Modeling Musculoskeletal Disorders in Zebrafish: Advancements in Muscle and Bone Research
Abstract
1. Introduction
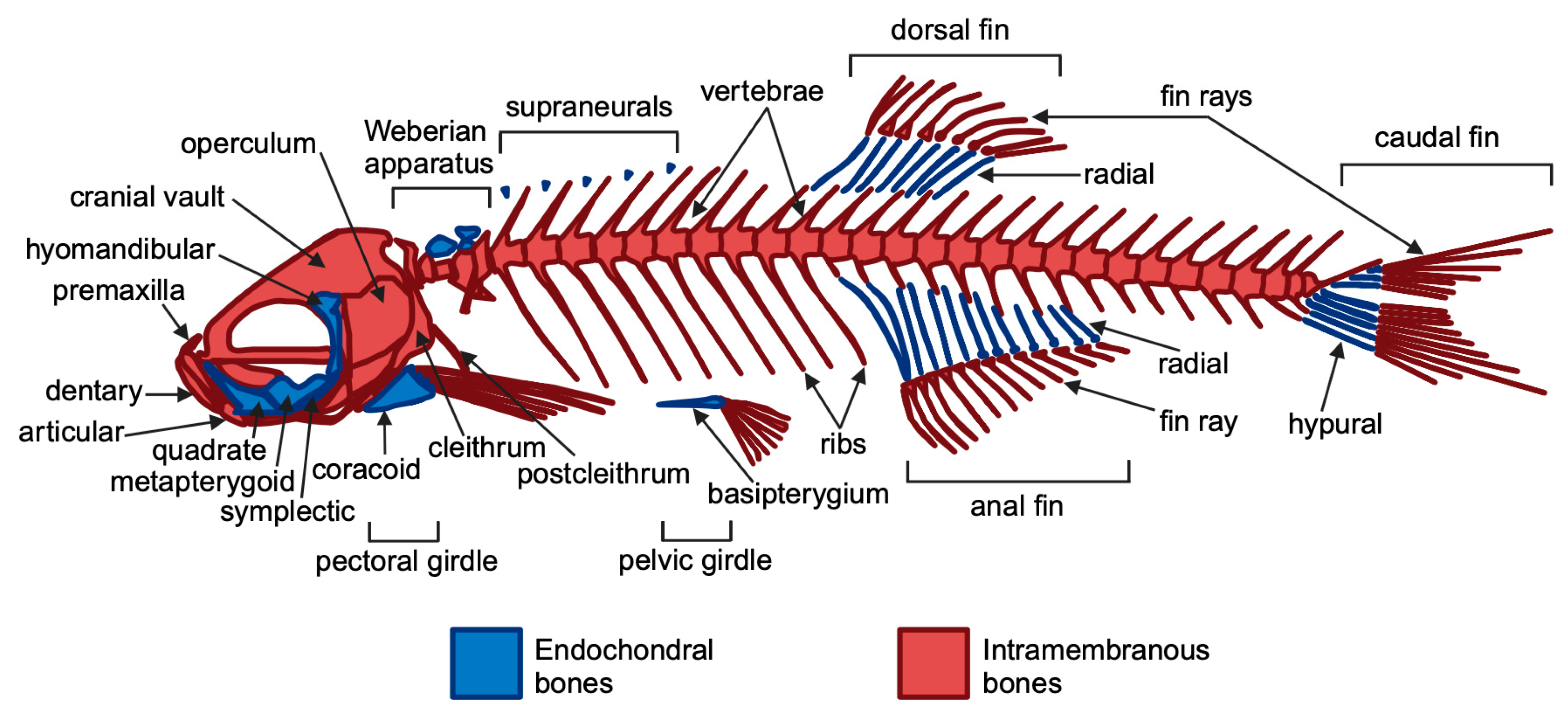
2. Zebrafish as a Model for Musculoskeletal Disorders
2.1. Bone Disorders
2.2. Muscle Disorders
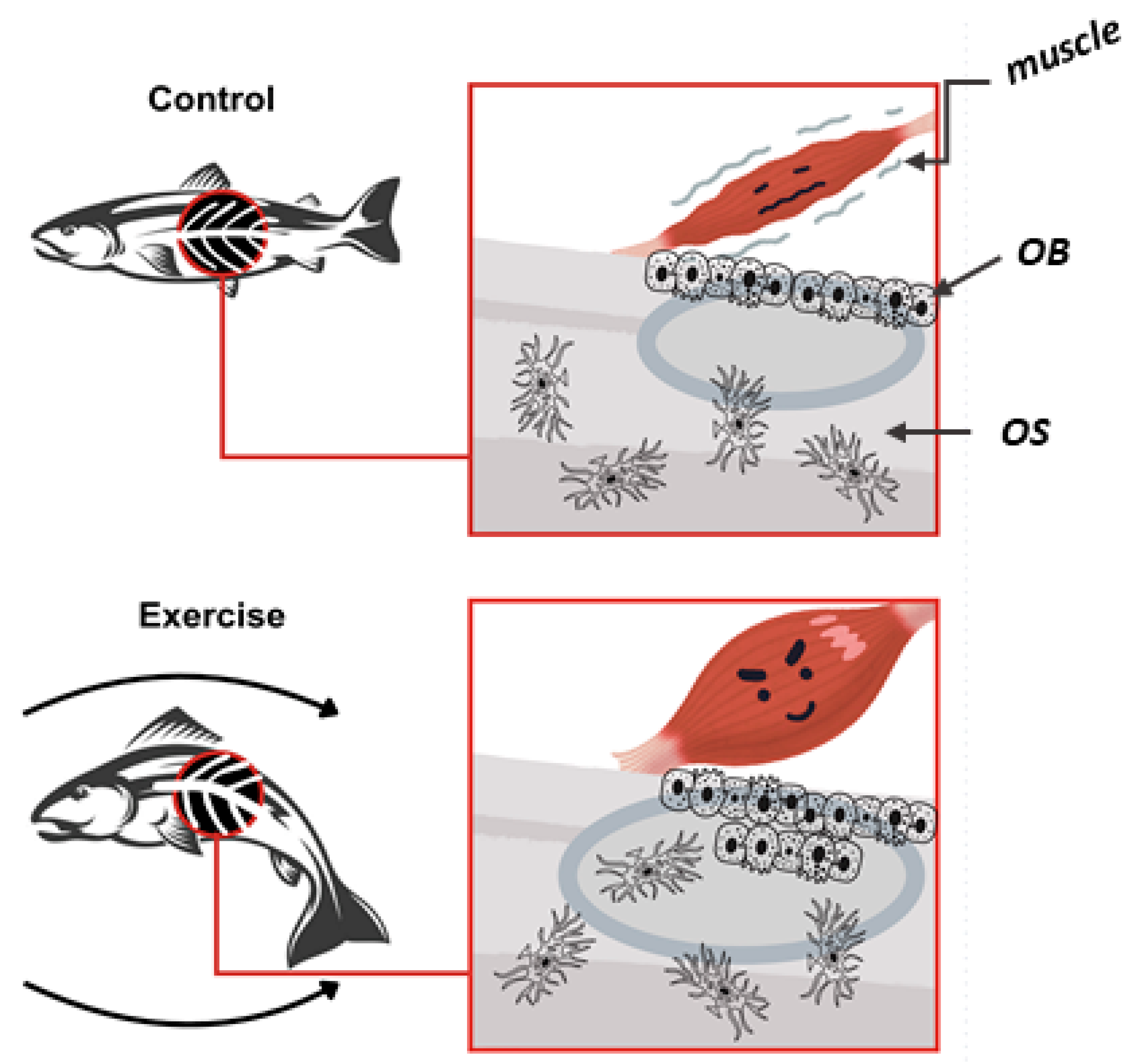
3. Muscle–Bone Crosstalk
4. Aging and the Musculoskeletal System
5. Zebrafish and Therapeutic Strategies
6. Limitations of the Zebrafish Model
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Parker, B. Origin and Evolution of Vertebrates; Scientific e-Resources: New Delhi, India, 2019. [Google Scholar]
- Ribas, L.; Piferrer, F. The zebrafish (Danio rerio) as a model organism, with emphasis on applications for finfish aquaculture research. Rev. Aquac. 2014, 6, 209–240. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Crouzier, L.; Richard, E.M.; Sourbron, J.; Lagae, L.; Maurice, T.; Delprat, B. Use of zebrafish models to boost research in rare genetic diseases. Int. J. Mol. Sci. 2021, 22, 13356. [Google Scholar] [CrossRef] [PubMed]
- Fishman, M.C.; Stainier, D.Y.; Breitbart, R.E.; Westerfield, M. Zebrafish: Genetic and embryological methods in a transparent vertebrate embryo. Methods Cell Biol. 1997, 52, 67–82. [Google Scholar] [PubMed]
- Yalcin, H.C.; Amindari, A.; Butcher, J.T.; Althani, A.; Yacoub, M. Heart function and hemodynamic analysis for zebrafish embryos. Dev. Dyn. 2017, 246, 868–880. [Google Scholar] [CrossRef]
- Yang, L.; Ho, N.Y.; Alshut, R.; Legradi, J.; Weiss, C.; Reischl, M.; Mikut, R.; Liebel, U.; Müller, F.; Strähle, U. Zebrafish embryos as models for embryotoxic and teratological effects of chemicals. Reprod. Toxicol. 2009, 28, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Singleman, C.; Holtzman, N.G. Growth and maturation in the zebrafish, Danio rerio: A staging tool for teaching and research. Zebrafish 2014, 11, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, H.M.; Jaias, I.; Mattoo, A.I.; Akram, T.; Akram, W.; Ganai, N.A. Breeding of Zebrafish and Its Life Cycle. In Zebrafish as a Model for Parkinson’s Disease; CRC Press: Boca Raton, FL, USA, 2024; pp. 29–42. [Google Scholar]
- Mcdonald, G. Functional Studies of High Bone Mass Associated Genes in Both In Vitro and In Vivo Skeletal Systems. Doctoral Dissertation, University of Bristol, Bristol, UK, 2024. [Google Scholar]
- Valenti, M.T.; Marchetto, G.; Mottes, M.; Dalle Carbonare, L. Zebrafish: A suitable tool for the study of cell signaling in bone. Cells 2020, 9, 1911. [Google Scholar] [CrossRef] [PubMed]
- Kirk, B.; Feehan, J.; Lombardi, G.; Duque, G. Muscle, bone, and fat crosstalk: The biological role of myokines, osteokines, and adipokines. Curr. Osteoporos. Rep. 2020, 18, 388–400. [Google Scholar] [CrossRef]
- Herrmann, M.; Engelke, K.; Ebert, R.; Müller-Deubert, S.; Rudert, M.; Ziouti, F.; Jundt, F.; Felsenberg, D.; Jakob, F. Interactions between muscle and bone—Where physics meets biology. Biomolecules 2020, 10, 432. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Yan, K.; Guan, Q.; Guo, Q.; Zhao, C. Mechanism and physical activities in bone-skeletal muscle crosstalk. Front. Endocrinol. 2024, 14, 1287972. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Sim, M.; Dalla Via, J.; Levinger, I.; Duque, G. The Interconnection Between Muscle and Bone: A Common Clinical Management Pathway. Calcif. Tissue Int. 2024, 114, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Mou, K.; Chan, S.M.; Vlahos, R. Musculoskeletal crosstalk in chronic obstructive pulmonary disease and comorbidities: Emerging roles and therapeutic potentials. Pharmacol. Ther. 2024, 108635. [Google Scholar] [CrossRef] [PubMed]
- Colaianni, G.; Storlino, G.; Sanesi, L.; Colucci, S.; Grano, M. Myokines and osteokines in the pathogenesis of muscle and bone diseases. Curr. Osteoporos. Rep. 2020, 18, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Cariati, I.; Bonanni, R.; Onorato, F.; Mastrogregori, A.; Rossi, D.; Iundusi, R.; Gasbarra, E.; Tancredi, V.; Tarantino, U. Role of physical activity in bone–muscle crosstalk: Biological aspects and clinical implications. J. Funct. Morphol. Kinesiol. 2021, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Goodman, C.A.; Hornberger, T.A.; Robling, A.G. Bone and skeletal muscle: Key players in mechanotransduction and potential overlapping mechanisms. Bone 2015, 80, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yuan, H.; Ma, G.; Cao, H. Bone-muscle crosstalk under physiological and pathological conditions. Cell. Mol. Life Sci. 2024, 81, 310. [Google Scholar] [CrossRef] [PubMed]
- Sui, H.; Dou, J.; Shi, B.; Cheng, X. The reciprocity of skeletal muscle and bone: An evolving view from mechanical coupling, secretory crosstalk to stem cell exchange. Front. Physiol. 2024, 15, 1349253. [Google Scholar] [CrossRef] [PubMed]
- Majeed, S.; Mumtaz, S.; Aslam, M.; Kousar, H.; Asif, M.; Gulzada, M.; Shahzad, K.; Lodhi, K. Age-Related Changes in Bone Density, Muscle Mass, And Joint Integrity, And Their Implications for the Development of Age-Related Conditions like Osteoporosis and Sarcopenia. Eur. Chem. Bull. 2023, 12, 4380–4386. [Google Scholar]
- Jin, Z.; Mao, Y.; Guo, Q.; Yin, Y.; Kiram, A.; Zhou, D.; Yang, J.; Zhou, Z.; Xue, J.; Feng, Z. Imbalanced Skeletal Muscle Mitochondrial Proteostasis Causes Bone Loss. Research 2024, 7, 0465. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.M.; Shields, M.D.; Watters, J.; Hamilton, A.; Beringer, T.; Elliott, M.; Quinlivan, R.; Tirupathi, S.; Blackwood, B. Interventions to prevent and treat corticosteroid-induced osteoporosis and prevent osteoporotic fractures in Duchenne muscular dystrophy. Cochrane Database Syst. Rev. 2017, 1, CD010899. [Google Scholar] [CrossRef] [PubMed]
- Watson, C.J.; Kwon, R.Y. Osteogenic programs during zebrafish fin regeneration. BoneKEy Rep. 2015, 4, 745. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lowes, L.P.; Hay, K. Musculoskeletal development and adaptation. In Campbell’s Physical Therapy forChildren Expert Consult-E-Book; Elsevier: Amsterdam, The Netherlands, 2016; Volume 99. [Google Scholar]
- Li, G.; Niu, W. Challenges toward musculoskeletal injuries and diseases. In Nanoengineering in Musculoskeletal Regeneration; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–41. [Google Scholar]
- Adamson, K.I.; Sheridan, E.; Grierson, A.J. Use of zebrafish models to investigate rare human disease. J. Med. Genet. 2018, 55, 641–649. [Google Scholar] [CrossRef]
- Phillips, J.B.; Westerfield, M. Zebrafish as a model to understand human genetic diseases. In The Zebrafish in Biomedical Research; Elsevier: Amsterdam, The Netherlands, 2020; pp. 619–626. [Google Scholar]
