Multiplexed Digital PCR Reference Gene Measurement for Genomic and Cell-Free DNA Analysis
Abstract
Highlights
- A five-gene multiplex digital PCR (dPCR) reference gene panel was successfully developed and validated across synthetic gene fragments, genomic DNA, and cell-free DNA, showing robust linearity, precision, and wide dynamic range.
- Both the hydrolysis probe and universal (Rainbow™) probe chemistries performed comparably, and the multiplex approach proved superior to single reference gene targets by mitigating bias from genomic instability.
- The multiplex reference gene panel offers a more reliable method for total DNA quantification, which is crucial for precision medicine applications such as NGS library preparations and copy number variation analysis.
- This method provides a pathway to establish traceable calibration standards, im-proving quality control and comparability of DNA measurements across laboratories and clinical diagnostics.
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.1.1. Human gDNA Restriction Digestion
2.1.2. gBlocksTM Preparation, Mixing, and Dilution Series
2.1.3. Cell-Free DNA
2.2. Digital PCR
2.2.1. Oligonucleotides
2.2.2. QIAcuity Protocol
2.3. Data Acquisition and Analysis
2.4. Statistical Analysis
3. Results
3.1. Assay Chemistry Performance
3.2. Linearity and Chemistry Comparison
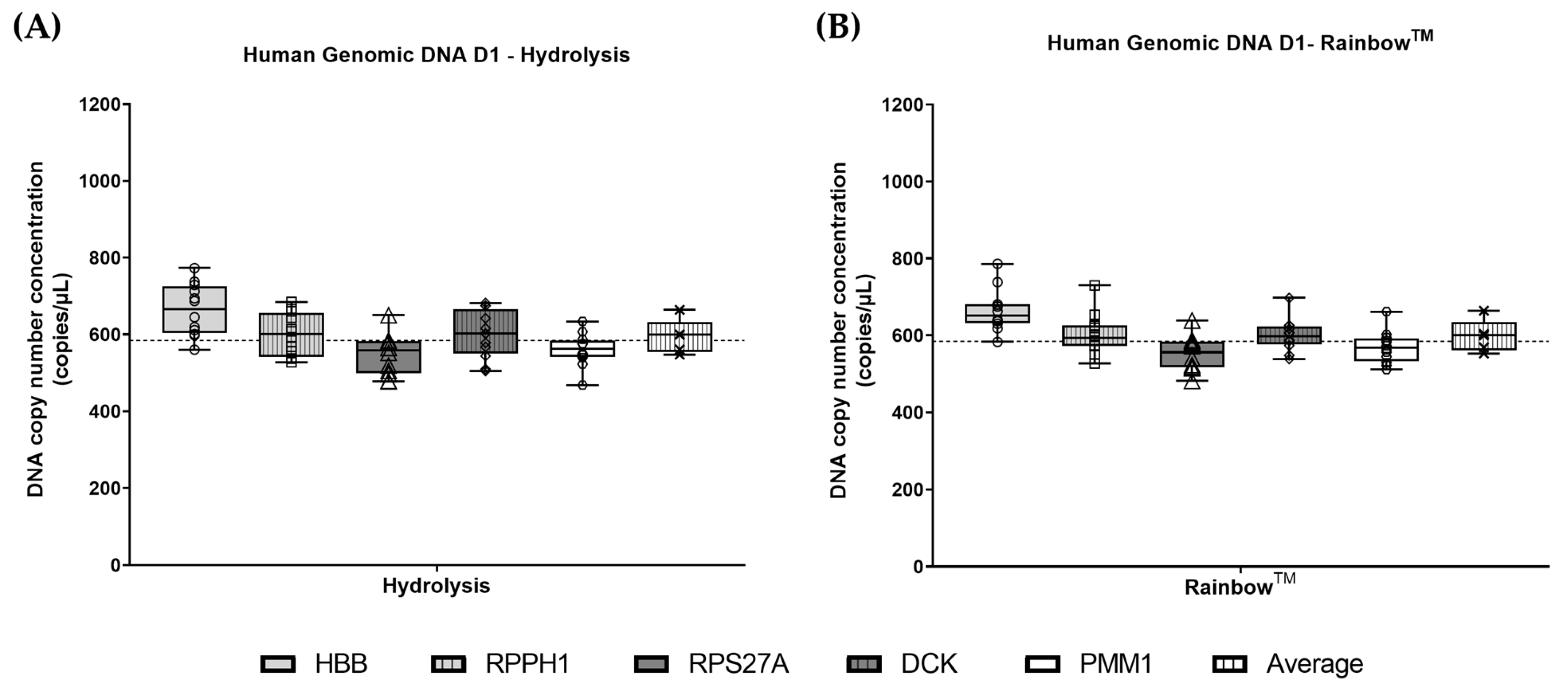
3.3. Comparison Between Reference Genes
3.4. Measurement Uncertainty (Analysis of gDNA)
3.5. Analysis of cfDNA Extracted from Plasma
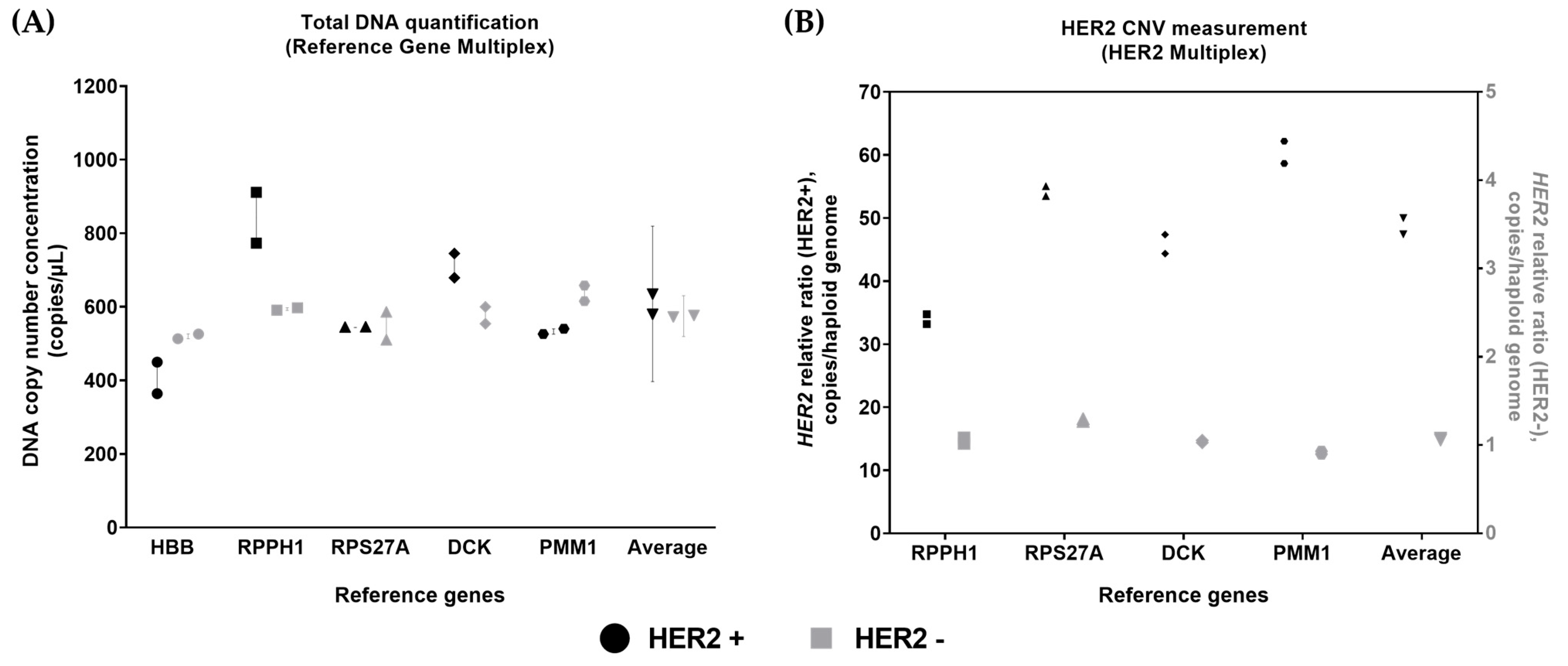
3.6. Analysis of Genomic DNA Material from Breast Carcinoma Cell Line
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| dPCR | Digital polymerase chain reaction |
| cfDNA | Cell-free DNA |
| CNV | Copy number variation |
| gDNA | Genomic DNA |
| GE | Genome equivalent |
| hgDNA | Human genomic DNA |
| NGS | Next generation sequencing |
| qPCR | Real-time quantitative PCR |
References
