. https://doi.org/10.3390/cells14020147
Chicago/Turabian StyleJiang, Chenxing S., and Michael Schrader. 2025. "Modelling Peroxisomal Disorders in Zebrafish" Cells 14, no. 2: 147. https://doi.org/10.3390/cells14020147
APA StyleJiang, C. S., & Schrader, M. (2025). Modelling Peroxisomal Disorders in Zebrafish. Cells, 14(2), 147. https://doi.org/10.3390/cells14020147