- González-Rosa, J.M. Zebrafish models of cardiac disease: From fortuitous mutants to precision medicine. Circ. Res. 2022, 130, 1803–1826. [Google Scholar] [CrossRef] [PubMed]
- Hino, H.; Kondo, S.; Kuroda, J. In vivo imaging of bone collagen dynamics in zebrafish. Bone Rep. 2024, 20, 101748. [Google Scholar] [CrossRef] [PubMed]
- Kwon, R.Y.; Watson, C.J.; Karasik, D. Using zebrafish to study skeletal genomics. Bone 2019, 126, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Howe, D.G.; Bradford, Y.M.; Eagle, A.; Fashena, D.; Frazer, K.; Kalita, P.; Mani, P.; Martin, R.; Moxon, S.T.; Paddock, H. The Zebrafish Model Organism Database: New support for human disease models, mutation details, gene expression phenotypes and searching. Nucleic Acids Res. 2017, 45, D758–D768. [Google Scholar] [CrossRef]
- Kettleborough, R.N.; Busch-Nentwich, E.M.; Harvey, S.A.; Dooley, C.M.; De Bruijn, E.; Van Eeden, F.; Sealy, I.; White, R.J.; Herd, C.; Nijman, I.J. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 2013, 496, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.F.; Gerstenfeld, L.C. The bone organ system: Form and function. In Marcus and Feldman’s Osteoporosis; Elsevier: Amsterdam, The Netherlands, 2021; pp. 15–35. [Google Scholar]
- DuMez, R.; Skromne, I. Bone Biology: From a to Zebrafish Authors and reviewers. Front. Young Minds 2023, 11. [Google Scholar] [CrossRef]
- Tonelli, F.; Bek, J.W.; Besio, R.; De Clercq, A.; Leoni, L.; Salmon, P.; Coucke, P.J.; Willaert, A.; Forlino, A. Zebrafish: A resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 2020, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, K.; Fiedler, I.A.; Kurzyukova, A.; López-Delgado, A.C.; McGowan, L.M.; Geurtzen, K.; Hammond, C.L.; Busse, B.; Knopf, F. Skeletal biology and disease modeling in zebrafish. J. Bone Miner. Res. 2020, 36, 436–458. [Google Scholar] [CrossRef] [PubMed]
- Blumer, M.J. Bone tissue and histological and molecular events during development of the long bones. Ann. Anat. -Anat. Anz. 2021, 235, 151704. [Google Scholar] [CrossRef] [PubMed]
- Le Pabic, P.; Dranow, D.B.; Hoyle, D.J.; Schilling, T.F. Zebrafish endochondral growth zones as they relate to human bone size, shape and disease. Front. Endocrinol. 2022, 13, 1060187. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Kraus, J.; Galloway, J.; Youngstrom, D.; Henke, K.; Farmer, D. Genetically Engineered Zebrafish as Models of Skeletal Development and Regeneration; University of California: California, CA, USA, 2023. [Google Scholar]
- Fisher, S.; Jagadeeswaran, P.; Halpern, M.E. Radiographic analysis of zebrafish skeletal defects. Dev. Biol. 2003, 264, 64–76. [Google Scholar] [CrossRef]
- Gerhard, G.S. Comparative aspects of zebrafish (Danio rerio) as a model for aging research. Exp. Gerontol. 2003, 38, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Nandi, A.; Sinha, A.; Patel, P.; Mohanty, S.; Jha, E.; Jena, S.; Kumari, P.; Ghosh, A.; Jerman, I. The posterity of Zebrafish in paradigm of in vivo molecular toxicological profiling. Biomed. Pharmacother. 2024, 171, 116160. [Google Scholar] [CrossRef] [PubMed]
- Uemoto, T.; Abe, G.; Tamura, K. Regrowth of zebrafish caudal fin regeneration is determined by the amputated length. Sci. Rep. 2020, 10, 649. [Google Scholar] [CrossRef] [PubMed]
- Pfefferli, C.; Jaźwińska, A. The art of fin regeneration in zebrafish. Regeneration 2015, 2, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Sehring, I.M.; Weidinger, G. Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip. Rev. Dev. Biol. 2020, 9, e367. [Google Scholar] [CrossRef]
- Van der Meulen, T.; Kranenbarg, S.; Schipper, H.; Samallo, J.; Van Leeuwen, J.; Franssen, H. Identification and characterisation of two runx2 homologues in zebrafish with different expression patterns. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 2005, 1729, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.V.; Tsang, V.W.K.; Hu, W.; Kalev-Zylinska, M.; Postlethwait, J.; Crosier, P.; Crosier, K.; Fisher, S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr. Patterns 2004, 4, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-C.; Tsai, C.-C.; Liao, Y.-F.; Fu, H.-C.; Tsay, H.-J.; Huang, T.-F.; Chen, Y.-H.; Hung, S.-C. Twist controls skeletal development and dorsoventral patterning by regulating runx2 in zebrafish. PLoS ONE 2011, 6, e27324. [Google Scholar] [CrossRef] [PubMed]
- Gistelinck, C.; Gioia, R.; Gagliardi, A.; Tonelli, F.; Marchese, L.; Bianchi, L.; Landi, C.; Bini, L.; Huysseune, A.; Witten, P.E. Zebrafish collagen type I: Molecular and biochemical characterization of the major structural protein in bone and skin. Sci. Rep. 2016, 6, 21540. [Google Scholar] [CrossRef] [PubMed]
- Harris, W.H.; Heaney, R.P. Skeletal renewal and metabolic bone disease. N. Engl. J. Med. 1969, 280, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Parfitt, A.M. The Coupling of Bone Formation to Bone Resorption: A Critical Analysis of the Concept and of Its Relevance to the Pathogenesis of Osteoporosis; Elsevier: Amsterdam, The Netherlands, 1982; Volume 4, pp. 1–6. [Google Scholar]
- Martin, T.J.; Rodan, G.A. Coupling of bone resorption and formation during bone remodeling. In Osteoporosis; Elsevier: Amsterdam, The Netherlands, 2001; pp. 361–371. [Google Scholar]
- Silva, I.; Branco, J.C. Rank/Rankl/opg: Literature review. Acta Reumatol. Port. 2011, 36. [Google Scholar]
- Negishi-Koga, T.; Shinohara, M.; Komatsu, N.; Bito, H.; Kodama, T.; Friedel, R.H.; Takayanagi, H. Suppression of bone formation by osteoclastic expression of semaphorin 4D. Nat. Med. 2011, 17, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Bergen, D.J.; Kague, E.; Hammond, C.L. Zebrafish as an emerging model for osteoporosis: A primary testing platform for screening new osteo-active compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Bek, J.W.; Shochat, C.; De Clercq, A.; De Saffel, H.; Boel, A.; Metz, J.; Rodenburg, F.; Karasik, D.; Willaert, A.; Coucke, P.J. Lrp5 mutant and Crispant zebrafish faithfully model human osteoporosis, establishing the zebrafish as a platform for CRISPR-based functional screening of osteoporosis candidate genes. J. Bone Miner. Res. 2020, 36, 1749–1764. [Google Scholar] [CrossRef]