- Richman, S.D.; Southward, K.; Chambers, P.; Cross, D.; Barrett, J.; Hemmings, G.; Taylor, M.; Wood, H.; Hutchins, G.; Foster, J.M.; et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: Analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J. Pathol. 2016, 238, 562–570. [Google Scholar] [CrossRef]
- Gevensleben, H.; Garcia-Murillas, I.; Graeser, M.K.; Schiavon, G.; Osin, P.; Parton, M.; Smith, I.E.; Ashworth, A.; Turner, N.C. Noninvasive detection of HER2 amplification with plasma DNA digital PCR. Clin. Cancer Res. 2013, 19, 3276–3284. [Google Scholar] [CrossRef]
- Chen, M.; Linstra, R.; van Vugt, M. Genomic instability, inflammatory signaling and response to cancer immunotherapy. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188661. [Google Scholar] [CrossRef]
- Aguilera, A.; Garcia-Muse, T. Causes of genome instability. Annu. Rev. Genet. 2013, 47, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; De Spiegelaere, W.; Vynck, M.; Trypsteen, W.; Gleerup, D.; Vandesompele, J.; Thas, O. Flexible methods for uncertainty estimation of digital PCR data. iScience 2025, 28, 111772. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer. 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Devonshire, A.S.; Whale, A.S.; Gutteridge, A.; Jones, G.; Cowen, S.; Foy, C.A.; Huggett, J.F. Towards standardisation of cell-free DNA measurement in plasma: Controls for extraction efficiency, fragment size bias and quantification. Anal. Bioanal. Chem. 2014, 406, 6499–6512. [Google Scholar] [CrossRef]
- Burke, D.; Pinheiro, L.; Somerville Glover, E.; Moon, F.; Deans, Z.; Corner, A. Between Laboratory Reproducibility of DNA Extraction from Human Blood and Fresh Frozen Tissue. J. Mol. Diagn. 2022, 24, 1041–1049. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Genetics 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Whale, A.S.; Jones, G.M.; Pavsic, J.; Dreo, T.; Redshaw, N.; Akyurek, S.; Akgoz, M.; Divieto, C.; Sassi, M.P.; He, H.J.; et al. Assessment of Digital PCR as a Primary Reference Measurement Procedure to Support Advances in Precision Medicine. Clin. Chem. 2018, 64, 1296–1307. [Google Scholar] [CrossRef]