- Marques-Pinheiro, A.; Levasseur, R.; Cormier, C.; Bonneau, J.; Boileau, C.; Varret, M.; Abifadel, M.; Allanore, Y. Novel LRP5 gene mutation in a patient with osteoporosis-pseudoglioma syndrome. Jt. Bone Spine 2010, 77, 151–153. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, K.-i.; Takahira, K.; Inari, M.; Satoh, Y.; Hayakawa, K.; Tabuchi, Y.; Ogai, K.; Nishiuchi, T.; Kondo, T.; Mikuni-Takagaki, Y. Zebrafish scales respond differently to in vitro dynamic and static acceleration: Analysis of interaction between osteoblasts and osteoclasts. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2013, 166, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Masiero, C.; Aresi, C.; Forlino, A.; Tonelli, F. Zebrafish Models for Skeletal and Extraskeletal Osteogenesis Imperfecta Features: Unveiling Pathophysiology and Paving the Way for Drug Discovery. Calcif. Tissue Int. 2024, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, F.; Cotti, S.; Leoni, L.; Besio, R.; Gioia, R.; Marchese, L.; Giorgetti, S.; Villani, S.; Gistelinck, C.; Wagener, R. Crtap and p3h1 knock out zebrafish support defective collagen chaperoning as the cause of their osteogenesis imperfecta phenotype. Matrix Biol. 2020, 90, 40–60. [Google Scholar] [CrossRef] [PubMed]
- Daponte, V.; Tonelli, F.; Masiero, C.; Syx, D.; Exbrayat-Héritier, C.; Biggiogera, M.; Willaert, A.; Rossi, A.; Coucke, P.J.; Ruggiero, F. Cell differentiation and matrix organization are differentially affected during bone formation in osteogenesis imperfecta zebrafish models with different genetic defects impacting collagen type I structure. Matrix Biol. 2023, 121, 105–126. [Google Scholar] [CrossRef] [PubMed]
- Debaenst, S.; Jarayseh, T.; De Saffel, H.; Bek, J.W.; Boone, M.; Josipovic, I.; Kibleur, P.; Kwon, R.Y.; Coucke, P.; Willaert, A. Crispant analysis in zebrafish as a tool for rapid functional screening of disease-causing genes for bone fragility. bioRxiv 2024. [Google Scholar]
- Xie, H.; Li, M.; Kang, Y.; Zhang, J.; Zhao, C. Zebrafish: An important model for understanding scoliosis. Cell. Mol. Life Sci. 2022, 79, 506. [Google Scholar] [CrossRef] [PubMed]
- Gorman, K.F.; Breden, F. Idiopathic-type scoliosis is not exclusive to bipedalism. Med. Hypotheses 2009, 72, 348–352. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Janssen, M.M.; de Wilde, R.F.; Kouwenhoven, J.-W.M.; Castelein, R.M. Experimental animal models in scoliosis research: A review of the literature. Spine J. 2011, 11, 347–358. [Google Scholar] [CrossRef]
- Bobyn, J.D.; Little, D.G.; Gray, R.; Schindeler, A. Animal models of scoliosis. J. Orthop. Res. 2015, 33, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Nie, C.-H.; Wan, S.-M.; Chen, Y.-L.; Huysseune, A.; Wu, Y.-M.; Zhou, J.-J.; Hilsdorf, A.W.S.; Wang, W.-M.; Witten, P.E.; Lin, Q. Single-cell transcriptomes and runx2b−/− mutants reveal the genetic signatures of intermuscular bone formation in zebrafish. Natl. Sci. Rev. 2022, 9, nwac152. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, F. OI Zebrafish Models and Their Use to Develop New Pharmacological Treatments. Ph.D. Thesis, Università Di Pavia, Pavia, Italy, 2018. [Google Scholar]
- Jarayseh, T.; Debaenst, S.; De Saffel, H.; Rosseel, T.; Milazzo, M.; Bek, J.W.; Hudson, D.M.; Van Nieuwerburgh, F.; Gansemans, Y.; Josipovic, I. Bmpr1aa modulates the severity of the skeletal phenotype in an fkbp10-deficient Bruck syndrome zebrafish model. J. Bone Miner. Res. 2024, zjae185. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhou, Y.; Zhang, R.; Wang, Z.; Xu, M.; Zhang, D.; Huang, J.; Luo, F.; Li, F.; Ni, Z. Dstyk mutation leads to congenital scoliosis-like vertebral malformations in zebrafish via dysregulated mTORC1/TFEB pathway. Nat. Commun. 2020, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Gray, R.S.; Wilm, T.P.; Smith, J.; Bagnat, M.; Dale, R.M.; Topczewski, J.; Johnson, S.L.; Solnica-Krezel, L. Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations. Dev. Biol. 2014, 386, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Lleras-Forero, L.; Newham, E.; Teufel, S.; Kawakami, K.; Hartmann, C.; Hammond, C.L.; Knight, R.D.; Schulte-Merker, S. Muscle defects due to perturbed somite segmentation contribute to late adult scoliosis. Aging 2020, 12, 18603. [Google Scholar] [CrossRef] [PubMed]
- Henke, K.; Daane, J.M.; Hawkins, M.B.; Dooley, C.M.; Busch-Nentwich, E.M.; Stemple, D.L.; Harris, M.P. Genetic screen for postembryonic development in the zebrafish (Danio rerio): Dominant mutations affecting adult form. Genetics 2017, 207, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sepich, D.S.; Solnica-Krezel, L. Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 2017, 13, e1006564. [Google Scholar] [CrossRef] [PubMed]
- Goebel, H.H.; Dittmayer, C.; Stenzel, W. Congenital myopathies: The current status. Indian J. Pathol. Microbiol. 2022, 65, S271–S276. [Google Scholar]
- Chiodo, A. Acquired myopathy/dystrophies. PMR 2013, 5, S74–S80. [Google Scholar] [CrossRef]
- Danielsson, O.; Häggqvist, B. Skeletal muscle immunohistochemistry of acquired and hereditary myopathies. Curr. Opin. Rheumatol. 2021, 33, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Emery, A.E. The muscular dystrophies. Lancet 2002, 359, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Shilling, C.; Leslie, N.D.; Flanigan, K.M.; al-Dahhak, R.; Gastier-Foster, J.; Kneile, K.; Dunn, D.M.; Duval, B.; Aoyagi, A. Evidence-based path to newborn screening for Duchenne muscular dystrophy. Ann. Neurol. 2012, 71, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Cai, A.; Kong, X. Development of CRISPR-mediated systems in the study of Duchenne muscular dystrophy. Hum. Gene Ther. Methods 2019, 30, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Wang, Y.X.; Von Maltzahn, J.; Pasut, A.; Bentzinger, C.F.; Brun, C.E.; Rudnicki, M.A. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat. Med. 2015, 21, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.E.; Nowak, K.J. Molecular mechanisms of muscular dystrophies: Old and new players. Nat. Rev. Mol. Cell Biol. 2006, 7, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.P.; Brown, R.H., Jr.; Kunkel, L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Guyon, J.R.; Goswami, J.; Jun, S.J.; Thorne, M.; Howell, M.; Pusack, T.; Kawahara, G.; Steffen, L.S.; Galdzicki, M.; Kunkel, L.M. Genetic isolation and characterization of a splicing mutant of zebrafish dystrophin. Hum. Mol. Genet. 2009, 18, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.I.; Bryson-Richardson, R.J.; Daggett, D.F.; Gautier, P.; Keenan, D.G.; Currie, P.D. Dystrophin is required for the formation of stable muscle attachments in the zebrafish embryo. Development 2003, 130, 5851–5860. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, L.M.; Bachrach, E.; Bennett, R.R.; Guyon, J.; Steffen, L. Diagnosis and cell-based therapy for Duchenne muscular dystrophy in humans, mice, and zebrafish. J. Hum. Genet. 2006, 51, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Granato, M.; Eeden, F.J.v.; Schach, U.; Trowe, T.; Brand, M.; Furutani-Seiki, M.; Haffter, P.; Hammerschmidt, M.; Heisenberg, C.-P.; Jiang, Y.-J. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Development 1996, 123, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, G.; Karpf, J.A.; Myers, J.A.; Alexander, M.S.; Guyon, J.R.; Kunkel, L.M. Drug screening in a zebrafish model of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 2011, 108, 5331–5336. [Google Scholar] [CrossRef] [PubMed]
- Waugh, T.A.; Horstick, E.; Hur, J.; Jackson, S.W.; Davidson, A.E.; Li, X.; Dowling, J.J. Fluoxetine prevents dystrophic changes in a zebrafish model of Duchenne muscular dystrophy. Hum. Mol. Genet. 2014, 23, 4651–4662. [Google Scholar] [CrossRef] [PubMed]
- Widrick, J.J.; Alexander, M.S.; Sanchez, B.; Gibbs, D.E.; Kawahara, G.; Beggs, A.H.; Kunkel, L.M. Muscle dysfunction in a zebrafish model of Duchenne muscular dystrophy. Physiol. Genom. 2016, 48, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Martínez, P.; Sayed, R.K.; Fernández-Martínez, J.; Ramírez-Casas, Y.; Yang, Y.; Escames, G.; Acuña-Castroviejo, D. Zebrafish as a Human Muscle Model for Studying Age-Dependent Sarcopenia and Frailty. Int. J. Mol. Sci. 2024, 25, 6166. [Google Scholar] [CrossRef]
- Kilroy, E.A.; Ignacz, A.C.; Brann, K.L.; Schaffer, C.E.; Varney, D.; Alrowaished, S.S.; Silknitter, K.J.; Miner, J.N.; Almaghasilah, A.; Spellen, T.L. Beneficial impacts of neuromuscular electrical stimulation on muscle structure and function in the zebrafish model of Duchenne muscular dystrophy. Elife 2022, 11, e62760. [Google Scholar] [CrossRef]
- Lin, Y.-Y. Muscle diseases in the zebrafish. Neuromuscul. Disord. 2012, 22, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.E.; Bryson-Richardson, R.J.; Berger, S.; Jacoby, A.S.; Cole, N.J.; Hollway, G.E.; Berger, J.; Currie, P.D. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin α2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. USA 2007, 104, 7092–7097. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, M.W.; Beggs, A.H.; Buj-Bello, A.; Childers, M.K.; Dowling, J.J.; James, E.S.; Meng, H.; Moore, S.A.; Prasad, S.; Schoser, B. Skeletal muscle pathology in X-linked myotubular myopathy: Review with cross-species comparisons. J. Neuropathol. Exp. Neurol. 2016, 75, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Jungbluth, H.; Gautel, M. Pathogenic mechanisms in centronuclear myopathies. Front. Aging Neurosci. 2014, 6, 339. [Google Scholar] [CrossRef] [PubMed]
- Todd, P.K.; Ackall, F.Y.; Hur, J.; Sharma, K.; Paulson, H.L.; Dowling, J.J. Transcriptional changes and developmental abnormalities in a zebrafish model of myotonic dystrophy type 1. Dis. Models Mech. 2014, 7, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Machuca-Tzili, L.E.; Buxton, S.; Thorpe, A.; Timson, C.M.; Wigmore, P.; Luther, P.K.; Brook, J.D. Zebrafish deficient for Muscleblind-like 2 exhibit features of myotonic dystrophy. Dis. Models Mech. 2011, 4, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, E.M.; Horstick, E.J.; Dowling, J.J. Swimming into prominence: The zebrafish as a valuable tool for studying human myopathies and muscular dystrophies. FEBS J. 2013, 280, 4187–4197. [Google Scholar] [CrossRef] [PubMed]
- Vacaru, A.M.; Unlu, G.; Spitzner, M.; Mione, M.; Knapik, E.W.; Sadler, K.C. In vivo cell biology in zebrafish–providing insights into vertebrate development and disease. J. Cell Sci. 2014, 127, 485–495. [Google Scholar] [CrossRef]
- Vinci, P.; Panizon, E.; Tosoni, L.M.; Cerrato, C.; Pellicori, F.; Mearelli, F.; Biasinutto, C.; Fiotti, N.; Di Girolamo, F.G.; Biolo, G. Statin-associated myopathy: Emphasis on mechanisms and targeted therapy. Int. J. Mol. Sci. 2021, 22, 11687. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, J.L.; Doitsidou, M.; Ho, S.-Y.; Raz, E.; Farber, S.A. Germ cell migration in zebrafish is dependent on HMGCoA reductase activity and prenylation. Dev. Cell 2004, 6, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.M.; Rios, E.A.; Guapyassu, L.; Midlej, V.; Atella, G.C.; Herculano-Houzel, S.; Benchimol, M.; Mermelstein, C.; Costa, M.L. Alterations in zebrafish development induced by simvastatin: Comprehensive morphological and physiological study, focusing on muscle. Exp. Biol. Med. 2016, 241, 1950–1960. [Google Scholar] [CrossRef]
- Baek, J.S.; Fang, L.; Li, A.C.; Miller, Y.I. Ezetimibe and Simvastatin Reduce Cholesterol Levels in Zebrafish Larvae Fed a High-Cholesterol Diet. Cholesterol 2012, 2012, 564705. [Google Scholar] [CrossRef] [PubMed]
- Palmio, J.; Udd, B. Borderlines between sarcopenia and mild late-onset muscle disease. Front. Aging Neurosci. 2014, 6, 267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Widrick, J.J.; Kawahara, G.; Alexander, M.S.; Beggs, A.H.; Kunkel, L.M. Discovery of novel therapeutics for muscular dystrophies using zebrafish phenotypic screens. J. Neuromuscul. Dis. 2019, 6, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Sztal, T.E.; Zhao, M.; Williams, C.; Oorschot, V.; Parslow, A.C.; Giousoh, A.; Yuen, M.; Hall, T.E.; Costin, A.; Ramm, G. Zebrafish models for nemaline myopathy reveal a spectrum of nemaline bodies contributing to reduced muscle function. Acta Neuropathol. 2015, 130, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, A.S.; Busch-Nentwich, E.; Bryson-Richardson, R.J.; Hall, T.E.; Berger, J.; Berger, S.; Sonntag, C.; Sachs, C.; Geisler, R.; Stemple, D.L. The zebrafish dystrophic mutant softy maintains muscle fibre viability despite basement membrane rupture and muscle detachment. Development 2009, 136, 3367–3376. [Google Scholar] [CrossRef]
- Dowling, J.J.; Vreede, A.P.; Low, S.E.; Gibbs, E.M.; Kuwada, J.Y.; Bonnemann, C.G.; Feldman, E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009, 5, e1000372. [Google Scholar] [CrossRef] [PubMed]
- Guyon, J.; Mosley, A.; Zhou, Y.; O’brien, K.; Sheng, X.; Chiang, K.; Davidson, A.; Volinski, J.; Zon, L.; Kunkel, L. The dystrophin associated protein complex in zebrafish. Hum. Mol. Genet. 2003, 12, 601–615. [Google Scholar] [CrossRef] [PubMed]
- Telfer, W.; Busta, A.; Bonnemann, C.; Feldman, E.; Dowling, J. Zebrafish models of collagen VI-related myopathies. Hum. Mol. Genet. 2010, 19, 2433–2444. [Google Scholar] [CrossRef]