- Wang, X.; Xing, D.; Liu, Z.; Zhang, Y.; Cheng, B.; Sun, S.; Wang, Q.; Dong, L. Establishment and evaluation of digital PCR methods for HER2 copy number variation in breast cancer. Anal. Bioanal. Chem. 2023, 415, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Whale, A.S.; Huggett, J.F.; Tzonev, S. Fundamentals of multiplexing with digital PCR. Biomol. Detect. Quantif. 2016, 10, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shao, L.; Fuksenko, T.; Liu, H.; Shi, R.; Dinh, A.; Highfill, S.L.; Zhang, N.; Panch, S.R.; Somerville, R.P.; et al. Reference gene selection for clinical chimeric antigen receptor T-cell product vector copy number assays. Cytotherapy 2023, 25, 598–604. [Google Scholar] [CrossRef]
- Heredia, N.J.; Belgrader, P.; Wang, S.; Koehler, R.; Regan, J.; Cosman, A.M.; Saxonov, S.; Hindson, B.; Tanner, S.C.; Brown, A.S.; et al. Droplet Digital PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods 2013, 59, S20–S23. [Google Scholar] [CrossRef] [PubMed]
- Vynck, M.; Vandesompele, J.; Nijs, N.; Menten, B.; De Ganck, A.; Thas, O. Flexible analysis of digital PCR experiments using generalized linear mixed models. Biomol. Detect. Quantif. 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Wils, G.; Hamerlinck, L.; Trypsteen, W.; Van Den Eeckhaut, C.; Weiss, J.; Nour, A.A.; Vergult, S.; Vandesompele, J. Digital PCR-Based Gene Expression Analysis Using a Highly Multiplexed Assay with Universal Detection Probes to Study Induced Pluripotent Stem Cell Differentiation into Cranial Neural Crest Cells. Methods Mol. Biol. 2025, 2880, 17–47. [Google Scholar] [CrossRef]
- JCGM 100:2008; Evaluation of the Measurement Data-Guide to the Expression of Uncertainty in Measurement. BIPM: Paris, France, 2008.