- Roostalu, U.; Strähle, U. In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell 2012, 22, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-Y.; White, R.J.; Torelli, S.; Cirak, S.; Muntoni, F.; Stemple, D.L. Zebrafish Fukutin family proteins link the unfolded protein response with dystroglycanopathies. Hum. Mol. Genet. 2011, 20, 1763–1775. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, P.; Bassett, D.; Lochmüller, H.; Bushby, K.; Straub, V. Developmental defects in a zebrafish model for muscular dystrophies associated with the loss of fukutin-related protein (FKRP). Brain 2008, 131, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, G.; Guyon, J.R.; Nakamura, Y.; Kunkel, L.M. Zebrafish models for human FKRP muscular dystrophies. Hum. Mol. Genet. 2010, 19, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Watanabe, T.; Hatakeyama, J.; Sprague, S.M.; Saint-Amant, L.; Nagashima, A.; Cui, W.W.; Zhou, W.; Kuwada, J.Y. Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development 2007, 134, 2771–2781. [Google Scholar] [CrossRef]
- Callegari, S.; Mirzaei, F.; Agbaria, L.; Shariff, S.; Kantawala, B.; Moronge, D.; Ogendi, B.M. Zebrafish as an Emerging Model for Sarcopenia: Considerations, Current Insights, and Future Directions. Int. J. Mol. Sci. 2023, 24, 17018. [Google Scholar] [CrossRef] [PubMed]
- Rutkove, S.B.; Chen, Z.-Z.; Pandeya, S.; Callegari, S.; Mourey, T.; Nagy, J.A.; Nath, A.K. Surface Electrical Impedance Myography Detects Skeletal Muscle Atrophy in Aged Wildtype Zebrafish and Aged gpr27 Knockout Zebrafish. Biomedicines 2023, 11, 1938. [Google Scholar] [CrossRef] [PubMed]
- Rutkove, S.B.; Callegari, S.; Concepcion, H.; Mourey, T.; Widrick, J.; Nagy, J.A.; Nath, A.K. Electrical impedance myography detects age-related skeletal muscle atrophy in adult zebrafish. Sci. Rep. 2023, 13, 7191. [Google Scholar] [CrossRef] [PubMed]
- Henderson, T.D.; Choi, J.; Leonard, S.W.; Head, B.; Tanguay, R.L.; Barton, C.L.; Traber, M.G. Chronic Vitamin E deficiency dysregulates purine, phospholipid, and amino acid metabolism in aging zebrafish skeletal muscle. Antioxidants 2023, 12, 1160. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.-Y.; Chen, Z.-L.; Sun, C.-C.; Yang, D.; Zhou, Z.-Q.; Xiao, Q.; Peng, X.-Y.; Tang, C.-F. A high-fat diet induces muscle mitochondrial dysfunction and impairs swimming capacity in zebrafish: A new model of sarcopenic obesity. Nutrients 2022, 14, 1975. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-L.; Guo, C.; Zou, Y.-Y.; Feng, C.; Yang, D.-X.; Sun, C.-C.; Wen, W.; Jian, Z.-J.; Zhao, Z.; Xiao, Q. Aerobic exercise enhances mitochondrial homeostasis to counteract D-galactose-induced sarcopenia in zebrafish. Exp. Gerontol. 2023, 180, 112265. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.C.; Yang, D.; Chen, Z.L.; Xiao, J.L.; Xiao, Q.; Li, C.L.; Zhou, Z.Q.; Peng, X.Y.; Tang, C.F.; Zheng, L. Exercise intervention mitigates zebrafish age-related sarcopenia via alleviating mitochondrial dysfunction. FEBS J. 2023, 290, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Dalle Carbonare, L.; Minoia, A.; Zouari, S.; Piritore, F.C.; Vareschi, A.; Romanelli, M.G.; Valenti, M.T. Crosstalk between bone and muscles during physical activity. Cells 2023, 12, 2088. [Google Scholar] [CrossRef] [PubMed]
- Rosa, N.; Simoes, R.; Magalhães, F.D.; Marques, A.T. From mechanical stimulus to bone formation: A review. Med. Eng. Phys. 2015, 37, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; You, X.; Zhang, L.; Zhang, C.; Zou, W. Mechanical regulation of bone remodeling. Bone Res. 2022, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Guarino, V.; Causa, F.; Ambrosio, L. Bioactive scaffolds for bone and ligament tissue. Expert Rev. Med. Devices 2007, 4, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Schwartzman, J.D.; McCall, M.; Ghattas, Y.; Pugazhendhi, A.S.; Wei, F.; Ngo, C.; Ruiz, J.; Seal, S.; Coathup, M.J. Multifunctional Scaffolds for Bone Repair following Age-Related Biological Decline: Promising Prospects for Smart Biomaterial-Driven Technologies. Biomaterials 2024, 122683. [Google Scholar] [CrossRef] [PubMed]
- Gargalionis, A.N.; Adamopoulos, C.; Vottis, C.T.; Papavassiliou, A.G.; Basdra, E.K. Runx2 and Polycystins in Bone Mechanotransduction: Challenges for Therapeutic Opportunities. Int. J. Mol. Sci. 2024, 25, 5291. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.M.; O’Sullivan, L.M.; Allison, H.; Casey, V.J.; Schiavi-Tritz, J.; McNamara, L.M. Altered extracellular matrix and mechanotransduction gene expression in rat bone tissue following long-term estrogen deficiency. JBMR Plus 2024, 8, ziae098. [Google Scholar] [CrossRef] [PubMed]
- Kjaer, M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol. Rev. 2004, 84, 649–698. [Google Scholar] [CrossRef] [PubMed]
- Hart, N.H.; Nimphius, S.; Rantalainen, T.; Ireland, A.; Siafarikas, A.; Newton, R. Mechanical basis of bone strength: Influence of bone material, bone structure and muscle action. J. Musculoskelet. Neuronal Interact. 2017, 17, 114. [Google Scholar] [PubMed]
- Brüggemann, G.-P.; Brüggemann, L.; Heinrich, K.; Müller, M.; Niehoff, A. Biological tissue response to impact like mechanical loading. Footwear Sci. 2011, 3, 13–22. [Google Scholar] [CrossRef]
- Mead, A.F.; Kennedy, G.G.; Palmer, B.M.; Ebert, A.M.; Warshaw, D.M. Mechanical characteristics of ultrafast zebrafish larval swimming muscles. Biophys. J. 2020, 119, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Saghiv, M.S.; Sagiv, M.S.; Saghiv, M.S.; Sagiv, M.S. Skeletal Muscles. In Basic Exercise Physiology: Clinical and Laboratory Perspectives; Springer: Cham, Switzerland, 2020; pp. 407–436. [Google Scholar]
- Suniaga, S.; Rolvien, T.; Vom Scheidt, A.; Fiedler, I.A.; Bale, H.A.; Huysseune, A.; Witten, P.E.; Amling, M.; Busse, B. Increased mechanical loading through controlled swimming exercise induces bone formation and mineralization in adult zebrafish. Sci. Rep. 2018, 8, 3646. [Google Scholar] [CrossRef] [PubMed]
- Carnovali, M.; Banfi, G.; Mariotti, M. Zebrafish models of human skeletal disorders: Embryo and adult swimming together. BioMed Res. Int. 2019, 2019, 1253710. [Google Scholar] [CrossRef]
- Yavropoulou, M.P.; Yovos, J. The molecular basis of bone mechanotransduction. J. Musculoskelet. Neuronal Interact. 2016, 16, 221. [Google Scholar] [PubMed]
- Valenti, M.T.; Deiana, M.; Cheri, S.; Dotta, M.; Zamboni, F.; Gabbiani, D.; Schena, F.; Dalle Carbonare, L.; Mottes, M. Physical exercise modulates miR-21-5p, miR-129-5p, miR-378-5p, and miR-188-5p expression in progenitor cells promoting osteogenesis. Cells 2019, 8, 742. [Google Scholar] [CrossRef] [PubMed]