- ISO 15193:2009; In Vitro Diagnostic Medical Devices—Measurement of Quantities in Samples of Biological Origin—Requirements for Content and Presentation of Reference Measurement Procedures. ISO: Geneva, Switzerland, 2009.
- Romsos, E.L.; Kline, M.C.; Duewer, D.L.; Toman, B.; Farkas, N. Certification of Standard Reference Material® 2372a Human DNA Quantitation Standard; NIST Special Publication: Gaithersburg, MD, USA, 2018. [Google Scholar] [CrossRef]
- Dong, L.; Wang, X.; Devonshire, A.; Huggett, J.; Ellison, S.; Gonzalez, A.F.; French, D.; Morris, P.; Temisak, S.; Bae, Y.; et al. CCQM-K176 Breast cancer biomarker HER2 copy number variation (CNV) measurement. Metrologia 2024, 61, 08017. [Google Scholar] [CrossRef]
- The dMIQE Group; Huggett, J.F. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org (accessed on 24 September 2025).
- Pinheiro, J.C.; Bates, D.M. Mixed-Effects Models in S and S-PLUS; Springer: New York, NY, USA, 2000; 528p. [Google Scholar]
- Navarro, E.; Serrano-Heras, G.; Castano, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 439, 231–250. [Google Scholar] [CrossRef]
- Turner, A.; Sasse, J.; Varadi, A. Rapid detection of pathological mutations and deletions of the haemoglobin beta gene (HBB) by High Resolution Melting (HRM) analysis and Gene Ratio Analysis Copy Enumeration PCR (GRACE-PCR). BMC Med. Genet. 2016, 17, 75. [Google Scholar] [CrossRef]
- Lockwood, C.M.; Borsu, L.; Cankovic, M.; Earle, J.S.L.; Gocke, C.D.; Hameed, M.; Jordan, D.; Lopategui, J.R.; Pullambhatla, M.; Reuther, J.; et al. Recommendations for Cell-Free DNA Assay Validations: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2023, 25, 876–897. [Google Scholar] [CrossRef]
- Udomruk, S.; Phanphaisarn, A.; Kanthawang, T.; Sangphukieo, A.; Sutthitthasakul, S.; Tongjai, S.; Teeyakasem, P.; Thongkumkoon, P.; Orrapin, S.; Moonmuang, S.; et al. Characterization of Cell-Free DNA Size Distribution in Osteosarcoma Patients. Clin. Cancer Res. 2023, 29, 2085–2094. [Google Scholar] [CrossRef]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef]
- Zhou, Q.; Kang, G.; Jiang, P.; Qiao, R.; Lam, W.K.J.; Yu, S.C.Y.; Ma, M.L.; Ji, L.; Cheng, S.H.; Gai, W.; et al. Epigenetic analysis of cell-free DNA by fragmentomic profiling. Proc. Natl. Acad. Sci. USA 2022, 119, e2209852119. [Google Scholar] [CrossRef]
- Kline, M.C.; Romsos, E.L.; Duewer, D.L. Evaluating Digital PCR for the Quantification of Human Genomic DNA: Accessible Amplifiable Targets. Anal. Chem. 2016, 88, 2132–2139. [Google Scholar] [CrossRef] [PubMed]