- Carina, V.; Della Bella, E.; Costa, V.; Bellavia, D.; Veronesi, F.; Cepollaro, S.; Fini, M.; Giavaresi, G. Bone’s response to mechanical loading in aging and osteoporosis: Molecular mechanisms. Calcif. Tissue Int. 2020, 107, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Rolvien, T.; Amling, M. Disuse osteoporosis: Clinical and mechanistic insights. Calcif. Tissue Int. 2022, 110, 592–604. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K. Bone and Muscle: Structure, Force, and Motion; Britannica Educational Publishing: New York, NY, USA, 2010. [Google Scholar]
- Zatsiorsky, V.M.; Prilutsky, B.I. Biomechanics of Skeletal Muscles; Human Kinetics: Champaign, IL, USA, 2012. [Google Scholar]
- Voesenek, C.J.; Muijres, F.T.; Van Leeuwen, J.L. Biomechanics of swimming in developing larval fish. J. Exp. Biol. 2018, 221, jeb149583. [Google Scholar] [CrossRef] [PubMed]
- Müller, U.K. Swimming and muscle. In Fish Larval Physiology; CRC Press: Boca Raton, FL, USA, 2020; pp. 523–549. [Google Scholar]
- Daya, A.; Donaka, R.; Karasik, D. Zebrafish models of sarcopenia. Dis. Models Mech. 2020, 13, dmm042689. [Google Scholar] [CrossRef]
- Qin, L.; He, T.; Chen, S.; Yang, D.; Yi, W.; Cao, H.; Xiao, G. Roles of mechanosensitive channel Piezo1/2 proteins in skeleton and other tissues. Bone Res. 2021, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Stassen, O.M.; Ristori, T.; Sahlgren, C.M. Notch in mechanotransduction–from molecular mechanosensitivity to tissue mechanostasis. J. Cell Sci. 2020, 133, jcs250738. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Sakiyama, K.; Kitamura, K.; Yamamoto, Y.; Takagi, T.; Sekiya, S.; Watanabe, G.; Taniguchi, S.; Ogawa, Y.; Ishizuka, S. Development and regeneration of muscle, tendon, and myotendinous junctions in striated skeletal muscle. Int. J. Mol. Sci. 2022, 23, 3006. [Google Scholar] [CrossRef] [PubMed]
- Vila Pouca, M.C.; Parente, M.P.; Jorge, R.M.N.; Ashton-Miller, J.A. Injuries in muscle-tendon-bone units: A systematic review considering the role of passive tissue fatigue. Orthop. J. Sports Med. 2021, 9, 23259671211020731. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, N. Strain Distribution Across the Myotendinous Junction and its Role in the Regeneration of the Junction. Ph.D. Thesis, University of Portsmouth, Portsmouth, UK, 2024. [Google Scholar]
- Amemiya, H.; Yamamoto, M.; Higa, K.; Watanabe, G.; Taniguchi, S.; Kitamura, K.; Jeong, J.; Yanagisawa, N.; Fukuda, K.-i.; Abe, S. Effects of myostatin on nuclear morphology at the myotendinous junction. Int. J. Mol. Sci. 2023, 24, 6634. [Google Scholar] [CrossRef] [PubMed]
- Snow, C.J.; Henry, C.A. Dynamic formation of microenvironments at the myotendinous junction correlates with muscle fiber morphogenesis in zebrafish. Gene Expr. Patterns 2009, 9, 37–42. [Google Scholar] [CrossRef]
- Malbouyres, M.; Guiraud, A.; Lefrançois, C.; Salamito, M.; Nauroy, P.; Bernard, L.; Sohm, F.; Allard, B.; Ruggiero, F. Lack of the myotendinous junction marker col22a1 results in posture and locomotion disabilities in zebrafish. Matrix Biol. 2022, 109, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Cai, Y.; Wei, Y.; Yang, J.; Gao, J.; Yang, Y. Sarcopenic obesity and osteoporosis: Research progress and hot spots. Exp. Gerontol. 2024, 195, 112544. [Google Scholar] [CrossRef] [PubMed]
- De Toni, L.; Di Nisio, A.; Rocca, M.; De Rocco Ponce, M.; Ferlin, A.; Foresta, C. Osteocalcin, a bone-derived hormone with important andrological implications. Andrology 2017, 5, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Functions of osteocalcin in bone, pancreas, testis, and muscle. Int. J. Mol. Sci. 2020, 21, 7513. [Google Scholar] [CrossRef]
- Mera, P.; Laue, K.; Wei, J.; Berger, J.M.; Karsenty, G. Osteocalcin is necessary and sufficient to maintain muscle mass in older mice. Mol. Metab. 2016, 5, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI (3) K/Akt/mTOR and PI (3) K/Akt/GSK3 pathways. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Brotto, M.; Johnson, M.L. Endocrine crosstalk between muscle and bone. Curr. Osteoporos. Rep. 2014, 12, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Whang, H.; Wu, Z.; Jiang, S.; Chen, L. Deletion of Asb15b gene can lead to a significant decrease in zebrafish intermuscular bone. Gene 2024, 923, 148561. [Google Scholar] [CrossRef] [PubMed]
- Leyhr, J.; Sanchez, S.; Dollman, K.N.; Tafforeau, P.; Haitina, T. Enhanced contrast synchrotron X-ray microtomography for describing skeleton-associated soft tissue defects in zebrafish mutants. Front. Endocrinol. 2023, 14, 1108916. [Google Scholar] [CrossRef] [PubMed]
- Kague, E.; Hughes, S.M.; Lawrence, E.A.; Cross, S.; Martin-Silverstone, E.; Hammond, C.L.; Hinits, Y. Scleraxis genes are required for normal musculoskeletal development and for rib growth and mineralization in zebrafish. FASEB J. 2019, 33, 9116. [Google Scholar] [CrossRef]
- Maurya, A.K.; Tan, H.; Souren, M.; Wang, X.; Wittbrodt, J.; Ingham, P.W. Integration of Hedgehog and BMP signalling by the engrailed2a gene in the zebrafish myotome. Development 2011, 138, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Kaliya-Perumal, A.-K.; Ingham, P.W. Musculoskeletal regeneration: A zebrafish perspective. Biochimie 2022, 196, 171–181. [Google Scholar] [CrossRef]
- Takeo, M.; Lee, W.; Ito, M. Wound healing and skin regeneration. Cold Spring Harb. Perspect. Med. 2015, 5, a023267. [Google Scholar] [CrossRef] [PubMed]
- Van Haele, M.; Snoeck, J.; Roskams, T. Human liver regeneration: An etiology dependent process. Int. J. Mol. Sci. 2019, 20, 2332. [Google Scholar] [CrossRef] [PubMed]
- Daponte, V.; Tylzanowski, P.; Forlino, A. Appendage regeneration in vertebrates: What makes this possible? Cells 2021, 10, 242. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J.R.; Mabrouk, A.; Garla, V.V. Fracture healing overview. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Walmsley, G.G.; Ransom, R.C.; Zielins, E.R.; Leavitt, T.; Flacco, J.S.; Hu, M.S.; Lee, A.S.; Longaker, M.T.; Wan, D.C. Stem cells in bone regeneration. Stem Cell Rev. Rep. 2016, 12, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Armanios, M.; Blackburn, E.H. The telomere syndromes. Nat. Rev. Genet. 2012, 13, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A.; Lee, H.-W.; Hande, M.P.; Samper, E.; Lansdorp, P.M.; DePinho, R.A.; Greider, C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 1997, 91, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, K.L.; Chang, S.; Lee, H.-W.; Blasco, M.; Gottlieb, G.J.; Greider, C.; DePinho, R.A. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 1999, 96, 701–712. [Google Scholar] [CrossRef]