- Kline, M.C.; Duewer, D.L. Evaluating digital PCR for the quantification of human nuclear DNA: Determining target strandedness. Anal. Bioanal. Chem. 2020, 412, 4749–4760. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.B.; Park, S.R.; Dong, L.; Wang, J.; Sui, Z.; Pavsic, J.; Milavec, M.; Akgoz, M.; Mozioglu, E.; Corbisier, P.; et al. International Comparison of Enumeration-Based Quantification of DNA Copy-Concentration Using Flow Cytometric Counting and Digital Polymerase Chain Reaction. Anal. Chem. 2016, 88, 12169–12176. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Meng, Y.; Wang, J.; Liu, Y. Evaluation of droplet digital PCR for characterizing plasmid reference material used for quantifying ammonia oxidizers and denitrifiers. Anal. Bioanal. Chem. 2014, 406, 1701–1712. [Google Scholar] [CrossRef]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef]
- Vandesompele, J. Accurate normalization of real-time quantitative RT-PCR data by genometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- Duewer, D.L.; Kline, M.C.; Romsos, E.L.; Toman, B. Evaluating droplet digital PCR for the quantification of human genomic DNA: Converting copies per nanoliter to nanograms nuclear DNA per microliter. Anal. Bioanal. Chem. 2018, 410, 2879–2887. [Google Scholar] [CrossRef]

| gBlock™ Mix | hgDNA | |||||
|---|---|---|---|---|---|---|
| Reference Genes | Hydrolysis R2 | Rainbow™ R2 | p-Values | Hydrolysis R2 | Rainbow™ R2 | p-Values |
| HBB | 0.996 | 0.994 | 0.546 | 0.988 | 0.985 | 0.612 |
| RPPH1 | 0.997 | 0.996 | 0.949 | 0.983 | 0.988 | 0.134 |
| RPS27A | 0.997 | 0.996 | 0.752 | 0.976 | 0.985 | 0.997 |
| DCK | 0.998 | 0.996 | 0.466 | 0.989 | 0.986 | 0.234 |
| PMM1 | 0.996 | 0.996 | 0.195 | 0.984 | 0.987 | 0.809 |
| Dilution | Five Gene Average, GE/µL | Relative Standard Uncertainty (%)— Single Gene | Relative Standard Uncertainty (%)— Five Gene | Relative Expanded Uncertainty (%)— Five Gene * |
|---|---|---|---|---|
| D1 | 664 | 5.49 | 4.34 | 12.1 |
| D2 | 360 | 6.40 | 4.59 | 12.7 |
| D3 | 179 | 6.23 | 4.54 | 12.6 |
| D4 | 83 | 6.30 | 4.56 | 12.7 |
| D5 | 41 | 10.49 | 5.90 | 16.4 |
| D6 | 21 | 13.80 | 7.14 | 19.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yener, D.; Busby, E.J.; Vandesompele, J.; Wils, G.; Richman, S.D.; Wood, H.M.; Huggett, J.F.; Foy, C.A.; Devonshire, A.S. Multiplexed Digital PCR Reference Gene Measurement for Genomic and Cell-Free DNA Analysis. Cells 2025, 14, 1544. https://doi.org/10.3390/cells14191544
Yener D, Busby EJ, Vandesompele J, Wils G, Richman SD, Wood HM, Huggett JF, Foy CA, Devonshire AS. Multiplexed Digital PCR Reference Gene Measurement for Genomic and Cell-Free DNA Analysis. Cells. 2025; 14(19):1544. https://doi.org/10.3390/cells14191544
Chicago/Turabian StyleYener, Dilek, Eloise J. Busby, Jo Vandesompele, Gertjan Wils, Susan D. Richman, Henry M. Wood, Jim F. Huggett, Carole A. Foy, and Alison S. Devonshire. 2025. "Multiplexed Digital PCR Reference Gene Measurement for Genomic and Cell-Free DNA Analysis" Cells 14, no. 19: 1544. https://doi.org/10.3390/cells14191544
APA StyleYener, D., Busby, E. J., Vandesompele, J., Wils, G., Richman, S. D., Wood, H. M., Huggett, J. F., Foy, C. A., & Devonshire, A. S. (2025). Multiplexed Digital PCR Reference Gene Measurement for Genomic and Cell-Free DNA Analysis. Cells, 14(19), 1544. https://doi.org/10.3390/cells14191544