- Carneiro, M.C.; de Castro, I.P.; Ferreira, M.G. Telomeres in aging and disease: Lessons from zebrafish. Dis. Models Mech. 2016, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, M.C.; Henriques, C.M.; Nabais, J.; Ferreira, T.; Carvalho, T.; Ferreira, M.G. Short telomeres in key tissues initiate local and systemic aging in zebrafish. PLoS Genet. 2016, 12, e1005798. [Google Scholar] [CrossRef]
- Cruz-Jentoft, A.J.; Sayer, A.A. Sarcopenia. Lancet 2019, 393, 2636–2646. [Google Scholar] [CrossRef] [PubMed]
- Angulo, J.; El Assar, M.; Rodriguez-Manas, L. Frailty and sarcopenia as the basis for the phenotypic manifestation of chronic diseases in older adults. Mol. Asp. Med. 2016, 50, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Christian, C.J.; Benian, G.M. Animal models of sarcopenia. Aging Cell 2020, 19, e13223. [Google Scholar] [CrossRef] [PubMed]
- Kishi, S.; Slack, B.E.; Uchiyama, J.; Zhdanova, I.V. Zebrafish as a genetic model in biological and behavioral gerontology: Where development meets aging in vertebrates–a mini-review. Gerontology 2009, 55, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Kishi, S.; Bayliss, P.E.; Uchiyama, J.; Koshimizu, E.; Qi, J.; Nanjappa, P.; Imamura, S.; Islam, A.; Neuberg, D.; Amsterdam, A. The identification of zebrafish mutants showing alterations in senescence-associated biomarkers. PLoS Genet. 2008, 4, e1000152. [Google Scholar] [CrossRef] [PubMed]
- Montandon, M.; Currie, P.D.; Ruparelia, A.A. Examining muscle regeneration in zebrafish models of muscle disease. JoVE (J. Vis. Exp.) 2021, e62071. [Google Scholar] [CrossRef]
- Quach, T.K.; Taylor, M.F.; Currie, P.D.; Eynon, N.; Ruparelia, A.A. Skeletal muscle ageing: Lessons from teleosts. J. Gerontol. Ser. A: Biol. Sci. Med. Sci. 2024, glae052. [Google Scholar] [CrossRef] [PubMed]
- Gaitán-Peñas, H.; Pérez-Rius, C.; Muhaisen, A.; Castellanos, A.; Errasti-Murugarren, E.; Barrallo-Gimeno, A.; Alcaraz-Pérez, F.; Estévez, R. Characterization of ClC-1 chloride channels in zebrafish: A new model to study myotonia. J. Physiol. 2024, 602, 3975–3994. [Google Scholar] [CrossRef] [PubMed]
- Ichii, S.; Matsuoka, I.; Okazaki, F.; Shimada, Y. Zebrafish models for skeletal muscle senescence: Lessons from cell cultures and rodent models. Molecules 2022, 27, 8625. [Google Scholar] [CrossRef]
- Rios, J.L.; Sapède, D.; Djouad, F.; Rapp, A.E.; Lang, A.; Larkin, J.; Ladel, C.; Mobasheri, A. Animal Models of Osteoarthritis Part 1–Preclinical Small Animal Models: Challenges and Opportunities for Drug Development. Curr. Protoc. 2022, 2, e596. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Bahri, M.A.; Degueldre, C.; Caetano da Silva, C.; Sanchez, C.; Ostertag, A.; Collet, C.; Cohen-Solal, M.; Plenevaux, A.; Henrotin, Y. A Zebrafish Mutant in the Extracellular Matrix Protein Gene efemp1 as a Model for Spinal Osteoarthritis. Animals 2023, 14, 74. [Google Scholar] [CrossRef]
- Zheng, L.; He, S.; Wang, H.; Li, J.; Liu, Y.; Liu, S. Targeting cellular senescence in aging and age-related diseases: Challenges, considerations, and the emerging role of senolytic and senomorphic therapies. Aging Dis. 2024, 15, 2554. [Google Scholar] [PubMed]
- Morsli, S.; Henriques, C.M.; Ellis, P.S.; Mortiboys, H.; Baxendale, S.; Loynes, C.A.; Renshaw, S.A.; Bellantuono, I. A p21-GFP zebrafish model of senescence for rapid testing of senolytics in vivo. Aging Cell 2023, 22, e13835. [Google Scholar] [CrossRef] [PubMed]
- Maves, L. Recent advances using zebrafish animal models for muscle disease drug discovery. Expert Opin. Drug Discov. 2014, 9, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Parichy, D.M.; Elizondo, M.R.; Mills, M.G.; Gordon, T.N.; Engeszer, R.E. Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 2009, 238, 2975–3015. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Sehring, I.; Cederlund, M.; Mulaw, M.; Weidinger, G. NF-κB signaling negatively regulates osteoblast dedifferentiation during zebrafish bone regeneration. Dev. Cell 2020, 52, 167–182. e7. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, M.; Carnovali, M.; Banfi, G. Danio rerio: The Janus of the bone from embryo to scale. Clin. Cases Miner. Bone Metab. 2015, 12, 188. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.-C.A.; Wilk, K.; Lin, C.P.; Intini, G. In vivo 3D histomorphometry quantifies bone apposition and skeletal progenitor cell differentiation. Sci. Rep. 2018, 8, 5580. [Google Scholar] [CrossRef] [PubMed]
- Cotti, S.; Di Biagio, C.; Huysseune, A.; Koppe, W.; Forlino, A.; Witten, P.E. Matrix first, minerals later: Fine-tuned dietary phosphate increases bone formation in zebrafish. JBMR Plus 2024, ziae081. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-R.; Lai, Y.-H.; Tsai, J.-J.; Hsiao, C.-D. Live fluorescent staining platform for drug-screening and mechanism-analysis in zebrafish for bone mineralization. Molecules 2017, 22, 2068. [Google Scholar] [CrossRef]
- Geurtzen, K.; Vernet, A.; Freidin, A.; Rauner, M.; Hofbauer, L.C.; Schneider, J.E.; Brand, M.; Knopf, F. Immune suppressive and bone inhibitory effects of prednisolone in growing and regenerating zebrafish tissues. J. Bone Miner. Res. 2017, 32, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Bohns, F.R.; Akhtar, R.; Chuang, Y.-J.; Chen, P.-Y. Bone quality in zebrafish vertebrae improves after alendronate administration in a glucocorticoid-induced osteoporosis model. J. Mech. Behav. Biomed. Mater. 2024, 154, 106521. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Yang, X.; Shen, J.; Yang, K.; Lv, G.; Zhang, H.; Sun, J. The role of the PI3K/Akt pathway in adenosine’s mechanism of action in osteoporosis with oxidative stress: A wet-dry experimental approach strategy. J. Funct. Foods 2024, 120, 106366. [Google Scholar] [CrossRef]
- Zuo, Y.; Chen, C.; Liu, F.; Hu, H.; Dong, S.; Shen, Q.; Zeng, J.; Huang, L.; Liao, X.; Cao, Z. Pinoresinol diglucoside mitigates dexamethasone-induced osteoporosis and chondrodysplasia in zebrafish. Toxicol. Appl. Pharmacol. 2024, 484, 116884. [Google Scholar] [CrossRef] [PubMed]
- Khajuria, D.K.; Kumar, V.B.; Gigi, D.; Gedanken, A.; Karasik, D. Accelerated bone regeneration by nitrogen-doped carbon dots functionalized with hydroxyapatite nanoparticles. ACS Appl. Mater. Interfaces 2018, 10, 19373–19385. [Google Scholar] [CrossRef] [PubMed]
- Ratanavaraporn, J.; Kanokpanont, S.; Tabata, Y.; Damrongsakkul, S. Growth and osteogenic differentiation of adipose-derived and bone marrow-derived stem cells on chitosan and chitooligosaccharide films. Carbohydr. Polym. 2009, 78, 873–878. [Google Scholar] [CrossRef]
- Balagangadharan, K.; Trivedi, R.; Vairamani, M.; Selvamurugan, N. Sinapic acid-loaded chitosan nanoparticles in polycaprolactone electrospun fibers for bone regeneration in vitro and in vivo. Carbohydr. Polym. 2019, 216, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Vimalraj, S.; Govindarajan, D.; Sudhakar, S.; Suresh, R.; Palanivel, P.; Sekaran, S. Chitosan derived chito-oligosaccharides promote osteoblast differentiation and offer anti-osteoporotic potential: Molecular and morphological evidence from a zebrafish model. Int. J. Biol. Macromol. 2024, 259, 129250. [Google Scholar] [CrossRef] [PubMed]
- Modarresi Chahardehi, A.; Arsad, H.; Lim, V. Zebrafish as a successful animal model for screening toxicity of medicinal plants. Plants 2020, 9, 1345. [Google Scholar] [CrossRef]
- Laizé, V.; Gavaia, P.J.; Cancela, M.L. Fish: A suitable system to model human bone disorders and discover drugs with osteogenic or osteotoxic activities. Drug Discov. Today Dis. Models 2014, 13, 29–37. [Google Scholar] [CrossRef]
- Kabir, A.; Selvaraj, V.; Sudhakar, S. Protein Nano Coop Complexes Promote Fracture Healing and Bone Regeneration in a Zebrafish Osteoporosis Model. Biomacromolecules 2024. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, Y.; Qi, X.; Chen, J.; Chen, W.; Qiu, G.; Wu, Z.; Wu, N. CRISPR/Cas9 in zebrafish: An efficient combination for human genetic diseases modeling. Hum. Genet. 2017, 136, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Liu, B.; Du, H.; Zhao, S.; Li, Y.; Cheng, X.; Wang, S.; Lin, J.; Zhou, J.; Qiu, G. The progress of CRISPR/Cas9-mediated gene editing in generating mouse/zebrafish models of human skeletal diseases. Comput. Struct. Biotechnol. J. 2019, 17, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, G.; Gasperini, M.J.; Myers, J.A.; Widrick, J.J.; Eran, A.; Serafini, P.R.; Alexander, M.S.; Pletcher, M.T.; Morris, C.A.; Kunkel, L.M. Dystrophic muscle improvement in zebrafish via increased heme oxygenase signaling. Hum. Mol. Genet. 2014, 23, 1869–1878. [Google Scholar] [CrossRef] [PubMed]
- Cardeira, J.; Laizé, V.; Martins, G.; Cancela, M.; Gavaia, P. An overview on the teleost bone mechanophysiology. J. Appl. Ichthyol. 2018, 34, 440–448. [Google Scholar] [CrossRef]
- Renn, J.; Winkler, C.; Schartl, M.; Fischer, R.; Goerlich, R. Zebrafish and medaka as models for bone research including implications regarding space-related issues. Protoplasma 2006, 229, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.L.; Escaleira, R.C.; Rodrigues, V.B.; Manasfi, M.; Mermelstein, C.S. Some distinctive features of zebrafish myogenesis based on unexpected distributions of the muscle cytoskeletal proteins actin, myosin, desmin, α-actinin, troponin and titin. Mech. Dev. 2002, 116, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Bassett, D.; Currie, P.D. Identification of a zebrafish model of muscular dystrophy. Clin. Exp. Pharmacol. Physiol. 2004, 31, 537–540. [Google Scholar] [CrossRef]
- Amores, A.; Force, A.; Yan, Y.-L.; Joly, L.; Amemiya, C.; Fritz, A.; Ho, R.K.; Langeland, J.; Prince, V.; Wang, Y.-L. Zebrafish hox clusters and vertebrate genome evolution. Science 1998, 282, 1711–1714. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.H.; Yan, Y.-L.; Gates, M.A.; Horne, S.; Amores, A.; Brownlie, A.; Donovan, A.; Egan, E.S.; Force, A.; Gong, Z. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998, 18, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Guyon, J.R.; Steffen, L.S.; Howell, M.H.; Pusack, T.J.; Lawrence, C.; Kunkel, L.M. Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2007, 1772, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.H.; Dobrinski, K.P.; Lee, A.S.; Gokcumen, O.; Mills, R.E.; Shi, X.; Chong, W.W.; Chen, J.Y.H.; Yoo, P.; David, S. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc. Natl. Acad. Sci. USA 2012, 109, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Diogo, R. Comparative anatomy, homologies and evolution of mandibular, hyoid and hypobranchial muscles of bony fish and tetrapods: A new insight. Anim. Biol. 2008, 58, 123–172. [Google Scholar] [CrossRef]
| Zebrafish Model | Applications | Ref |
|---|---|---|
| LRP5 mutant or crispant | Osteoporosis | [59] |
| runx2b mutants | Bone development | [70] |
| Chihuahua (Chi/+) | Osteogenesis imperfecta | [63,64] |
| p3h1−/− model | Osteogenesis imperfecta | [63,70] |
| CRTAP mutant | Osteogenesis imperfecta | [71] |
| fkbp10a mutant | Bone fragility | [72] |
| aldh7a1, daam2, esr1 and sost crispant | Osteoporosis | [65] |
| creb3l1 ifitm5 mbtps2 sec24d serpinf1 and sparc crispant | Osteogenesis imperfecta | [65] |
| dstyk mutant | Scoliosis | [73] |
| col8a1a mutant | Scoliosis | [74] |
| tbx6 mutant | Scoliosis | [75] |
| myadm mutant | Scoliosis | [76] |
| col2a1a mutant | Scoliosis | [76] |
| stat3 mutant | Scoliosis | [77] |
| Zebrafish Model | Applications | Ref |
|---|---|---|
| sapje | Duchenne muscular dystrophy | [88,89,90,93,109] |
| sapje-like | Duchenne muscular dystrophy | [87] |
| candyfloss (lama2 mutant) | Congenital muscular dystrophy | [97] |
| Tg(ACTA1 D286G-eGFP) | Nemaline myopathy | [110] |
| softy (lamb2mutant) | Muscular dystrophy lamb2 mutation | [111] |
| mtm1 morpholin | Myotubular myopathy | [98,112] |
| dmd morpholin | Duchenne muscular dystrophy | [113] |
| col6a1 morpholin | Ullrich congenital muscular dystrophy; Bethlem myopathy | [114] |
| dysf morpholino | Myoshi myopathy; limb–girdle muscular dystrophy | [91,115] |
| fkrp morpholino | Multiple forms of dystroglycanopathy | [116,117,118] |
| ryr1b mutant | Multiple forms of congenital myopathy | [119] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dalle Carbonare, L.; Braggio, M.; Minoia, A.; Cominacini, M.; Romanelli, M.G.; Pessoa, J.; Tiso, N.; Valenti, M.T. Modeling Musculoskeletal Disorders in Zebrafish: Advancements in Muscle and Bone Research. Cells 2025, 14, 28. https://doi.org/10.3390/cells14010028
Dalle Carbonare L, Braggio M, Minoia A, Cominacini M, Romanelli MG, Pessoa J, Tiso N, Valenti MT. Modeling Musculoskeletal Disorders in Zebrafish: Advancements in Muscle and Bone Research. Cells. 2025; 14(1):28. https://doi.org/10.3390/cells14010028
Chicago/Turabian StyleDalle Carbonare, Luca, Michele Braggio, Arianna Minoia, Mattia Cominacini, Maria Grazia Romanelli, João Pessoa, Natascia Tiso, and Maria Teresa Valenti. 2025. "Modeling Musculoskeletal Disorders in Zebrafish: Advancements in Muscle and Bone Research" Cells 14, no. 1: 28. https://doi.org/10.3390/cells14010028
APA StyleDalle Carbonare, L., Braggio, M., Minoia, A., Cominacini, M., Romanelli, M. G., Pessoa, J., Tiso, N., & Valenti, M. T. (2025). Modeling Musculoskeletal Disorders in Zebrafish: Advancements in Muscle and Bone Research. Cells, 14(1), 28. https://doi.org/10.3390/cells14010028












